Inferring the demographic history underlying parallel genomic divergence among pairs of parasitic and nonparasitic lamprey ecotypes
Abstract
Understanding the evolutionary mechanisms generating parallel genomic divergence patterns among replicate ecotype pairs remains an important challenge in speciation research. We investigated the genomic divergence between the anadromous parasitic river lamprey (Lampetra fluviatilis) and the freshwater-resident nonparasitic brook lamprey (Lampetra planeri) in nine population pairs displaying variable levels of geographic connectivity. We genotyped 338 individuals with RAD sequencing and inferred the demographic divergence history of each population pair using a diffusion approximation method. Divergence patterns in geographically connected population pairs were better explained by introgression after secondary contact, whereas disconnected population pairs have retained a signal of ancient migration. In all ecotype pairs, models accounting for differential introgression among loci outperformed homogeneous migration models. Generating neutral predictions from the inferred divergence scenarios to detect highly differentiated markers identified greater proportions of outliers in disconnected population pairs than in connected pairs. However, increased similarity in the most divergent genomic regions was found among connected ecotype pairs, indicating that gene flow was instrumental in generating parallelism at the molecular level. These results suggest that heterogeneous genomic differentiation and parallelism among replicate ecotype pairs have partly emerged through restricted introgression in genomic islands.
Introduction
Understanding the mechanisms responsible for the build-up and maintenance of species divergence is of primary importance in evolutionary biology. In particular, it is now common to observe highly heterogeneous genomic differentiation patterns between divergent lineages (Harrison 1986; Nadeau et al. 2013; Tine et al. 2014; Fraïsse et al. 2015), ecotype pairs (Hohenlohe et al. 2010; Gagnaire et al. 2013; Westram et al. 2014; Ferchaud & Hansen 2016) or closely related species (Harr 2006; Ellegren et al. 2012; Renaut et al. 2013). However, determining whether genomic regions of increased differentiation harbour speciation genes that are resistant to gene flow (Harrison 1986; Wu 2001; Harrison & Larson 2014) or instead reveal the incidental consequences of selection reducing neutral diversity in low-recombining regions (Noor & Bennett 2009; Turner & Hahn 2010; Cruickshank & Hahn 2014) is not always straightforward. Therefore, it is a hard task to rewind the sequence of historical and selective events that underlie heterogeneous genomic differentiation patterns while taking into account the diversity of mechanisms which may be at play. Many studies have used genome scans to investigate patterns of heterogeneous differentiation and map the so-called genomic islands of differentiation (see reviews by Feder et al. 2012; Seehausen et al. 2014 and references therein). However, disentangling the relative influence of gene flow, historical processes and recombination rate variations on the genomic landscape of differentiation has remained highly challenging so far (Barrett & Hoekstra 2011; Nachman & Payseur 2012; Roesti et al. 2013; Ellegren 2014).
The study of replicated pairs of populations (e.g. Schluter & McPhail 1993; Nosil et al. 2002, 2009; Berner et al. 2009; Gagnaire et al. 2013) inhabiting different environments offers great opportunities to investigate these processes for which variable outcomes are expected (Welch & Jiggins 2014; Lindtke & Buerkle 2015). It is often hypothesized that these pairs offer independent replicates of a repeated evolutionary response to similar ecological constraints and thus provide ideal systems to study ecological speciation (e.g. Feder et al. 2012). More generally, parallel phenotypic and genetic divergence among population pairs is commonly attributed to the repeated action of natural selection driving speciation (Schluter & Nagel 1995; Johannesson 2001).
However, different scenarios can lead to such patterns of parallel genomic divergence (Johannesson et al. 2010; Bierne et al. 2013; Welch & Jiggins 2014). For instance, allopatric divergence followed by secondary contacts in multiple locations can generate genetic parallelism through differential introgression between neutral and selected regions (Bierne et al. 2013), a process known to be difficult to distinguish from primary differentiation (Endler 1977, 1982; Barton & Hewitt 1985). Therefore, understanding the origin of adaptive variation is becoming a key issue in studies of parallel evolution, as there is a need to determine whether divergence has been fuelled either by new mutations, standing variation which arose by mutations or gene flow in the ancestral population, or by recent secondary contact (Welch & Jiggins 2014). Reconstructing the demographic history of species divergence may help to identify the evolutionary scenarios underlying observed divergence patterns and therefore has the potential to reveal how much parallel divergence patterns reflect parallel evolutionary histories. Moreover, demographic events may confound the detection of selection in neutrality tests (Brandvain & Wright 2016; Flatt 2016). Therefore, understanding the demographic divergence history is a necessary requisite for building appropriate neutral divergence models in selection detection tests, which may contribute to make better sense of genome scans (Nielsen et al. 2007, 2009; Li et al. 2012).
Recent methods relying on full likelihood analysis (Li & Durbin 2011; Mailund et al. 2012), approximate Bayesian computation (Tavare et al. 1997; Beaumont et al. 2002; Beaumont 2010) or composite likelihood (Wiuf 2006; Gutenkunst et al. 2009) have greatly helped testing increasingly complex hypotheses about the demographic history. However, one particularly challenging task in reconstructing the history of species divergence is to integrate temporal variations in gene flow intensity as well as the possibility for variable amounts of gene flow across the genome. Only few studies have taken this heterogeneity into account (e.g. Roux et al. 2013, 2014; Sousa et al. 2013; Tine et al. 2014; Le Moan et al. 2016) to address the effect of genetic barriers reducing the effective migration rate at linked neutral loci (Barton & Bengtsson 1986; Feder & Nosil 2010). Here, our goal was to address whether parallel phenotypic differentiation between pairs of lamprey ecotypes was accompanied by parallel genetic differentiation, and whether these patterns have likely resulted from independent or shared divergence histories.
Lampreys are jawless vertebrates (agnathans) generally occurring as ‘paired’ species (or ecotypes) with drastically divergent life history strategies: adults are either parasitic anadromous or nonparasitic and freshwater resident. In Western Europe, the parasitic river lamprey (Lampetra fluviatilis, Linnaeus 1758) and the nonparasitic brook lamprey (Lampetra planeri, Bloch 1784) form replicate population pairs that are found from the Iberic peninsula (Tagus river) to Scandinavia and Balkans. These ecotype pairs exhibit varying levels of geographical connectivity, from complete geographical overlap during breeding (sympatric pairs) to varying degrees of spatial disconnection (parapatric and allopatric pairs). Relatively little is known about evolutionary divergence between parasitic and freshwater-resident ecotypes (Docker 2009). Most genetic studies have found little to moderate level of genetic differentiation between ecotypes (Schreiber & Engelhorn 1998; Espanhol et al. 2007; Blank et al. 2008; Pereira et al. 2010; Bracken et al. 2015), but they neither clearly distinguished sympatric and parapatric sites nor used enough markers to address the extent of divergence across the genome. Only the study by Mateus et al. (2013) used RAD-sequencing data and found a strong genomewide differentiation between ecotypes in a sympatric population pair from the southern limit of the distribution range. They provided a list of candidate genes putatively involved in adaptation to migratory vs. resident lifestyles, which could be implicated in reproductive isolation between ecotypes. However, this study focused on a single population pair provided limited insights into the historical, demographic and selective aspects underlying genetic divergence. More recently, Rougemont et al. (2015) studied 10 population pairs located in the northern part of the distribution range and varying in their level of geographic connectivity. They showed that within-river opportunities for gene flow have a strong influence on the average level of differentiation measured with microsatellite markers. The sympatric pairs displayed lower genetic differentiation than parapatric pairs, which were themselves less divergent than the southern sympatric pair described by Mateus et al. (2013). Incomplete reproductive isolation and gene flow may thus have allowed stronger genetic introgression in northern compared with southern sympatric pairs, with potentially important consequences for choosing the most relevant population pairs to disentangle the effects of selection from genetic drift during divergence. Indeed, in the populations connected by gene flow, only genomic regions involved in reproductive isolation and local adaptation are expected to resist the homogenizing effect of introgression (Harrison & Larson 2016). Thus, these populations may be more appropriate to study the genetic basis of reproductive isolation. Accordingly, Rougemont et al. (2016) attempted to infer the demographic history of divergence using microsatellite data using population pairs connected by gene flow. However, distinguishing between scenarios of primary divergence vs. secondary contact was difficult because both tend to converge to the same equilibrium with neutral markers (Bierne et al. 2013). Therefore, moving towards a genomic approach incorporating heterogeneous differentiation along the genome should improve our understanding of the divergence process in European lampreys.
In this study, we used a model-based approach to reconstruct the divergence history of L. fluviatilis and L. planeri using RAD-sequencing data. We took advantage of the original distribution of resident and migratory lampreys in both sympatry and parapatry in nine replicated pairs to document levels of gene flow between ecotypes and among population pairs. We then used these population pairs, to (i) compare alternative models of demographic divergence history, (ii) estimate the proportion of the genome experiencing reduced gene flow, (iii) identify genomic markers showing particularly high differentiation between ecotypes under the most likely divergence demographic model and (iv) evaluate the extent of parallelism among replicate pairs of ecotypes.
Materials and methods
Sampling
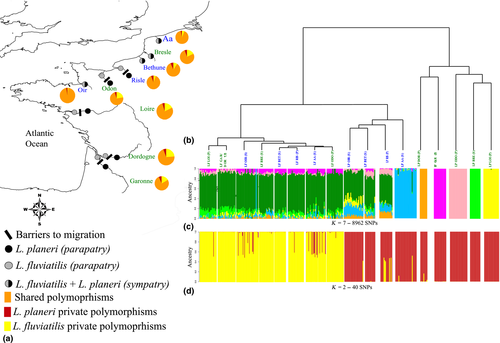
We focused on nine population pairs of lampreys sampled in France, including four sympatric and five parapatric pairs varying in their level of neutral genetic differentiation and geographic connectivity (Rougemont et al. 2015) (Fig. 1). Populations were collected in sympatry in the Aa, Oir and Bethune and very close to each other in Bresle, where sampling sites were located 8 km apart with no obstacle between them. Hence, brook lampreys could freely drift downstream (Dawson et al. 2015) and river lampreys move upstream leaving place for potential interbreeding of the two ecotypes. Pairs from the Risle and Odon River were collected on the same river but were separated by dams, with Lampetra planeri (Lp) from the Odon being captured far upstream in sites unlikely to be colonized by Lampetra fluviatilis (Lf), whereas Lp on the Risle might still be connected to Lf through passive drift during flood events. Pairs from the Loire-Cens, Dordogne-Jalles and Garonne-Saucats were strongly disconnected. In these cases, Lf were collected in mainstream areas before spawning, whereas Lp were collected in isolated upstream reaches not accessible to Lf. Details of sampling methods are provided in Rougemont et al. (2015). A total of 338 individuals were included in the present analysis (Table 1). Three sea lamprey samples (Petromyzon marinus) collected in 2012 on the Scorff River, France (DNA extracted from fin clips preserved in 95EtOH), were also included as an out-group to polarize SNPs.
| River | Situation | N Lf/N Lp | FST μsat | N SNPs | N hybrids | Obs Het Lf | Obs Het Lp | FST SNPs | FST max | FST = 1 | 90% FST quantile | Outliers |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OIR | S | 26/25 | 0.030 | 16 557 | 7 | 0.294 | 0.296 | 0.042 | 0.823 | 0 | 0.117 | 324 |
| BET | S | 14/12 | 0.028 | 14 199 | 0 | 0.297 | 0.293 | 0.053 | 1 | 21 | 0.168 | 643 |
| RIS | P | 19/15 | 0.040 | 16 672 | 6 | 0.294 | 0.296 | 0.065 | 1 | 2 | 0.166 | 958 |
| AA | S | 22/27 | 0.080 | 15 405 | 9 | 0.292 | 0.280 | 0.076 | 0.821 | 0 | 0.169 | 883 |
| BRE | S | 17/12 | 0.076 | 15 511 | 0 | 0.300 | 0.257 | 0.143 | 1 | 24 | 0.315 | 2150 |
| DOR | P | 22/8 | 0.190 | 17 330 | 0 | 0.293 | 0.237 | 0.150 | 1 | 23 | 0.312 | NA |
| GAR | P | 22/15 | 0.087 | 16 926 | 0 | 0.293 | 0.261 | 0.157 | 1 | 24 | 0.354 | 2483 |
| LOI | P | 21/19 | 0.150 | 16 407 | 1 | 0.300 | 0.254 | 0.153 | 1 | 5 | 0.327 | 2535 |
| ODO | P | 20/22 | 0.190 | 15 908 | 0 | 0.305 | 0.247 | 0.207 | 1 | 29 | 0.404 | 3317 |
- ‘Situation’ indicates the location of the two populations in the watershed (S, sympatry; P, parapatry, see Materials and methods for more information). Numbers of Lampetra fluviatilis and Lampetra planeri (N Lf/N Lp) are provided as well as estimates of genetic differentiation based on 13 microsatellite markers (Rougemont et al. 2015). N SNPs, Number of SNPs; N hybrids, Number of hybrids, Observed Heterozygosity (Obs Het) in each populations are indicated as well as genomewide FST, maximum FST value, number of FST reaching the maximum value of 1, the 90th quantile and the number of outliers found using either the 97% quantile method or the neutral model.
- The neutral expectations for the DOR population cannot be computed (see methods).
Library preparation and sequencing
Genomic DNA was extracted using individual extraction kits (NucleoSpin Tissue; Macherey Nagel) following manufacturer's recommended protocols. DNA quality was checked using a spectrophotometer (NanoDrop 2000; Thermo Scientific), quantified using a fluorimeter (qubit 2.0) and standardized to 22 ng/μL. DNA was then used to construct a total of 13 libraries, each composed of 48 randomly chosen individuals across all lamprey populations and following the protocol from Baird et al. (2008) using the restriction enzyme SbfI. Samples were individually barcoded and paired-end sequenced on eight lanes of an Illumina Hiseq 2500 (125-bp paired-end reads) and five lanes of an Illumina Hiseq 2000 (100-bp paired-end reads) at montpellier genomix and genotoul sequencing platforms, respectively.
Genotyping and bioinformatics
Raw reads were first demultiplexed using gbsx (Herten et al. 2015), filtered for overall quality, checked for the presence of barcode using cutadapt (Martin 2011) and trimmed to 85 bp. We then took advantage from paired-end sequences to identify and remove PCR duplicates using stacks V 1.24 program clone_filter (Catchen et al. 2013). We subsequently used the stacks V 1.24 pipeline to identify RAD loci from forward reads using all individuals from all populations. Due to the large divergence with P. marinus and an incompletely assembled genome for this species (Smith et al. 2013), we performed a de novo analysis using ustacks with a minimum stack depth (m) of four and a maximum of mismatches (M) of four. These parameters were determined using replicated individuals included at random in each sequencing platform. More specifically, we tested all combinations of m and M parameters between 2 and 10 using the replicated individuals. The same operation was repeated for each replicated individual. Optimal m and M values were chosen so as to obtain minimal differences in the total number of SNPs between replicates (see Table S1, Supporting information). This allowed us to minimize the presence of paralogs in the data set while maximizing the genetic diversity obtained. A maximum of three mismatches between two given homozygote loci was allowed to consider them as homologues, resulting in a catalogue (build using cstacks) containing 282610 RAD tags. Each individual was finally genotyped after matching its RAD data against the catalogue using the sstacks program, and the genotypes were exported in VCF format using population program for further filtering in vcftools (Danecek et al. 2011). Different data sets were created: for population genetics analyses, we constructed nine data sets (one for each river) comprising both Lf and Lp populations from the same pair; for demographic analyses, we constructed another series of nine data sets with different filtering criteria, and we ultimately created a global data set to investigate patterns of global population structure including all populations and ecotypes. We applied a series of filtering steps that aimed at excluding as much as possible SNPs with genotyping errors and false SNPs resulting from the merging of paralogs. To do so, the starting data set was first split into nine data sets corresponding to each river pair. Each of them was further split into two data sets corresponding to each ecotype. We only kept markers genotyped in at least 80% of the individuals in each population, with a minimum sequencing depth of 10× and a maximum of 100× to avoid the inclusion of highly repetitive tags that could reflect paralogy. We then excluded loci deviating from Hardy–Weinberg equilibrium by keeping only those loci with a P-value higher than 0.05. To limit the presence of rare variants that are poorly informative (Roesti et al. 2012), we kept loci with a minor allele frequency (MAF) higher than 0.1 in at least one of the populations separately or with a MAF higher than 0.05 in each population pair data set for all analyses except for demographic inferences. This filtering step allowed us to retain variants occurring at low frequency in some Lf but monomorphic in Lp and vice versa, in order to obtain comparable representations of low-frequency polymorphisms present in each population. In a final filtering step, we excluded loci with an observed heterozygosity larger than 0.5 in both Lf and Lp in each river (Hohenlohe et al. 2011). The nine ‘pairwise’ data sets obtained were then merged into a global data set in which only markers genotyped in at least 70% of the individuals across all populations were kept to reduce the proportion of missing data. The different filters were applied using vcftools, R scripts and custom bash scripts.
Population genetic differentiation, diversity and individual clustering
For each locus, the level of genetic differentiation between ecotypes within each river and over all populations was estimated using FST (Weir & Cockerham 1984) in vcftools. Negative values were set to zero to compute the mean genomewide FST. Significance of FST values and 95% confidence intervals were computed in R using bootstrap methods as implemented in the stampp package (Pembleton et al. 2013). We further measured the proportion of shared polymorphisms independently for each pair. To investigate population clustering, we first computed a matrix of Nei's genetic distance using Da (Nei et al. 1983) in the stampp R package for each pair of populations. The distance matrix was then used to create an upgma dendrogram using the ggplot2 R package (Wickham 2009) to describe broad patterns of genetic structure. Second, we used admixture v 1.23 (Alexander et al. 2009) to perform individual clustering and to estimate individual admixture proportions. Given the strong geographical component of genetic differentiation in L. planeri (see Results), we were not able to distinguish the two ecotypes in all populations based on this full data set. However, we were concerned about distinguishing putatively recent hybrids from pure individuals to avoid biases in our demographic inferences. We therefore used the 10% most highly differentiated SNPs that were shared between the four least differentiated pairs (i.e. Aa, Bethune, Oir and Risle), based on the distribution of between-ecotypes FST, to improve the ability to discriminate between the two ecotypes and identify potential hybrids. This resulted in a subset of 40 SNPs that were also shared between the remaining population pairs (see Results). We then tested the ability of these markers to assign individuals to their respective ecotypes and to identify putative hybrid genotypes (i.e. possible F1 and backcrosses) using structure 2.3.3 (Pritchard et al. 2000) and newhybrids 1.1 (Anderson & Thompson 2002). Details of structure and newhybrids settings are presented in Supporting Information. The robustness of the inference based on newhybrids was evaluated using simulated data with hybridlab 1.3 (Nielsen et al. 2006). Briefly, individuals with a q-value (structure analysis) or assignment probability (newhybrids) >0.9 of being either Lf or Lp were used to generate individual genotypes belonging to four different hybrid classes (F1, F2 and first-generation backcross in both directions), which were then reclassified in newhybrids. As the subset of 40 SNPs offered a high power to identify hybrid genotypes (>99%), we excluded all individuals identified as hybrids from the real data set containing the nine populations pairs and reran the admixture analysis using the subset of 40 markers to better discriminate ecotypes.
Demographic history of divergence
We used a modified version of the diffusion approximation method implemented in δaδi (Gutenkunst et al. 2009) to analyse the joint site frequency spectrum (JSFS) of Lf and Lp in each population pair. Different models of demographic divergence were compared for each pair, and the model providing the best fit to the observed JSFS was determined using the Akaike information criterion (AIC). The main changes to the original δaδi program that were implemented in the modified version by Tine et al. (2014) include (i) the addition of two simulated annealing optimization phases prior to the Broyden–Fletcher–Goldfarb–Shanno optimization to better explore the likelihood landscape and improve global convergence; (ii) the introduction of a parameter that accounts for the proportion of mis-oriented SNPs when using unfolded JSFS; and (iii) most importantly, the incorporation of varying migration rates across the genome to account for semi-permeability due to localized barriers to gene flow (Wu 2001) as in Tine et al. (2014). This last change was performed by defining two categories of loci occurring in proportions P and 1-P across the genome, the first one containing neutral loci that are exchanged between populations with gross migration rates M12 and M21, and the second category comprising loci that experience reduced effective migration rates Me12 and Me21.
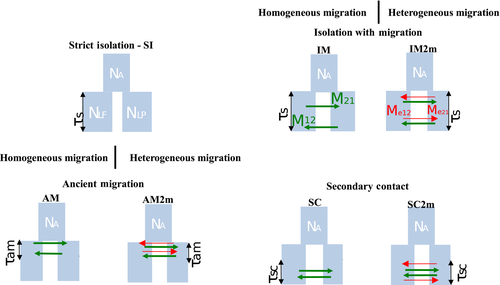
Seven alternatives models of speciation were fitted to the observed JSFS and compared (Fig. 2), including the model of strict isolation (SI), three different models incorporating homogeneous migration along the genome (IM, AM and SC) and three models incorporating heterogeneous migration along the genome (IM2m, AM2m and SC2m). Gene flow was supposed to be either ongoing during the whole divergence history (isolation-with-migration models IM and IM2m), only present at the beginning of divergence (ancient migration models AM and AM2m) or starting after a period of complete isolation (secondary contact models SC and SC2m).
The observed JSFS were built using SNP data sets with no MAF threshold filter with the function implemented in δaδi. For each of the nine pairs, we randomly sampled a single SNP per RAD tag to avoid as much as possible to include linked SNPs in the spectrum, which produced JSFS composed of 5000–10 000 SNPs depending on pairs. Computing levels of linkage disequilibrium to remove strongly linked variants should be ideally performed to follow δaδi's assumptions. However, our model comparison procedure should not be strongly affected by our approach based on keeping a single SNP per RAD. We excluded the population pair from the Dordogne River as it contained only seven individuals suitable for this analysis (one individual had a too low genotyping rate). We also excluded individuals of hybrid origin to avoid the confounding effect of contemporary hybridization on the inference of long-term gene flow. A total of 25 independent runs were performed for each model to check for convergence. The run providing the lowest AIC was kept for each model to make comparisons among models and estimate model parameters. Demographic parameters were all scaled by theta in the ancestral population and were not converted into biological estimates as relevant estimates of mutation rate are not available in lampreys. However, some parameter ratios which do not depend on theta or on the mutation rate can be used for (careful) interpretation. We used the folded JSFS for model selection because the P. marinus out-group was too distant which resulted in a highly reduced number of orientable polymorphisms. Nevertheless, we evaluated the diffusion prediction accuracy for estimating migration rates (m) and the proportion of neutral loci (P) using the unfolded JSFS. In these cases, the previously estimated time for split, secondary contact or migration stop and the effective population size were fixed using estimated values obtained from the folded JSFS.
Detecting selection and measuring parallel differentiation
Demographic investigations confirmed that some populations exchange genes while others are currently almost isolated (see Results). Consequently, the population pairs connected by gene flow were more appropriate for investigating the genetic basis of reproductive isolation as mainly genomic regions directly and indirectly involved in reproductive isolation are expected to resist the homogenizing effect of gene flow (Seehausen et al. 2014; Harrison & Larson 2016). In addition, the complex history inferred from δaδi analyses (see Results) challenges the use of classical model-based outlier detection tests that may result in high rates of false positives in such cases (Lotterhos & Whitlock 2014, 2015). To circumvent these problems, we took advantage of our previous analyses where model parameters (time of divergence, time of secondary contact, time of ancient migration, effective population size NLf and NLp and homogeneous migration rates M1 and M2) were inferred from the best model. Heterogeneous gene flow was, however, not incorporated in the simulated model as we were interested in simulating a neutral model in which genes were freely exchanged between populations. We thus used neutral gene flow estimates to simulate data sets with msstatsfst (Eckert et al. 2010) that uses the coalescent simulator ms (Hudson 2002), and subsequently computed the neutral envelop of Weir et Cockerham's FST. Markers with FST values lying above this envelop were considered as putatively under direct or indirect selection. Each simulated data set was generated for 200 000 SNPs using the same number of individuals as in the real corresponding data set.
To test for parallelism in divergence, we first counted the number of shared outliers (determined from our neutral model) between connected pairs (i.e. 10 comparisons among pairs) and performed randomization tests to test whether this number was greater than expected by chance. We subsequently constructed coplots of FST/FST between pairs of populations to better visualize the extent of outlier sharing and parallelism. To validate our hypothesis that the most disconnected populations were less suited to investigate genetic parallelism, we also performed coplots of FST/FST between pairs of disconnected populations based on outliers identified from our neutral model. To further gain insight into parallel allele frequency shifts observed at outlier loci, we determined the frequency of the derived allele (using the P. marinus as an out-group) when this information was available. We used a stringent approach in which the ancestral state of the allele was inferred only when fixed in P. marinus. We then tested for unbalanced derived allele frequencies between the two ecotypes using t-tests. All analyses were performed using CUSTOM r SCRIPTS. Finally, we used the sequences of the markers potentially under divergent selection between the two ecotypes in connected population pairs to perform blast (Altschul et al. 1990) analysis on the ncbi nr database using an e-value threshold of 1 × 10−8.
Results
Genomewide diversity
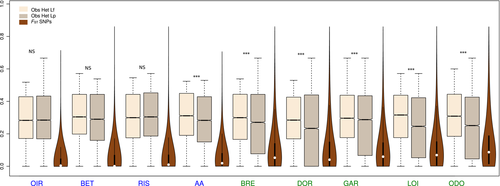
1 054 852 013 reads were obtained after demultiplexing, representing an average of 2 747 010 reads per individuals. Twenty-two individuals were excluded due to their low read numbers and 316 individuals were kept for subsequent analyses. After appropriate filtering, a total of 8962 SNPs were kept in the global data set containing all populations. The number of SNPs kept for the nine population pairs sub-data sets ranged from 14 201 to 17 335 (Table 1, Table S2, Supporting information). Average observed heterozygosity was 0.296 in Lf and 0.264 in Lp and was significantly higher in Lf compared with Lp for all parapatric pairs (t-test, P = 2.10−16 Fig. 3), except the Risle (t-test, P > 0.01). There were significant differences in observed heterozygosity in two sympatric pairs, the Aa and Bresle populations (t-test, P = 2.10−16). Overall, Lf displayed 2.7 times more private polymorphic sites than Lp. As for heterozygosity, this pattern hides a more complex situation (Fig. 1a) in which sympatric Lf populations had 1.8 times more private polymorphic loci than resident Lp, whereas parapatric Lf populations contained 3.2 times more polymorphic loci than isolated Lp populations.
Based on their genetic characteristics, the Risle and Bresle populations were two ‘outliers’ with respect to within-river connectivity between ecotypes. The number of private polymorphisms in Lf and Lp was similar in the Risle River (949 vs. 901) but strongly asymmetric between the sympatric populations from the Bresle River (2758 vs. 889 polymorphic only in Lf and Lp, respectively). In agreement with these results, minor allelic frequencies in isolated Lp populations displayed an L-shaped distribution, whereas allelic frequencies were more evenly distributed in connected Lp and in Lf populations, as illustrated by their JSFS (Fig. 5).
Population structure and introgression
Patterns of population structure inferred from the admixture analysis and neighbour-joining tree (Fig. 1b,c) revealed the effect of geographical isolation on the genomewide level of differentiation. First, the admixture analysis for K = 2 (Fig. S1, Supporting information) did not separate the two ecotypes as the most geographically isolated Lp populations (Lp Odon and Lp Loire) clustered separately from all other populations. Admixture analyses performed with increasing values of K confirmed a clustering by geographical origin of the geographically disconnected Lp populations rather than by ecotypes. The optimal number of clusters (K) determined using admixture cross-validation procedure (K = 7) allowed to discriminate each geographically disconnected Lp population (those for which genomewide Fst >0.10) from the remaining populations. The geographically connected populations from the Oir, Bethune and Risle rivers (Fst < 0.10) were admixed with Lf populations. Ultimately, for K = 10, all Lp populations but the Bethune River clustered separately and we found two Lf clusters corresponding to the channel and Atlantic areas (Fig. S2, Supporting information).
The subset of 40 highly differentiated markers displayed an average FST of 0.7 between Lp and Lf. structure analysis indicated that this marker subset was able to accurately discriminate the two ecotypes (mean q-value from structure was 0.03 and 0.05, in Lf and Lp, respectively, see also Fig. 2d). The structure analysis also identified a total of 22 individuals (five females, nine males, seven with undetermined sexes, Table S3, Supporting information) with mixed q-values resembling F1 and one individual of possible backcross origin. newhybrids analysis confirmed structure results and identified 20 F1 hybrids, 1 F2 and 1 backcross (Table S3, Supporting information). The majority of these hybrid individuals (64%) displayed a Lf phenotype (Table S3, Supporting information). Simulated parents, F1 and backcrosses with Lp individuals were always correctly assigned using hybridlab into their respective categories using an assignment probability threshold of 0.9, yielding a detection power of 100% for these three hybrid classes (Table S4, Supporting information). One individual backcrossed with Lf and 7 F2 was correctly classified but with a probability threshold below 0.9, hence yielding an assignment power of 99.6% and 97.6% for these two categories, respectively.
Genomewide differentiation patterns
After exclusion of hybrids, the global genomewide differentiation between the two ecotypes was 0.128 (95% CI = 0.124–0.131, P < 0.00001), but genetic differentiation was highly variable across pairs (Table 1). In general, parapatric pairs were more genetically differentiated than sympatric pairs (Table 1). In sympatry, the genomewide FST between ecotypes ranged from 0.045 in the Oir River to 0.135 in the Bresle River, whereas in parapatry, it ranged from 0.06 in the Risle River to 0.20 in the Odon River. Global FST among Lf populations was 0.01 (95% CI = 0.005–0.015, P < 0.0001, ranging from FST = 0–0.024 in pairwise comparisons). In contrast, Lp populations displayed a stronger genetic structure (global FST = 0.167, 95% CI = 0.162–0.173, P < 0.0001), with pairwise FST values ranging from 0.062 to 0.262. The genomewide variance in FST for ecotype pairs also increased with geographical isolation, and the frequency distribution of FST values was shifted towards its right tail (Fig. 3). Accordingly, the number of differentially fixed loci increased with genomewide divergence between ecotypes from 0 in Oir and Aa to more than 20 in the Bresle, Dordogne and Odon pairs. Surprisingly, 21 differentially fixed loci were found in the Bethune pair, despite a low genomewide divergence. Similarly, the 90% FST quantile increased with increasing genomewide divergence and geographical isolation in all cases but the Bresle, where Lf and Lp populations were moderately differentiated despite their close geographical proximity.
Demographic history
Detailed results for demographic inferences are provided in Table 2, Fig. 4 and Fig. S3 (Supporting information). In general, the strict isolation model (SI) was weakly supported as it never captured asymmetries in the observed JSFS and failed to recover the most differentiated SNPs. In comparison, the three models with homogeneous gene flow (AM, IM and SC) provided better fits to the data with good predictions of the JSFS’ asymmetry. However, in every case, incorporating heterogeneous gene flow (AM2m, IM2m, SC2m) largely improved the fit (Table 2). In connected populations, under the AM (and AM2m) model, the timing of ancient migration stop (τam) reached a value of zero, meaning that this model converged to the simple IM (and IM2m) model. Overall, we repeatedly found that the secondary contact model incorporating heterogeneous migration rates (SC2m) provided the best fit to the observed JSFS in the four population pairs for which FST was <0.10. In the sympatric population with a FST >0.10 (Bresle), the most likely scenario was IM2 m, but both the SC2m and AM2m produced very similar AICs (ΔAIC < 2). By contrast, the AM2m model unambiguously provided the best fit (ΔAIC >10) for the three remaining parapatric population pairs. In every pair, the effective population size was larger in Lf than in Lp as indicated by the ratio of (Ne Lf)/(Ne Lp) in which Ne was on average 8 and 19 times higher in Lf under the SC2 m and AM2m model, respectively. Using unfolded JSFS, we observed asymmetries in gene flow with ratios of m21/m12 indicating a stronger migration from Lf to Lp in all rivers except in the Oir where levels of gene flow were similar in both directions under the SC2m. Under the AM2m model, a similar trend was observed except for the Cens River. Ratios of secondary contact to divergence times (τsc/τsplit) indicated relatively short periods of introgression ranging from 7% to 22% of the total divergence time. Using the unfolded JSFS, estimates of the proportion of loci freely exchanged in connected populations ranged from 0.88 (Aa) to 0.94 (Risle and Oir), indicating that from 6% to 12% of loci displayed a reduced effective migration rate between the two ecotypes. The inferred proportions of freely exchanged loci in disconnected population pairs was lower, ranging from 0.83 (GAR) to 0.89 (ODO). In disconnected populations, the ratios of τam/τsplit also suggested a very recent arrest of gene flow after a much longer period of isolation.
| (a) Connected population pairs | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| River | Model | AIC | log lik | theta | N Lf | N Lp | m12 | m21 | me12 | me21 | Ts | Tsc/am | P | O |
| OIR | SI | 2217.3 | −1105.7 | 2475.6 | 19.00 | 0.44 | 0.03 | |||||||
| OIR | AM | 1697.5 | −842.8 | 4532.6 | 18.42 | 0.39 | 19.34 | 4.84 | 3.80 | 3.80 | ||||
| OIR | IM | 1740.7 | −865.3 | 1278.5 | 12.36 | 0.56 | 6.72 | 8.30 | 3.15 | |||||
| OIR | SC | 1733.6 | −860.8 | 1189.3 | 12.80 | 0.53 | 6.74 | 8.66 | 1.22 | 1.17 | ||||
| OIR | AM2m | 1706.4 | −844.2 | 381.7 | 7.82 | 2.06 | 0.09 | 37.77 | 1.32 | 2.17 | 9.95 | 9.95 | 0.27 | |
| OIR | IM2m | 1688.6 | −836.3 | 697.0 | 7.42 | 1.60 | 10.95 | 3.36 | 0.86 | 2.72 | 8.23 | 0.70 | ||
| OIR | SC2m | 1663.5 | −822.7 | 2125.5 | 1.97 | 0.55 | 41.29 | 8.85 | 3.48 | 7.44 | 0.49 | 0.14 | 0.67 | |
| OIR | SC2m | 789.7 | −384.8 | 421.6 | 1.95 | 0.55 | 8.67 | 7.17 | 0.29 | 0.39 | 0.55 | 0.13 | 0.94 | 0.98 |
| BET | SI | 1705.3 | −849.7 | 1575.4 | 10.84 | 0.83 | 0.08 | |||||||
| BET | AM | 1237.2 | −612.6 | 863.3 | 19.16 | 0.98 | 3.63 | 2.25 | 2.31 | 2.31 | ||||
| BET | IM | 1326.9 | −658.4 | 2027.9 | 8.69 | 0.43 | 8.86 | 3.22 | 3.80 | |||||
| BET | SC | 1199.5 | −593.7 | 191.7 | 9.70 | 4.34 | 1.24 | 0.91 | 8.82 | 0.84 | ||||
| BET | AM2m | 1051.5 | −516.7 | 668.2 | 19.38 | 0.58 | 0.63 | 39.03 | 4.99 | 2.71 | 9.56 | 9.54 | 0.14 | |
| BET | IM2m | 1067.2 | −525.6 | 994.1 | 2.75 | 0.24 | 0.00 | 26.51 | 0.03 | 0.90 | 0.93 | 0.95 | ||
| BET | SC2m | 1028.4 | −505.2 | 653.9 | 8.23 | 0.54 | 16.33 | 0.18 | 0.00 | 9.74 | 5.12 | 0.38 | 0.59 | |
| BET | SC2m | 563.3 | −271.6 | 77.4 | 8.15 | 0.53 | 3.89 | 9.86 | 0.26 | 0.37 | 5.07 | 0.37 | 0.90 | 0.98 |
| RIS | SI | 1478.4 | −736.2 | 2098.4 | 19.96 | 0.50 | 0.05 | |||||||
| RIS | AM | 1213.1 | −600.5 | 1009.5 | 19.69 | 1.14 | 4.05 | 2.29 | 3.05 | 3.05 | ||||
| RIS | IM | 1162.2 | −576.1 | 1965.8 | 9.98 | 0.36 | 8.43 | 7.72 | 0.91 | |||||
| RIS | SC | 1200.1 | −594.1 | 1630.8 | 1.69 | 0.25 | 3.10 | 19.67 | 0.42 | 0.07 | ||||
| RIS | AM2m | 1038.3 | −510.2 | 591.1 | 4.79 | 0.36 | 0.00 | 16.39 | 0.09 | 1.36 | 4.83 | 4.83 | 0.95 | |
| RIS | IM2m | 1033.3 | −508.7 | 1068.8 | 2.70 | 0.19 | 0.00 | 29.22 | 0.11 | 2.59 | 1.98 | 0.95 | ||
| RIS | SC2m | 992.9 | −487.4 | 2311.4 | 2.80 | 0.46 | 59.88 | 0.00 | 1.75 | 9.42 | 0.79 | 0.10 | 0.68 | |
| RIS | SC2m | 670.95 | −325.47 | 351.81 | 2.77 | 0.45 | 5.71 | 7.01 | 0.76 | 0.29 | 0.78 | 0.10 | 0.94 | 0.98 |
| AA | SI | 2215.7 | −1104.9 | 1624.8 | 5.33 | 0.58 | 0.07 | |||||||
| AA | AM | 1642.7 | −815.4 | 2679.0 | 16.67 | 0.36 | 8.59 | 2.46 | 7.38 | 7.38 | ||||
| AA | IM | 1756.8 | −873.4 | 595.3 | 4.14 | 0.62 | 0.93 | 4.68 | 2.59 | |||||
| AA | SC | 1677.0 | −832.5 | 1016.0 | 2.20 | 0.86 | 5.22 | 2.40 | 0.78 | 0.20 | ||||
| AA | AM2m | 1501.5 | −741.8 | 575.6 | 4.60 | 0.40 | 0.36 | 9.94 | 0.17 | 0.51 | 2.69 | 2.69 | 0.95 | |
| AA | IM2m | 1489.6 | −736.8 | 1177.4 | 12.94 | 0.55 | 11.17 | 0.82 | 0.00 | 4.44 | 6.33 | 0.87 | ||
| AA | SC2m | 1475.1 | −728.6 | 658.0 | 5.72 | 1.09 | 0.59 | 3.16 | 9.72 | 0.00 | 3.23 | 0.44 | 0.45 | |
| AA | SC2m | 528.2 | −254.1 | 102.4 | 5.66 | 1.08 | 3.00 | 7.87 | 0.58 | 0.31 | 3.19 | 0.44 | 0.88 | 0.98 |
| (b) Disconnected population pairs | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| River | Model | AIC | log lik | theta | N Lf | N Lp | m12 | m21 | me12 | me21 | Ts | Tsc/am | P | O |
| BRE | SI | 1699.8 | −846.9 | 1820.8 | 4.66 | 0.16 | 0.05 | |||||||
| BRE | AM | 1465.6 | −726.8 | 480.9 | 4.87 | 0.23 | 0.11 | 6.57 | 5.55 | 0.01 | ||||
| BRE | IM | 1475.5 | −732.7 | 319.4 | 7.19 | 0.39 | 0.12 | 3.40 | 8.95 | |||||
| BRE | SC | 1475.3 | −731.6 | 1364.1 | 1.66 | 0.08 | 0.60 | 16.96 | 0.66 | 0.18 | ||||
| BRE | AM2m | 1394.6 | −688.3 | 1312.7 | 1.76 | 0.11 | 0.88 | 14.56 | 0.03 | 1.67 | 0.95 | 0.00 | 0.95 | |
| BRE | IM2m | 1393.9 | −688.9 | 418.4 | 5.30 | 0.39 | 0.02 | 0.50 | 0.36 | 3.84 | 6.53 | 0.05 | ||
| BRE | IM2m | 635.3 | −308.7 | 219.5 | 5.25 | 0.39 | 7.28 | 7.10 | 0.70 | 0.55 | 6.47 | 0.94 | 0.98 | |
| BRE | SC2m | 1395.6 | −688.8 | 1359.8 | 1.65 | 0.12 | 1.22 | 12.29 | 0.00 | 1.46 | 0.00 | 0.86 | 0.95 | |
| SAU | SI | 2129.8 | −1061.9 | 2586.6 | 1.73 | 0.16 | 0.04 | |||||||
| SAU | AM | 1946.4 | −967.2 | 656.3 | 4.73 | 0.37 | 0.25 | 5.34 | 6.10 | 0.03 | ||||
| SAU | IM | 2098.0 | −1044.0 | 2560.0 | 1.52 | 0.17 | 0.80 | 3.65 | 0.07 | |||||
| SAU | SC | 2099.9 | −1044.0 | 2560.6 | 1.55 | 0.16 | 0.72 | 3.63 | 0.00 | 0.06 | ||||
| SAU | AM2m | 1863.6 | −922.8 | 1621.2 | 1.91 | 0.15 | 0.93 | 15.28 | 0.15 | 2.10 | 1.52 | 0.01 | 0.95 | |
| SAU | AM2m | 1601.3 | −790.7 | 474.0 | 1.89 | 0.15 | 3.61 | 5.18 | 0.46 | 0.71 | 1.51 | 0.01 | 0.83 | 0.98 |
| SAU | IM2m | 1975.8 | −979.9 | 1287.3 | 2.29 | 0.28 | 0.17 | 0.49 | 1.00 | 4.98 | 2.33 | 0.05 | ||
| SAU | SC2m | 1973.5 | −977.7 | 2041.3 | 1.50 | 0.16 | 1.38 | 8.78 | 0.17 | 0.89 | 0.00 | 0.69 | 0.95 | |
| CEN | SI | 2228.3 | −1111.2 | 2675.7 | 2.05 | 0.10 | 0.03 | |||||||
| CEN | AM | 2015.7 | −1001.9 | 484.9 | 7.01 | 0.25 | 0.12 | 7.09 | 8.69 | 0.02 | ||||
| CEN | IM | 2176.0 | −1083.0 | 531.0 | 6.18 | 0.36 | 0.33 | 3.09 | 7.90 | |||||
| CEN | SC | 2178.1 | −1083.0 | 1562.5 | 2.11 | 0.12 | 0.96 | 9.20 | 0.06 | 1.64 | ||||
| CEN | AM2m | 1889.3 | −935.7 | 461.6 | 7.41 | 0.42 | 0.01 | 0.53 | 0.62 | 4.99 | 9.34 | 0.04 | 0.05 | |
| CEN | AM2m | 513.7 | −246.9 | 320.0 | 7.33 | 0.41 | 6.10 | 4.55 | 0.62 | 0.69 | 9.24 | 0.04 | 0.85 | 0.98 |
| CEN | IM2m | 2000.0 | −992.0 | 2206.9 | 1.51 | 0.13 | 2.25 | 9.51 | 0.01 | 0.80 | 0.58 | 0.95 | ||
| CEN | SC2m | 2003.8 | −992.9 | 2100.0 | 1.68 | 0.11 | 2.17 | 12.01 | 0.00 | 1.55 | 0.07 | 0.64 | 0.95 | |
| ODO | SI | 3095.1 | −1544.5 | 2855.6 | 1.27 | 0.06 | 0.02 | |||||||
| ODO | AM | 2724.1 | −1356.1 | 1109.8 | 3.28 | 0.08 | 0.12 | 14.89 | 3.23 | 0.01 | ||||
| ODO | IM | 2916.3 | −1453.1 | 619.1 | 5.77 | 0.17 | 0.15 | 4.92 | 6.84 | |||||
| ODO | SC | 2893.6 | −1440.8 | 527.6 | 5.75 | 0.15 | 0.19 | 5.50 | 8.66 | 0.59 | ||||
| ODO | AM2m | 2648.1 | −1315.1 | 538.3 | 7.75 | 0.27 | 22.30 | 0.00 | 0.25 | 4.38 | 9.41 | 0.02 | 0.15 | |
| ODO | AM2m | 1129.2 | −554.6 | 232.7 | 7.67 | 0.26 | 2.93 | 5.90 | 0.58 | 0.64 | 9.31 | 0.02 | 0.89 | 0.98 |
| ODO | IM2m | 2791.8 | −1387.9 | 2105.0 | 1.75 | 0.05 | 0.84 | 17.43 | 0.08 | 1.53 | 0.79 | 0.95 | ||
| ODO | SC2m | 2781.2 | −1381.6 | 2263.8 | 1.56 | 0.05 | 1.34 | 19.70 | 0.00 | 1.87 | 0.43 | 0.11 | 0.95 | |
- AIC, Akaike information criterion; log lik, maximum likelihood; theta, 4 Nrefμ, effective mutation rate of the reference population, which here corresponds to the ancestral population; N Lf and NLp, effective population size of Lampetra fluviatilis and Lampetra planeri; m12 and m21, migration from L. planeri to L. fluviatilis and from L. fluviatilis to L. planeri, respectively; me12 and me21, effective migration rate estimated in the most differentiated regions of the genome from L. planeri to L. fluviatilis and from L. fluviatilis to L. planeri, respectively; τs, time of split of the ancestral population in the two daughter species; τsc and τam, duration of the secondary contact (SC and SC2m) and of ancestral migration (AM and AM2m) episodes, respectively; P, proportion of the genome freely exchanged (1-P provides the proportion of the genome non-neutrally exchanged); JSFS, joint site frequency spectrum; SI, strict isolation. (O) proportion of correctly oriented ancestral allelic state.
- The best models are in bold. An additional line (in italic) provides the results of parameter estimation using unfolded JSFS for each best model. The duration of allopatric or sympatric divergence (τs), secondary contact (τsc), ancient migration (τam) and effective population size (NLf, NLp) were fixed based on values inferred from the folded JSFS.
- For each best model, m12, m21, me12, me21 and P were estimated using the unfolded JSFS.

Outlier detection and extent of parallel differentiation
Outlier SNPs represented from 1.96% to 5.75% of the markers in each connected pair (Oir, Bethune, Risle, Aa). The Bresle pair was again an ‘outlier’ (with regard to the level of connectivity) with a high number of SNPs departing from neutral predictions (13.9%). Conversely, we consistently observed a larger proportion of outlier SNPs in parapatry: from 14.7% to 20.9% of the data sets in disconnected pairs.
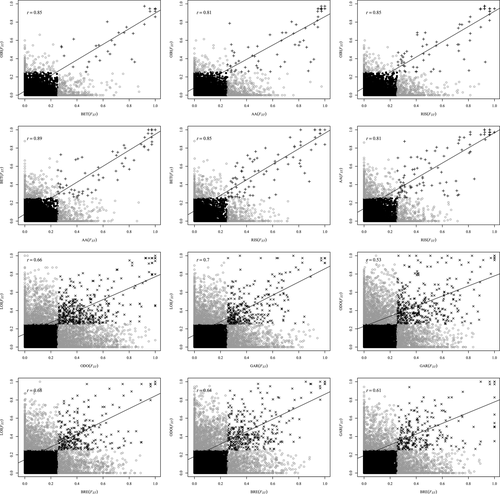
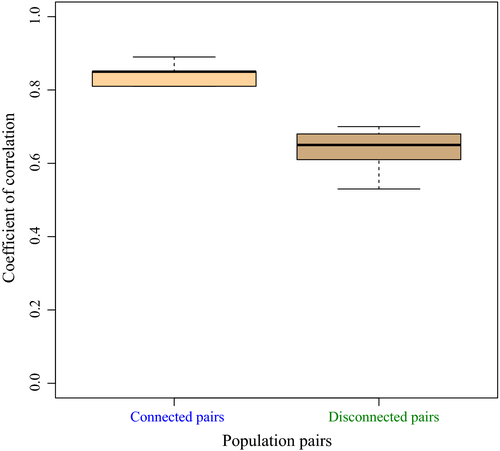
We investigated the proportions of outlier loci that were shared across the four connected population pairs. In the six FST/FST pairwise comparisons, the majority of putatively ‘neutral’ loci were shared across the pairs (mean = 10 667). Simulations under the neutral model yielded a total of 28 outliers (24 independent loci) shared across the six comparisons and an average of 100 outliers SNPs (46–172) shared in all possible population pairs. In all cases, these loci displayed high correlations in their FST values (r = 0.76–0.89, P-value <0.0001, Fig. 5) in FST/FST coplots. This amount of sharing was higher than expected by chance alone (1000 permutations, all P-values <0.0001). Inferences from our neutral models in disconnected populations revealed a similar level of outlier sharing (Fig. 5). However, these populations displayed significantly lower values of pairwise correlation coefficients (r = 0.53–0.70 P-value <0.0001, Fig. 5), as compared to the connected ones (Wilcoxon test W = 60, P-value <0.001; Fig. 6). Using data from P. marinus allowed us to identify the derived allele frequency (DAF) for some outliers in the connected pairs. The DAF was systematically higher in Lp than in Lf for these outlier loci (Fig. 7, paired t-test, all P-values <0.001).

blast analysis
blast analysis of the 28 shared outliers from connected pairs (see Appendix S1) resulted in the identification of two sequences with significant hits (e-value <10e−8, sequence identity higher than 93%) (Table S5, Supporting information). One matched a region of 90 kb known to play key roles in osmoregulation and in the expression of the GnRh developmental hormone. The second sequence-matched genes involved in the immune system: an axial patterning gene, a pineal gland specific opsin gene and a sodium channel gene that was also known to play a role in osmoregulation (Table S5, Supporting information).
Discussion
Disentangling the relative influence of demographic and selective processes at the genomic level is a challenging issue in speciation genomics. In particular, most studies that tested whether parallel phenotypic divergence is mirrored by genetic parallelism did not attempt to infer historical divergence scenarios to explicitly account for the confounding effects of demography in the detection of selection (Hohenlohe et al. 2010; Kaeuffer et al. 2012; Gagnaire et al. 2013; Roda et al. 2013; Westram et al. 2014; Ravinet et al. 2016). Here, we compared contrasted demographic divergence models to determine the most likely evolutionary scenario underlying genomewide patterns of divergence among nine pairs of L. fluviatilis and L. planeri displaying variable levels of geographic connectivity. Gene flow was strong in four connected pairs, whereas the most geographically isolated populations were highly genetically differentiated and showed reduced contemporary gene flow. The most connected populations consistently revealed a signal of historical divergence followed by a recent secondary contact between L. fluviatilis and L. planeri. In addition, there was a higher degree of parallelism in the differentiation level of shared outliers in connected populations than in disconnected ones. Altogether, these results suggest that correlated genomic differentiation patterns among replicate ecotype pairs result from a common history of divergence and gene flow rather than independent gene reuse due to the repeated action of natural selection (Conte et al. 2012; Roesti et al. 2015).
Connectivity determines the rate of hybridization
Before analysing genomic differentiation patterns and demography, a prerequisite was to confirm that RAD-sequencing analysis of population genetic diversity and structure corroborates recent findings based on neutral markers (Rougemont et al. 2015), and then to determine the most appropriate population pairs for understanding the history of ecotypic divergence. First, we validated the high levels of gene flow among L. fluviatilis populations (Bracken et al. 2015; Rougemont et al. 2015) suggesting no homing behaviour, as observed in other migratory lamprey systems (Spice et al. 2012; Hess et al. 2013). Second, despite a relatively low level of polymorphism observed using microsatellite data in European lampreys (Bracken et al. 2015; Rougemont et al. 2015; Mateus et al. 2016) and in other lampreys (Spice et al. 2012), we found a reasonably higher level of polymorphism based on SNPs data, which should provide higher power to address questions related to divergence processes and history. Third, the admixture analysis (Fig. 1c) confirmed that L. planeri populations could be broadly classified into two categories: populations located in downstream areas and highly connected by gene flow to L. fluviatilis and isolated upstream populations that currently do not exchange genes with L. fluviatilis. Ecotype pairs consisting of well-connected populations of L. fluviatilis and L. planeri consistently displayed low levels of genetic differentiation (genomewide FST <0.10). In these pairs, populations of L. planeri also showed similar levels of heterozygosity compared with the migratory ecotype (Table 1, Fig 1) and clustered together with L. fluviatilis in the population tree (Fig. 1b). Contrastingly, the most disconnected pairs displayed higher levels of genetic differentiation (genomewide FST >0.10). Disconnected populations of L. planeri had a lower heterozygosity, shared less polymorphism with river lamprey and formed a separate cluster on the population tree (Fig. 1b). Interestingly, two L. planeri populations (the Bresle and Risle) were outliers with respect to the geographic context (geographical connection or not), as they displayed lower polymorphism and high differentiation in sympatry (Bresle), or higher polymorphism and low differentiation in parapatry (Risle) compared with other populations in comparable geographical contexts. This suggests that the spatial distribution of these populations has changed through time, or, in the case of Risle, that gene flow may occur despite the barriers to migration (e.g. through downstream migration of juvenile Lp or occasional upstream migration of adult Lf). The current geographical distribution of these populations might thus not reflect the demographic history that has shaped their genetic structure (Bierne et al. 2013).
Using a subset of 40 highly differentiated SNPs (FST = 0.7) shared among connected population pairs (identified as such prior to demographic analyses) allowed us to circumvent geographical effects and to distinguish the two ecotypes in all nine population pairs. The low genetic differentiation generally observed between L. fluviatilis and L. planeri has been either taken as an evidence for the role of phenotypic plasticity in ecomorphological differentiation between ecotypes (Beamish 1987; Yamazaki et al. 2006; April et al. 2011; Docker et al. 2012), or as an evidence for a very recent divergence between ecotypes (Docker et al. 1999; Espanhol et al. 2007; Okada et al. 2010). However, these interpretations mostly relied on mitochondrial variation patterns (the most widely used genetic marker in lamprey studies so far), which can be obscured by introgression (Shaw 2002). Our results clearly refute the hypothesis of phenotypic plasticity within a single homogeneous gene pool and demonstrate that parallel genetic differences exist among replicate pairs of brook–river lamprey ecotypes. The subset of highly discriminating markers also allowed us to document geographical hybridization patterns. In general, sympatric populations displayed higher proportions of hybrids compared with parapatric pairs. However, the Bresle River (i.e. the ‘outlier’ sympatric pair) did not contain any hybrid whereas the Risle River (i.e. the ‘outlier’ parapatric pair) contained a relatively high proportion of hybrids. Genome-wide differentiation levels between L. fluviatilis and L. planeri thus appeared to be mostly determined by the degree of geographic connectivity in ecotype pairs, which directly influences the opportunity for gene exchange through hybridization. Among the four highly connected pairs, the Aa and Oir rivers displayed the largest proportions of F1 hybrids (Aa: 16%: Oir 13%). Evidence for frequent hybridization in these rivers may be exacerbated by sampling effects, especially if individuals were collected in places where hybrids tend to occur at higher densities (Vines et al. 2016) as it can be the case during the simultaneous sampling of individuals in the same spawning nest. The comparatively small proportions of later generation hybrids that were detected (one backcross and one F2) may suggest some form of hybrid breakdown (i.e. selection against hybrids or reduced fertility), a hypothesis that would require a more extensive sampling in these hybrid zones to be validated. These results confirm the interest of these populations for the study of speciation in lampreys (Barton & Hewitt 1985; Abbott et al. 2013; Harrison & Larson 2014) and raise the necessity of investigating the effect of gene flow during the long-term divergence history of lamprey ecotypes.
A globally shared history of divergence
Our demographic inferences provided new insights into the recent history of divergence of European lampreys, revealing a relatively longer period of divergence in allopatry compared with the subsequent recent episode of secondary contact. In accordance with previous studies (Roux et al. 2013, 2014; Tine et al. 2014; Le Moan et al. 2016), we found that integrating the heterogeneity of migration rate strongly improves models’ prediction accuracy, thus supporting the importance of taking this source of variation into account for inferring the history of gene flow during speciation. Our analysis revealed two well-supported scenarios (ΔAIC >10) related to the degree of connectivity between L. fluviatilis and L. planeri populations. Connected populations pairs generally held a signal of secondary contact with heterogeneous introgression rates (SC2m), whereas disconnected populations pairs held a signal of recent divergence preceded by heterogeneous migration (AM2m). These contrasted divergence models may, however, not necessarily reflect radically different divergence histories. Indeed, the signal of a past secondary contact may have been lost or obscured by recent drift in parapatric populations. In any case, our results do not support the hypothesis of divergence with ongoing migration (IM/IM2m), except in the case of the Bresle River where it could not be excluded. However in this river, all models including heterogeneous migration rates displayed nearly similar supports (ΔAIC <10 for AM2m and SC2 m compared with IM2m). A possible explanation is that a large difference in effective population sizes during the initial phase of the secondary contact has facilitated gene swamping from the largest population into the small introgressed population.
In the connected populations where the secondary contact scenario was unambiguously detected, only regions involved in divergence are expected to resist the homogenizing effect of gene flow. This makes these population pairs good models to study the evolution of reproductive isolation. These results, together with the finding of hybrids, confirm that RI remains partial as suggested in a previous study (Rougemont et al. 2015). Thus, the genetic architecture of divergence between lamprey ecotypes seems mostly consistent with the existence of barrier loci that locally reduce the effective migration rate along the genome (Barton & Bengtsson 1986; Feder & Nosil 2010). Parameters estimates in connected populations further suggest that the period of allopatry has been (on average) eight times longer than the duration of secondary contact, which matches well the idea of a differential erosion of past genetic differentiation outside the direct vicinity of barrier loci. This seems also consistent with the overall low level of reproductive isolation measured experimentally by Rougemont et al. (2015). This interpretation is also compatible with the lack of divergence observed with mitochondrial markers (Espanhol et al. 2007; Blank et al. 2008) and the moderate level of divergence observed at neutral loci (Bracken et al. 2015; Rougemont et al. 2015). We also repeatedly found higher effective population size in L. fluviatilis compared with L. planeri validating recent findings by Rougemont et al. (2016) based on approximate Bayesian computations using microsatellite data. Such differences may contribute to increase genome swamping in L. planeri. In these conditions, the signal of adaptation and reproductive isolation held by divergent loci can quickly be lost as may be the case in the least differentiated populations (e.g. Oir) (Yeaman 2015).
Recent gene flow and the origin of genetic parallelism
Our genome scan approach based on the inferred neutral model revealed a high proportion (between 2% and 14%) of loci departing from neutrality in each ecotype pair. The level of parallelism was significantly larger in connected pairs compared with disconnected pairs (Figs 5 and 6), which is the exact pattern expected after differential introgression (Bierne et al. 2013). Thus, our results support that recent gene flow has played a key role in generating genetic parallelism, because effective migration is strongly reduced in the same regions of the genome in the different population pairs. These genomic regions experiencing restricted introgression, which are believed to harbour speciation genes, are best revealed in connected population pairs where the confounding effect of drift is less important than in disconnected populations. We found 28 loci shared across the five connected pairs. Such level of parallelism was greater than expected by chance, and in most cases stronger than that observed in several other systems (Gagnaire et al. 2013; Perrier et al. 2013; Westram et al. 2014; Ravinet et al. 2016) but similar to levels of parallelism observed in European anchovies (Le Moan et al. 2016). In addition, our demographic inferences suggested that about 6–12% of the sampled genome was not exchanged neutrally in connected population pairs against 11–17% in disconnected ones, in which our modelling approach may underestimate the neutral variance in differentiation values.
All the genetic differences between ecotypes were not found in parallel among replicate pairs, and different hypotheses can explain this partial genetic parallelism (black and maroon points in Fig. 5). First, the ‘private’ outliers found in a single ecotype pair may correspond to genomic regions with a peak-valley-peak signature between different freshwater-derived populations. This type of chromosomal differentiation pattern may be left by a global selective sweep or by the recent spatial recolonization of the resident ecotype (Bierne 2010; Kim & Maruki 2011; Roesti et al. 2014). Second, partial genetic parallelism may reflect different outcomes of the coupling process (Barton & de Cara 2009) between incompatibilities (e.g. Dobzhansky–Muller incompatibilities) and local adaptation loci following secondary contact (Gagnaire et al. 2013). Depending on the timing and duration of secondary contact, historical differentiation can be also differentially eroded by gene flow among rivers, hence contributing to partial parallelism among different pairs. While we suggest that differential introgression is the most likely process involved in generating parallelism, incomplete parallelism can be explained by all the aforementioned hypotheses (i.e. a global sweep in structured populations, recolonization by brook lamprey ecotypes, variable outcomes of the coupling process and differential erosion of islands among pairs following multiple secondary contacts). All these factors may act jointly at different spatiotemporal scales, and disentangling the respective effect of each is still challenging. In addition, the very high number of nonshared outliers in disconnected populations can be more simply explained by the effect of drift acting independently in each river. In these conditions, genome scans are probably not suited to detect the genomic regions that were influenced by selection. On the other hand, these disconnected populations were extremely useful to highlight the instrumental role of gene flow in generating partial parallelism in the connected population pairs.
We also aimed at investigating the origin of genetic variation that contributes to parallel divergence patterns at the molecular level. The modelling approach indicated that parallelism was not generated by independent de novo mutations arising during divergence with continuous gene flow (IM model). Similarly, it is unlikely that parallelism was generated by recent de novo mutations that arose since the beginning of the secondary contact. Other possible scenarios may involve (i) secondary contact from multiple freshwater refugia, (ii) parallel gene reuse from standing variation present in the parasitic population or (iii) secondary contact between parasitic and nonparasitic populations having diverged in different refugia, with a subsequent spread (spatial reassortment) of alleles involved in nonparasitism in the neighbouring rivers (i.e. the transporter hypothesis; Schluter & Conte 2009; Bierne et al. 2013; Welch & Jiggins 2014). The scenario (ii) would imply divergence with gene flow, a model that was not supported in our demographic inferences. Without further support for the multiple refugia hypothesis, the hypothesis of a spatial reassortment of ancestral variation by migration between rivers appears more parsimonious. This raises the question of whether L. planeri alleles are segregating at low frequency in L. fluviatilis populations at linkage equilibrium, or instead have spread among rivers using the bridge of hybrid genotypes. In any case, the repeated colonization of new rivers by a few individuals with freshwater residency-adapted alleles is expected to have driven rare mutations to high frequencies. This allele surfing effect (Travis et al. 2007; Excoffier & Ray 2008; Lehe et al. 2012) may have been detected here because we found an excess of derived mutations reaching higher frequencies in L. planeri than L. fluviatilis populations for the most highly differentiated loci (Fig. 7). This last line of evidence supports a scenario involving a spread of founder genotypes from river to river during the postglacial recolonization of rivers by the L. planeri ecotype.
The 28 strongly differentiated loci, repeatedly found across nine population pairs, could be considered as good candidates implied in the divergence between L. planeri and L. fluviatilis. Accordingly, some of these outliers were located in genomic regions containing key genes of the GnRh2 family involved in maturation and growth, another gene involved in fast skeletal development and two genes involved in immunity. Interestingly, these genes were also identified as outliers between L. planeri and L. fluviatilis by Mateus et al. (2013) in a population pair from Portugal, which further suggests a shared history of divergence and adaptation at a larger spatial scale. In the future, it would be interesting to implement a global modelling approach that was not feasible here using two-dimensional sfs, in order to test the likelihood of the single secondary contact scenario using the global data set. However, such a global approach is still challenging for the existing demographic inference methods, so our pairwise approach represents a significant first step towards understanding the divergence history of lampreys.
A nondirectly addressed question was whether the heterogeneous patterns of differentiation found here between L. planeri and L. fluviatilis were exclusively compatible with the hypothesis of a semipermeable barrier to gene flow (Wu 2001). An alternative hypothesis is that heterogeneous differentiation has been shaped by selective effects that are not directly involved in reproductive isolation. This may arise during periods of geographical isolation due to positive or background selection acting independently in each population, and reducing genetic diversity at linked neutral sites. This mechanism called postspeciation selection at linked sites can locally increases FST by reducing within-population diversity in some regions of the genome (Cruickshank & Hahn 2014). Here, our demographic inferences were consolidated by the finding of hybrids, which collectively support a role for gene flow in eroding historical differentiation. This interpretation is, however, not incompatible with recombination rate variations favouring the accumulation of differentiation in regions of low recombination (Noor & Bennett 2009; Turner & Hahn 2010; Tine et al. 2014). However, given the high level of genetic differentiation observed in some regions, which contrast to the low levels of differentiation elsewhere, it appears unlikely that linked selection in low-recombining regions would produce such heterogeneity alone. The two processes may act jointly to shape genomic divergence in lampreys and would be better evaluated using a genetic map.
Conclusion and perspectives
Our results support that parallel patterns of genetic divergence between L. planeri and L. fluviatilis were likely caused by a common history of divergence initiated in allopatry and then followed by secondary gene flow eroding past divergence at variable rates across the genome. The level of geographic connectivity between population pairs was a strong determinant of observed divergence patterns, with direct impacts on both demographic inference and genome scans. In particular, stronger drift in populations of small effective size could obscure signals of divergence and result in smaller level of parallelism. Genetic swamping and the evolution of further ecological divergence after secondary contact can also act to, respectively, erode divergent regions or create new regions of divergence that will not bear any signal of parallel divergence. Overall, our data support the idea that the speciation process is best studied in population pairs experiencing high levels of gene flow. The mechanisms underlying divergence between L. fluviatilis and L. planeri were thus best described using sympatric population pairs. In addition, the use of replicated pairs enabled us to identify candidate regions under the direct or indirect effect of selection. Further investigations of these genomic regions should be performed in the future to determine their role in the evolution of life history divergence between L. planeri and L. fluviatilis. Finally, it would be particularly interesting to accurately quantify the relative contribution and interactions between recombination rate variations, selection at linked site (Charlesworth & Campos 2014) and differential introgression (Harrison 1986) on the heterogeneous patterns of differentiation along the genome. Integrating chromosomal variation in effective population size in the demographic divergence models could help to address these issues.
Acknowledgements
We thank A. Oger for his help with sample collection and DNA extraction. We thank our colleagues from U3E INRA staff F. Marchand and J. Tremblay, as well as A. Gaigher, V. Dolo, Y. Salaville, R. Lemasquerier, V. Lauronce, C. Taverny, B. Rigault, C. Rigaud, Y. Perraud, G. Sanson, J.-L. Fagard and P. Domalain who helped us to collect the samples. We are grateful to the Genotoul bioinformatics platform Toulouse Midi-Pyrenees for providing computing and storage resources. We also wish to thank the organizers of this special issue as well as two anonymous reviewers for their insightful comments on the manuscript. This study was funded by the European Regional Development Fund (Transnational programme Interreg IV, Atlantic Aquatic Resource Conservation Project) and by the Office National de l'Eau et des Milieux Aquatiques (ONEMA).
References
Author Contributions:
Q.R., G.E. and S.L. conceived the study. Q.R., A.L.B. and C.G. performed laboratory analyses.
Q.R., P.A.G., C.P. performed bioinformatic and statistical analyses. QR wrote the manuscript with contributions of P.A.G., C.P., S.L., G.E. All authors read and approved the final manuscript.
Data accessibility
Raw demupliplexed fastq.gz files are availables at NCBI SRA accession number SRP074287. VCF files and jsfs spectra are available at: doi:10.5061/dryad.5p5c0. Pipelines for RAD-sequencing analysis as well as the modified version of dadi are available on the first author github pages:
https://github.com/QuentinRougemont/DemographicInference https://github.com/QuentinRougemont/RADseqAnalysis




