Acquisition and excretion of Bartonella quintana by the cat flea, Ctenocephalides felis felis
Abstract
Bartonella quintana is transmitted by the infected faeces of body lice. Recently, this bacterium was detected in cat fleas (Ctenocephalides felis) and in two humans with chronic adenopathy whose only risk factor was contact with cat fleas. In this study, a total of 960 C. felis were divided into 12 groups (2 control groups and 10 infected groups) each containing 80 fleas. The fleas were fed B. quintana-inoculated human blood at different dilutions (≈3.6 × 104 − 8.4 × 109 bacteria) for 4 days via an artificial membrane. Subsequently, all flea groups were fed uninfected blood until day 13 postinfection (dpi). On day 3 pi, B. quintana was detected with two specific genes by quantitative PCR in 60–100% of randomly chosen fleas per dilution: 52% (26/50) in the infected fleas in Trial 1 and 90% (45/50) of the fleas in Trial 2. B. quintana was also identified by molecular and culture assays in flea faeces. The average number of B. quintana as determined by qPCR decreased until the 11th dpi and was absent in both trials at the 13th dpi. Bacteria were localized only in the flea gastrointestinal gut by specific immunohistochemistry. Our results indicate that cat fleas can acquire B. quintana by feeding and release viable organisms into their faeces. Therefore, fleas may play a role as vectors of trench fever or other clinical manifestations that are caused by B. quintana. However, the biological role of C. felis in the transmission of B. quintana under natural conditions is yet to be defined.
Introduction
Bartonella quintana, the causative agent of trench fever, is a fastidious Gram-negative bacterium and is a reemerging human pathogen (Anderson & Neuman 1997). Trench fever was described during World Wars I and II when thousands of soldiers suffered from the disease (McNee et al. 1916; Kostrzewski 1949). In 1990, B. quintana was identified as an agent of bacillary angiomatosis in patients with AIDS (Relman et al. 1990), endocarditis (Drancourt et al. 1995; Spach et al. 1995a), chronic bacteraemia (Spach et al. 1995b; Brouqui et al. 1999) and chronic lymphadenopathy (Raoult et al. 1994). As early as 1920, B. quintana has been observed in body lice, which are considered the main vector of this bacterium (Byam & Lloyd 1920). B. quintana has recently been found in head lice (Bonilla et al. 2009; Angelakis et al. 2011), fleas (Rolain et al. 2003b, 2005), ticks (Chang et al. 2001) and mites (Melter et al. 2012). However, most of these studies detected B. quintana by the use of molecular tools but lacked proof of arthropod vector competence.
Several flea species, including Pulex irritans, Ctenocephalides felis felis, C. canis, Ceratophyllus gallinae, Ceratophyllus columbae and Archaeopsylla erinacei, may infest humans (Rolain et al. 2005). The presence of B. quintana DNA has been reported in cat fleas (C. felis) that were collected from various regions in France and in human fleas (P. irritans) that were collected from a pet monkey (Cercopithecus cephus) in Gabon, Africa (Rolain et al. 2003b, 2005). These results confirm that B. quintana may be found in fleas and may explain two clinical reports of chronic adenopathy that were attributed to B. quintana infection for which the only epidemiologic risk factor that was identified was the presence of fleas (Raoult et al. 1994; Drancourt et al. 1996). Some studies suggest that few animals (such as dogs and cynomolgus and rhesus macaques) can serve as reservoir hosts for B. quintana (Chomel et al. 2006; Huang et al. 2011; Li et al. 2013). Our objective was to evaluate whether cat fleas could acquire B. quintana and excrete viable bacteria in their faeces, thus constituting a potential vector for B. quintana transmission. For this purpose, we used an experimental model of cat flea infection.
Materials and methods
Rearing fleas
Since 2010, cat fleas (C. felis felis, strain Bristol) have been maintained by our team at the Rickettsioses laboratory of the Medicine Faculty in Marseille, France. Adult fleas were fed in vitro using human citrated blood via an artificial membrane of Parafilm®M (Sigma-Aldrich, Saint-Louis, Missouri, USA). The flea larvae were maintained at 80% humidity in containers with 40 g of sand, 3 g of spray-dried human blood, 20 g of rat food and 2 g of brewer's yeast (T. Kernif, K. Stafford, G. C. Coles, I. Bitam, P. Kassim, J. Chironi, D. Raoult & P. Parola, submitted). The fleas and their faeces were proven to be free from B. quintana using a quantitative real-time PCR (qPCR) assay with primers that are described below (section: 2.5). To prevent contamination, the feeder machine as described by Wade and Georgi (1988) was introduced in a clear acrylic glove box, reference number ‘830 ABD/EXP/SP’ (Fisher® Scientific, USA), to raise fleas in adapted containment.
Bartonella quintana strain
Culturing of B. quintana and all procedures involving experimental infections of fleas were conducted in a Biosafety Level 2 (BSL2) room. The B. quintana strain Oklahoma (ATCC 49793) was used. The strain was cultured on Columbia sheep blood agar plates (5%, BioMerieux®, Marcy l'Etoile, France) and incubated at 37°°C under a 5% CO2 atmosphere as described previously (Rolain et al. 2003a). Between 8 and 12 days after inoculation of blood culture, the bacteria were harvested by adding 400 μL of phosphate-buffered saline (PBS), pH 7.2 (BioMerieux®, Craponne, France), followed by a serial dilution of the isolates from 10 to 10−4. Subsequently, 200 μL of the pure bacterial suspension and each dilution was mixed with 2 mL of whole blood and used as bloodmeals for fleas. We tested the inocula by qPCR to ensure the presence of bacteria. In addition, we cultured 150 μL of the initial dilutions and other suspensions (up to 10−10) on sheep blood agar plates to estimate the number of colony forming bacteria per microlitre and the viability of the bacteria.
Infection of fleas and sampling strategy
Two separate trials were conducted using fleas drawn from the same colony and with the same age. We formed 6 groups for each trial (including 1 control group and 5 infected batches) that consisted of 80 fleas in each group with approximately 25 males and 55 females. The concentration of B. quintana in pure suspension was 1.8 × 106 bacteria in group 1 (G1) and 4.2 × 107 bacteria in group 1′ (G1′) per μL of PBS in Trials 1 and 2, respectively, and this bacterial suspension was diluted to a concentration of 10−4. Each group of fleas (G1 to G5 of Trial 1 and G1′ to G5′ of Trial 2) was fed for 4 days with infected blood containing 2 mL of blood and 200 μL of the bacterial suspension at the different dilutions (≈3.6 × 104 − 8.4 × 109 bacteria). The control groups were fed 2 mL of uninfected blood containing 200 μL of PBS. Subsequently, all flea groups were fed uninfected blood beginning on the 3rd day postinfection (dpi) until the 13th dpi. At the 3rd dpi, 10 viable fleas and approximately 50 mg of faeces from each group were recovered for analysis by qPCR and culturing to determine the acquisition and viability of the B. quintana in cat fleas. Five fleas from each dilution were immunohistochemically analysed to determine bacterial localization in the fleas (Table 1 and Fig. 1). We recovered the faeces from fleas that had been infected by inocula at ≈8.4 × 109 bacteria every 48 h to monitor the excretion of B. quintana (Table 2). At the end of the experiment (13th dpi), 10 fleas of each dilution group were analysed by qPCR.
| Trials | Group of Fleas (No) | Sampling (Quantity) | Day 3 Postinfection (P.I.) | Day 13 P.I. | |||
|---|---|---|---|---|---|---|---|
| qPCR (fabF3) | Culture | Immunohistochemistry | qPCR (fabF3) | ||||
| Bacteria(≈) | No. positive (%) | ||||||
| Trial 1 | Group 1 (80) | Bloodmeala (2 mL) | 3.6 × 108 | + | |||
| Fleas (10) | 6 × 102 to 9.6 × 103 | 10 (100%) | +c | +b | − | ||
| Faeces (≈ 50 mg) | 1.9 × 106 | − | − | ||||
| G 2 (80) | Bloodmeala(2 mL) | 3.6 × 107 | + | ||||
| Fleas (10) | 5.4 × 10 to 3.4 × 103 | 10 (100%) | ND | +b | − | ||
| Faeces (≈ 50 mg) | 1.1 × 103 | − | − | ||||
| G 3 (80) | Bloodmeala(2 mL) | 3.6 × 106 | + | ||||
| Fleas (10) | 3.8 × 10 to 7.6 × 10 | 6 (60%) | ND | +b | − | ||
| Faeces (≈ 50 mg) | 2.1 × 103 | − | − | ||||
| G4 (80) | Bloodmeala(2 mL) | 3.6 × 105 | + | ||||
| Fleas (10) | − | 0 (0%) | ND | +b | − | ||
| Faeces (≈ 50 mg) | 1.4 × 102 | − | − | ||||
| G5 (80) | Bloodmeala(2 mL) | 3.6 × 104 | + | ||||
| Fleas (10) | − | 0 (0%) | ND | +b | − | ||
| Faeces (≈ 50 mg) | − | − | − | ||||
| Group Control (80) | Bloodmeala(2 mL) | − | − | − | |||
| Fleas (10) | − | 0 (0%) | ND | − | − | ||
| Faeces (≈ 50 mg) | − | − | − | − | |||
| Trial 2 | Group 1′ (80) | Bloodmeala(2 mL) | 8.4 × 109 | + | |||
| Fleas (10) | 1.5 × 102 to 7 × 104 | 10 (100%) | ND | ND | − | ||
| Faeces (≈ 50 mg) | 9.7 × 106 | +c | − | ||||
| G 2′ (80) | Bloodmeala(2 mL) | 8.4 × 108 | + | ||||
| Fleas (10) | 3.8 × 10 to 9.6 × 103 | 10 (100%) | ND | ND | − | ||
| Faeces (≈ 50 mg) | 3 × 105 | − | − | ||||
| G3′ (80) | Bloodmeala(2 mL) | 8.4 × 107 | + | ||||
| Fleas (10) | 7.6 × 10 to 4.5 × 103 | 9 (90%) | ND | ND | − | ||
| Faeces (≈ 50 mg) | 1.9 × 104 | − | − | ||||
| G4′ (80) | Bloodmeala(2 mL) | 8.4 × 106 | + | ||||
| Fleas (10) | 7.6 × 10 to 4.5 × 103 | 8 (80%) | ND | ND | − | ||
| Faeces (≈ 50 mg) | 6 × 102 | − | − | ||||
| G 5′ (80) | Bloodmeala(2 mL) | 8.4 × 105 | + | ||||
| Fleas (10) | 3.8 × 10 to 3 × 102 | 8 (80%) | ND | ND | − | ||
| Faeces (≈ 50 mg) | 2.4 × 103 | − | − | ||||
| Group Control' (80) | Bloodmeala(2 mL) | − | − | ||||
| Fleas (10) | 0 (0%) | 0 (0%) | ND | ND | − | ||
| Faeces (≈ 50 mg) | − | − | − | ||||
- No, number of fleas; (+) positive; (−) negative; ND, not done; (≈), approximately; qPCR, quantitative real-time polymerase chain reaction; fabF3, 3-oxoacyl-[acyl-carrier-protein] synthase.
- a Infected or uninfected blood.
- b Observed in gut.
- c Confirmation by qPCR.
| Day Postinfection | qPCR (fabF3 ‘bacteria’) | Culture | ||
|---|---|---|---|---|
| G1 (Trial 1) | G1′ (Trial 2) | G1 (T1) | G1′(T2) | |
| D3 | 1.9 × 106 | 9.7 × 106 | − | + |
| D5 | ND | 2.5 × 106 | ND | ND |
| D7 | ND | 3.3 × 102 | ND | ND |
| D9 | ND | 1.5 × 102 | ND | ND |
| D11 | 3.8 × 10 | 7.6 × 10 | − | − |
| D13 | − | − | ND | ND |
| Control groups | Trial 1 | Trial 2 | T1 | T2 |
|---|---|---|---|---|
| D3 | − | − | − | − |
| D11 | − | − | − | − |
- D, day; (+) positive; (−) negative; ND, not done.
Extraction of DNA from samples
The fleas from each dilution group were individually surface-decontaminated by 5-min immersion in ethanol alcohol (COOPER®, Paris, France), followed by three 5-min immersions in sterile PBS as described previously (La Scola et al. 2001). Each flea was incised using a scalpel and then incubated overnight at 56°°C in 180 μL of buffer G2 (30 mm Tris-Cl; 30 mm EDTA; 5% Tween 20; 0.5% Triton X-100; 800 mm GuHCl) containing 20 μL of proteinase K (activity = 600 mAU/mL solution or 40 mAU/mg of protein) until lysis and then homogenized. In addition, 50 mg of faeces was homogenized in 200 μL of PBS. After the prelysing steps, DNA extraction for 200 μL of both homogenization types was performed using an automatic EZ1 robot (QIAGEN-BioRobot® EZ1, Tokyo, Japan), according to the manufacturer's instructions (EZ1 DNA Tissue Kit, QIAGEN®, Hilden, Germany).
Real-time PCR amplification
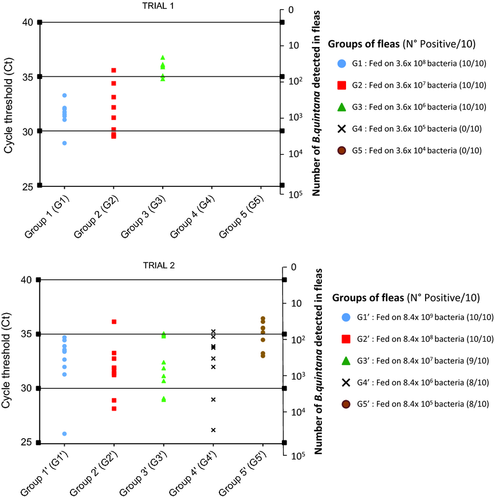
DNA was the template in qPCR assays targeting two specific B. quintana genes that encoded 3-oxoacyl-[acyl-carrier-protein] synthase (fabF3) and a hypothetical intracellular effector (yopP). The primer sequences were as follows: Bqui05300F/FabF3/5′-GCT-GGC-CTT-GCT-CTT-GAT-GA-3′, Bqui05300R/FabF3/5′-GCT-ACT-CTG-CGT-GCC-TTG-GA-3′, probed with Bqui05300P/FabF3/6-FAM-TGC-AGC-AGG-TGG-AGG-AGA-ACG-TG-TAMRA, and Bqui11580F/yopP/50-TAA-ACC-TCG-GGG-GAA-GCA-GA-30, Bqui11580R/yopP/5′-TTT-CG T-CCT-CAA-CCC-CAT-CA-3′, probed with Bqui11580P /yopP/6-FAM-CGT-TGC-CGA-CAA-GAC-GTC-CTT-G C-TAMRA (Angelakis et al. 2011). Briefly, a 20-μL qPCR reaction mixture was set up containing 2 × Reaction MasterMix for Fast qPCR with Taq DNA polymerase (Eurogentec®), 20 μm of each primer, 2.5 μm of each probe, DNase⁄RNase-free dH2O and 5 μL of DNA from each sample. The reaction components were mixed in 96-well plates, and the assay was performed using the CFX96® qPCR Detection System (Bio-Rad, France). The qPCR conditions were as follows: the reaction mixtures were kept at 50°°C for 2 min, then 95°°C for 5 min and subsequently put through 40 cycles of 95°°C for 1 s and 60°°C for 35 s. The qPCR was considered positive when the cycle threshold (Ct) was lower than 36 (Fig. 2). The number of B. quintana in each sample was calculated based on the DNA copy numbers of B. quintana. A qPCR standard curve was obtained by analysing the fabF3 primers in serial dilutions of B. quintana culture, and the standard value was determined for duplicate trials. The B. quintana infection density was quantified as the ratio of the log of the transformed fabF3 copy numbers per individual flea, faeces and bloodmeal (Table 1 and Fig. 2).
Culture sampling
Bartonella quintana colonies are typically small, creamy white colonies that allow them to be readily distinguished from those formed by other flea gut commensals, but some colonies are very large and invasive. Approximately 300 μL of homogenized faeces (50 mg in 500 μL of PBS) from infected and uninfected fleas in 5% sheep's blood was filtered using a 0.8-μm filter (Millex Ø 33 mm, Dominique Dutscher®) before culturing. These filtrates were grown on agar plates (sheep blood 5%, BioMerieux®) and incubated at 37°°C under an atmosphere of 5% CO2. After formation of the first colonies on the agar plate, the culturing procedure was repeated to isolate the bacterial species and for qPCR confirmation. Ten fleas fed the ≈3.6 × 108 bacteria dilution were decontaminated for 5 min in ethanol alcohol (COOPER®, Paris, France) and rinsed three times for 5 min in sterile PBS as described elsewhere (La Scola et al. 2001). These fleas were subsequently crushed in 500 μL of PBS and 300 μL of the resulting suspension was cultured on blood agar plates; the remaining homogenate was analysed by qPCR.
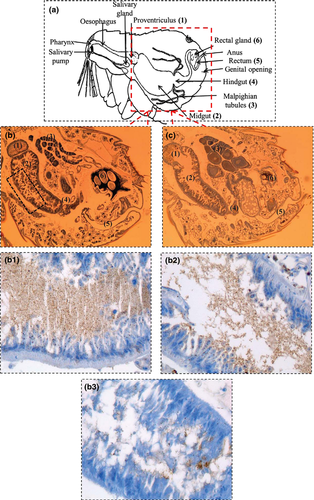
Immunohistological analysis
Immunohistochemistry was performed on 3-μm-thick, paraffin-embedded sections of formalin-fixed fleas using the Ventana Benchmark autostainer (Ventana Medical Systems, Inc.), and uninfected fleas were used as a negative control. After deparaffinization, each tissue section was incubated with polyclonal rabbit anti-B. quintana antibody, diluted 1:5000 as previously described (Lepidi et al. 2000).
Statistical analysis
To determine whether B. quintana influenced flea mortality, statistical analysis was performed with SPSS, version 17.2, for Windows. The number of dead fleas in uninfected and infected groups was compared using the chi-squared (χ²) and Fisher's exact tests. One-way ANOVA was performed to analyse the differences in the mean scores of bacterial DNA copy number per individual flea in ten groups for each gene (fabF3 and yopP) using Open Epi version 3.01. The differences were considered statistically significant at P-value <0.05 for all analyses.
Results
Bartonella quintana in bloodmeal and surviving fleas fed on infected blood
Using culture-based techniques, we confirmed that B. quintana grew 8 to 10 days postplating on blood agar, and DNA extraction from these colonies resulted in positive qPCR products. The average number of B. quintana in each sample of bloodmeal was calculated (Table 1).
To determine whether B. quintana influenced flea mortality, 80 fleas infected with inocula at 3.6 × 108 bacteria were compared with 80 uninfected fleas at 3, 11 and 13 dpi. The mortality rates of infected fleas were 5/80 (10 viable fleas recovered for analysis), 12/65 and 21/53 fleas at 3 dpi, 11 dpi and 13 dpi, respectively. The mortality rates for uninfected fleas were 3/80 (10 viable fleas recovered for analysis), 13/67 and 18/54 fleas at 3 dpi, 11 dpi and 13 dpi, respectively. We did not observe a difference in the mortality rates of infected and control fleas using chi-squared (χ²) test (P > 0.13).
Acquisition of B. quintana by cat fleas
Fleas were exposed to B. quintana-infected bloodmeal for 4 days. On the 3rd dpi, the B. quintana infection load was examined at the whole individual flea level in both trials. We detected B. quintana in 52% (26/50) in the infected fleas in Trial 1 and in 90% (45/50) of the fleas in Trial 2. The control fleas were negative for the bacterium in both trials. We detected B. quintana-positive fleas in all groups (G1′ to G5′) of Trial 2 and flea groups of G1, G2 and G3 for Trial 1 (Table 1). The greatest B. quintana infection loads for each group of fleas in both trials are reported in Table 1. Fleas that were infected with pure inoculum (G1; G1′) exhibited the greatest quantities of B. quintana per individual flea sample (6 × 102 to 9.6 × 103 bacteria in Trial 1 and 1.5 × 102 to 7 × 104 bacteria in Trial 2). Fleas from both trials had significantly different B. quintana infection loads, and those values decreased in the groups of fleas that were fed pure to 10−4 dilutions of B. quintana. The fleas in the two last groups (G4 and G5) of Trial 1 were negative for B. quintana after they were fed bloodmeal with < 8 × 105 bacteria. The quantities of B. quintana in each group per individual flea sample per trial analysed by qPCR of the fabF3 gene are represented with bacteria (3.8 × 10 to 7 × 104) and a Ct value ranging from 25.92 to 36.5 in Fig. 2 (yopP gene results are shown in Table S1a,b, Supporting Information). Finally, there was no significant difference in the mean DNA copy number per individual flea in the ten groups using the fabF3 gene (P = 0.76244) or the yopP gene (P = 0.51222).
Localization of B. quintana in the bodies of Ctenocephalides felis felis
Immunohistochemical results of the 5 tested fleas (from the 3rd dpi) from Trial 1 demonstrated the presence of B. quintana as dense clusters of immunopositive microorganisms in the infected flea gut tract (Fig. 1). Immunopositive microorganisms were found in all flea dilution groups except for the control fleas; however, the density of the clusters was low in fleas of the G4 and G5 from Trial 1.
Evaluation of B. quintana in fleas and their faeces
Detection of B. quintana in faeces
On the 3rd dpi, we separately tested the faeces of the infected fleas and controls from both trials by qPCR. The results indicated the presence of B. quintana in C. felis felis faeces in all groups of Trial 2, and the levels decreased from fleas of group G1′ (9.7 × 106 bacteria) to G5′ (2.4 × 103 bacteria). In Trial 1, the presence of B. quintana in C. felis felis faeces was found in all groups except G5, and the levels ranged from 1.9 × 106 bacteria (G1) to 1.4 × 102 bacteria (G4) (Table 1).
Viability of B. quintana in fleas and their faeces
We used MALDI-TOF (matrix-assisted laser desorption/ionization time-of-flight mass spectrometry) analysis to detect faecal bacteria after culturing as described previously (Fournier et al. 2009). To avoid the detection of flea gut commensal bacteria such as Serratia marcescens (family: Enterobacteriaceae, Gram negative), we filtered the different dilutions of faeces using a 0.8-μm filter before culturing. On the 3rd dpi, we grew B. quintana from the faeces of fleas that had been fed inocula with 8.4 × 109 bacteria in Trial 2 on agar plates. Colonies formed after 10 days postplating; however, none colonies have grown in the other dilutions, including the group G1 (≈3.6 × 108 bacteria) of Trial 1 (Table 1). Furthermore, we only observed a positive culture from one of 10 group G1 fleas (infected with ≈3.6 × 108 bacteria) from Trial 1 at 30 days postplating. In Trial 2, the existence of B. quintana colonies was detected by culturing group G1′ flea faeces (infected with ≈8.4 × 109 bacteria) and was confirmed using a second culture step (direct and indirect cultures); these results were verified by qPCR for fabF3 and yopP genes.
Persistence of B. quintana in faeces
Using qPCR, we followed the presence and elimination of faecal bacteria in fleas that were fed pure suspensions (Trials 1 and 2, 3.6 × 108 bacteria and 8.4 × 109 bacteria, respectively). The results reported in Table 2 demonstrate that the average number of B. quintana in the flea faeces increased in Trials 1 and 2 until the 3rd dpi to a maximal number of 9.7 × 106 bacteria, and this value then decreased logarithmically until the 11th dpi to a minimum of 7.6 × 10 bacteria in both trials. At the 13th dpi, the faeces of all inocula-infected fleas were negative for B. quintana in both trials (Table 2). On the 13th dpi, we tested 10 viable fleas from each group by qPCR, and those fleas were negative for B. quintana, which indicates that the bacteria were completely eliminated from infected fleas (Table 1).
Discussion
The cat flea, C. felis felis, is found worldwide and has been reported to parasitize many species of wild and domestic animals (Rust & Dryden 1997). The cat flea is the only arthropod that has been demonstrated to date to biologically transmit Rickettsia felis (Reif & Macaluso 2009). In addition, the cat flea has been confirmed as a vector for Bartonella henselae, the causative agent of cat scratch disease (Zangwill et al. 1993; Chomel et al. 1996). Foil et al. (1998) demonstrated the experimental infection of cats with B. henselae after inoculation with cat flea faeces (Foil et al. 1998). Recently, this flea was suspected to be a potential vector for transmitting B. quintana, B. clarridgeiae, B. koehlerae, B. birtlesii and B. tribocorum to mammals (Bouhsira et al. 2013a).
A symbiont–host relationship has been observed in the human body louse Pediculus humanus corporis vector, which excretes the agent of trench fever, B. quintana, in its faeces (Higgins et al. 1996). The first clinical manifestation of trench fever was attributed to B. quintana and was characterized by a sudden onset of headache, pain in the shins, dizziness and fever. Thereafter, the primary infection resolved but had frequent relapses (Foucault et al. 2006). Chronic bacteraemia developed in some patients (Brouqui et al. 1999), and chronic asymptomatic bacteraemia in humans indicated that humans may be the natural reservoir of B. quintana (Foucault et al. 2002). Although humans are a primary reservoir host for B. quintana, some recent reports have found B. quintana in cat teeth (La et al. 2005), in dogs and in cynomolgus and rhesus macaques (Chomel et al. 2006; Huang et al. 2011; Li et al. 2012, 2013). Our results indicate that other arthropods, such as fleas, may acquire B. quintana and then excrete it in its faeces.
Several studies suggested that biting arthropods such as flies, lice, fleas or ticks can transmit Bartonella spp. during their bloodmeal (Tsai et al. 2011), but few studies have described the details of the growth kinetics and transmission of these bacteria. For example, human infection by B. quintana is presumed to ensue from the inoculation of faeces into louse bites by scratching (Maurin & Raoult 1996). Another organism, B. henselae, is also thought to be transmitted by contamination of a bite wound with infected faeces, and the C. felis flea is capable of ingesting B. henselae, supporting replication of these bacteria in their digestive tract and excreting viable organisms in their faeces (Higgins et al. 1996). As suggested by Morick et al. (2013), Bartonella spp. are likely not pathogenic to fleas and are well adapted to their vectors. Our study indicates that cat fleas could maintain B. quintana in their gastrointestinal tract and viable colonies could be produced from faeces upon inoculation on sheep blood agar plates. We suggest a similar scenario for the transmission of B. quintana by cat fleas to cats and humans.
Our experimental results corroborate data reported by Bouhsira et al. (2013a), which used artificial conditions for feeding Bartonella species including B. quintana, and found no difference in the persistence of the cat flea or of its faeces excretion up to the 3rd dpi. In a study by Seki et al. (2007), a quantitative analysis of bacterial multiplication rate was performed – the proliferation of B. quintana in body lice was observed to begin 4 days after ingestion. Furthermore, the bacteria were constantly excreted into the faeces for at least 3 weeks (Seki et al. 2007). However, in our study, viable B. quintana in faeces were observed in fleas that were fed a suspension of bacteria ≥4.2 × 109 per mL of blood and bacterial load in faeces decreased gradually after 3rd dpi. Our findings suggest that infectivity did not remain in a faecal environment for a prolonged time. In addition, we suggest that B. quintana was completely shed in flea faeces until the 13th dpi or B. quintana was disseminated in the different body cavity of the cat fleas. Nonetheless, we observed the absence of B. quintana in all fleas and in their faeces on day 13. The absence of the Bartonella was observed in previous studies: no B. quintana DNA was detected in louse faeces on either day 5, 7, 9 or 11 after infection (Seki et al. 2007) and no B. henselae DNA was detected in cat fleas and their faeces on day 9 (Bouhsira et al. 2013b). Further studies are required to determine whether the B. quintana was completely eliminated after 13 dpi.
Our immunohistological approach assessed B. quintana localization only in the flea gut at the 3rd dpi. Unfortunately, the immunohistological method was not used on cat fleas at 13 dpi to evaluate B. quintana dissemination from the gut to other cat flea organs. Recently, R. felis has been shown to replicate in the cat fleas' digestive tract, migrate to the hemolymph and then disseminate through the excretory system (such as the Malpighian tubules, hindgut and rectal ampulla) and reproductive tissues (Thepparit et al. 2013). However, the method of Bartonella migration (e.g. B. quintana or B. henselae) from the digestive tract of arthropods (such as the louse or flea) to other tissues has not yet been elucidated (Seki et al. 2007; Bouhsira et al. 2013b).
Although no clear replication pattern or dissemination of B. quintana in cat fleas was reported throughout the study, we showed that cat fleas can acquire B. quintana, a known human pathogen, by feeding and releasing viable organisms into their faeces. Therefore, fleas may play a role as vectors of trench fever or other infections caused by B. quintana. More studies are required to better understand B. quintana persistence in both fleas and their faeces and also to understand the role of fleas in B. quintana infections.
Acknowledgement
We thank Dr. Samir BENKOUITEN and Dr. Alpha Kabinet KEITA from URMITE for their technical assistance with the statistical analysis.
Conflict of interests
The authors declare that they have no competing interests.
References
K.T., C.S., B.I., R.D. and P.P. conceived and designed the experiments. K.T. and Leu. H. performed the experiments. K.T., Leu. H., C.S., R.J.-M., R.D. and P.P. analysed the data. Lep. H. contributed immunohistochemistry tools. C.S., B.J.-M. and R.D. contributed reagents/materials/analysis tools. K.T., C.S., R.J.-M. and P.P. wrote the paper. K.T., C.S., R.J.-M., R.D. and P.P. critically discussed the manuscript.
Data accessibility
The data for a Ct and copy number of the fabF3 and yopP genes of Bartonella quintana in individual fleas of each group at 3 days postinfection in Trial 1 and Trial 2 may be found as supplemental material in the online version of this article.




