Do pathogens reduce genetic diversity of their hosts? Variable effects of sylvatic plague in black-tailed prairie dogs
Abstract
Introduced diseases can cause dramatic declines in—and even the loss of—natural populations. Extirpations may be followed by low recolonization rates, leading to inbreeding and a loss of genetic variation, with consequences on population viability. Conversely, extirpations may create vacant habitat patches that individuals from multiple source populations can colonize, potentially leading to an influx of variation. We tested these alternative hypotheses by sampling 15 colonies in a prairie dog metapopulation during 7 years that encompassed an outbreak of sylvatic plague, providing the opportunity to monitor genetic diversity before, during and after the outbreak. Analysis of nine microsatellite loci revealed that within the metapopulation, there was no change in diversity. However, within extirpated colonies, patterns varied: In half of the colonies, allelic richness after recovery was less than the preplague conditions, and in the other half, richness was greater than the preplague conditions. Finally, analysis of variation within individuals revealed that prairie dogs present in recolonized colonies had higher heterozygosity than those present before plague. We confirmed plague survivorship in six founders; these individuals had significantly higher heterozygosity than expected by chance. Collectively, our results suggest that high immigration rates can maintain genetic variation at a regional scale despite simultaneous extirpations in spatially proximate populations. Thus, virulent diseases may increase genetic diversity of host populations by creating vacant habitats that allow an influx of genetic diversity. Furthermore, even highly virulent diseases may not eliminate individuals randomly; rather, they may selectively remove the most inbred individuals.
Introduction
Genetic variation is of fundamental importance to natural populations because it enables adaptation to novel conditions (Lavergne & Molofsky 2007), confers resistance to pathogens (de Bellocq et al. 2008) and protects against inbreeding depression (Reed & Frankham 2003). The level of genetic diversity within a population reflects the combined forces of drift and selection acting on variation generated by mutation, recombination and migration. Selection happens as a consequence of abiotic conditions and biotic interactions, including competition (Vellend 2008), predation (Dingemanse et al. 2009), parasitism (Lachish et al. 2011) and disease caused by pathogens (Hawley & Fleischer 2012).
Pathogens have long been recognized as potentially important influences on host evolutionary dynamics (Haldane 1949; Hamilton 1982). The effects of pathogens on host genetic diversity (e.g. Trudeau et al. 2004) may vary, in part, depending on pathogen virulence, (namely the incidence of host mortality due to pathogens; Lenski & May 1994) and whether there are nearby populations that can serve as source areas for recolonization (Teacher et al. 2009; Hoban et al. 2010). At low and intermediate virulence, pathogens may increase adaptive genetic diversity of their host populations due to diversifying selection at resistance loci (Schulte et al. 2010). In contrast, highly virulent pathogens can cause dramatic reductions in population size and population extirpation (Cully & Williams 2001; de Castro & Bolker 2005; Caillaud et al. 2006). In populations with drastic declines, genetic variation is rapidly eroded, thereby increasing the effects of inbreeding (Lachish et al. 2011) and susceptibility to other pathogens (Spielman et al. 2004; Valsecchi et al. 2004), potentially leading to a positive feedback cycle.
The effect of pathogens on genetic variation will also vary depending on the connectivity of populations in a complex landscape, which probably influences both rates of local extinction (hereafter, ‘extirpation’) and recolonization. Well-connected populations may have higher probabilities of extirpation because pathogen spread is rapid (Hess 1996; Collinge et al. 2005), resulting in reduction in variation. Alternatively, well-connected populations may facilitate recolonization by migration from multiple populations (Fahrig & Merriam 1994), a situation that increases variation. Less connected populations may have lower probabilities of disease incidence (Real & Biek 2007), but correspondingly fewer source populations for recolonization (Hanski 1998). There are a number of possible scenarios depending on rates of population extirpation by pathogens, rates of recolonization and the number of possible source populations for recolonization. If, for instance, extirpation rate is high, recolonization rate is low, and there are few source populations, disease outbreaks are expected to erode genetic variation (Harrison & Hastings 1996). By contrast, slow rates of extirpation, rapid recolonization and multiple source populations would buffer against the loss of genetic diversity. Therefore, landscape context can contribute to the effects of pathogens on genetic variation within and among populations (Biek & Real 2010).
One highly virulent pathogen infecting mammals is the bacterium Yersinia pestis, the agent of sylvatic plague. Plague is transmitted by fleas and was introduced to western North America from Asia around 1900 (Cully et al. 2000); its effects are particularly severe in naïve hosts such as prairie dogs (Cynomys spp; family Sciuridae). Prairie dogs comprise five species of social burrowing rodents inhabiting the grasslands of western North America; their populations are organized into colonies consisting of several groups of related individuals (Hoogland 1995). Prairie dog mortality from plague approaches 99% (Cully et al. 1997; Pauli et al. 2006; Biggins et al. 2010). Once Y. pestis is endemic within a region, prairie dog colonies experience metapopulation dynamics (Antolin et al. 2006) with plague-induced extirpations (Cully & Williams 2001) and subsequent recolonization (Roach et al. 2001). In Boulder County, Colorado, dispersal of prairie dogs among colonies is dependent on the intervening landscape matrix, where urbanization suppresses movement but other land types facilitate movement (Sackett et al. 2012). Because plague causes colony extirpation that is often, but not always, followed by recolonization, the disease is expected to have consequences on the distribution of neutral genetic variation within and among colonies.
Our research examines the effects of pathogen extirpations on neutral genetic variation at three scales. At the scale of the metapopulation, variation could either decrease due to population extirpation, or be maintained by recolonization (Whitlock & McCauley 1990; Bohonak 1999). At the scale of the colony, genetic diversity could decline due to founder effects (Leberg 1992; Tsutsui et al. 2000), which would be exacerbated if immigration occurs from only one or few source colonies (Pannell & Charlesworth 1999), as may be expected in isolated colonies or those surrounded by urbanization (Magle et al. 2010; Sackett et al. 2012). Alternatively, recolonization from multiple sources would replenish variation within a colony after recolonization (Slatkin 1977). At the scale of the individual, those remaining after widespread extirpations could be a random subset of the original population (for instance, if they were not exposed to the pathogen because of stochastic effects), leading to a loss of within-individual genetic diversity due to inbreeding that results from founder effects (Trudeau et al. 2004). Conversely, surviving individuals could have higher within-individual genetic diversity (heterozygosity) because in many systems, more inbred individuals suffer higher mortality from infection than less inbred individuals (Coltman et al. 1999; Valsecchi et al. 2004). Higher within-individual heterozygosity could also result from admixture following recolonization from multiple sources.
To test the hypotheses of how Y. pestis influences the distribution of neutral genetic diversity in black-tailed prairie dogs (Cynomys ludovicianus, hereafter ‘prairie dogs’) at different scales, we sampled 15 prairie dog colonies before, during and after a plague epizootic. Using nine microsatellite markers, we examined the effects of plague-induced colony extirpation and subsequent recolonization on neutral genetic diversity in prairie dogs at three scales: within the metapopulation, within colonies and within individuals. At the metapopulation scale, we investigated regional temporal changes in the degree of allelic richness and isolation by distance before extirpation and after recolonization. At the colony scale, we examined changes in allelic richness and temporal differentiation within colonies and estimated immigration rates into extirpated colonies. At the individual scale, we calculated observed heterozygosity (as the proportion of heterozygous loci) before plague and after recolonization. We found that within the metapopulation, plague did not change genetic diversity; within colonies, genetic diversity changed depending on the number of inferred source populations; and within individuals, genetic diversity increased.
Methods
Study location and sample collection
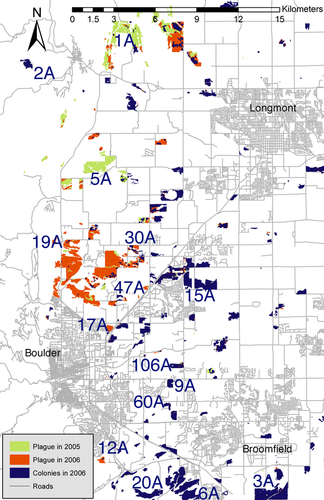
Prairie dog colonies occupy 1200 hectares of land in Boulder County; data were collected from 15 of these colonies from 2003 to 2009 (Fig. 1; Table S1, Supporting information). Study colonies were chosen to represent the spectrum of surrounding habitat types in the area (Johnson & Collinge 2004). Of these, six colonies were extirpated by plague in 2006–2007 and subsequently recolonized, four were extirpated and not recolonized, two could not be sampled after 2006 for logistical reasons, and three were not affected by plague. Colonies for which we obtained no postrecolonization data were excluded from analyses involving temporal comparisons. Because prairie dogs are diurnal and highly social, extirpations were easy to document when they occurred: they resulted in >99% reduction in prairie dog numbers (McClure & Collinge, unpublished manuscript) and active colony area (data from Boulder Open Space and Mountain Parks). Sampled colonies were separated by pairwise distances varying from 1.5 to 36 km and varied in their surrounding habitat matrix (Johnson & Collinge 2004; Collinge et al. 2005). At each colony, 49–104 Tomahawk traps were placed 20 m apart on a grid centred at the approximate centre of the colony, baited, set and left for up to 2.5 h. Prairie dogs were trapped for 1–2 weeks at each site by targeting active burrows with 1–4 traps (Hoogland 1995).
Prairie dog trapping and processing were conducted in accordance with protocols approved by the University of Colorado's Institutional Animal Care and Use Committee and are described in detail therein (available upon request). Captured prairie dogs were anaesthetized under close supervision with 1–4% isoflurane in oxygen using a precision, calibrated vaporizer to control the dosage (Heath et al. 1997). Processing involved collection of ear tissue for DNA; collection of blood for pathogen screening; and insertion of a passive integrated transponder (PIT) tag for future identification. Tissue for DNA analysis was collected using a 2-mm-diameter ear punch (Braintree Scientific) and stored frozen in a solution of EDTA-DMSO until DNA extraction. After processing, animals were placed back into the traps until the anaesthesia wore off and they became alert, at which time they were released at their capture locations.
Blood samples were sent to the Centres of Disease Control and Prevention in Fort Collins, CO for the detection of plague antibodies. The presence of these antibodies indicates exposure to Y. pestis and allows for the evaluation of survival of prairie dogs exposed to plague. DNA from prairie dogs was extracted using a Qiagen tissue kit, and individuals were genotyped at nine unlinked microsatellite loci (Jones et al. 2005; Sackett et al. 2009). Loci were examined for null alleles in the program Micro-checker (van Oosterhout et al. 2004). We tested all loci in all colonies for deviations from neutrality using the Fdist algorithm (Beaumont & Nichols 1996) implemented in Lositan (Antao et al. 2008) and following the protocol of Bryja et al. (2007). For all estimates of genetic diversity before plague and after recolonization, data from multiple years were pooled (i.e. ‘preplague’ constituted samples from 2003 to 2006 and ‘postrecolonization’ comprised samples from 2007 to 2009). Our rationale for pooling was threefold: (i) our goal was not to examine diversity over time per se, but to determine the effects of extirpation and recolonization on genetic diversity by comparing diversity at two time points, (ii) low sample sizes the first year after plague (e.g. four individuals genotyped in colony 47A) provided limited statistical power and (iii) recapture rates between years (mean = 34.62%) demonstrated that many prairie dogs sampled in 1 year were also present the next year. Individuals recaptured in multiple years were genotyped only once. To ensure that pooling years was justifiable, we tested for differentiation between years within colonies using 10 000 randomizations in fstat, and we subsequently performed anovas on allelic richness and heterozygosity among years within time points.
Analysis of genetic variation within the regional metapopulation
Genetic variation in the metapopulation (across six recolonized and three control colonies) was examined before and after the plague epizootic. We estimated the number of alleles per locus and allelic richness using permutation analysis in the program fstat (Goudet 1995) to control for unequal sample sizes, using the smallest number of individuals typed for a locus in each time period (before plague and after recolonization; Table S1, Supporting information). We then tested for changes in the number of alleles and allelic richness using paired Wilcoxon tests for differences in means, implemented in R (the R Project for Statistical Computing, www.r-project.org). We calculated spatial differentiation using theta, an unbiased estimator of FST (Weir & Cockerham 1984), in fstat and assessed the significance of differentiation by performing Fisher's exact tests in Genepop (Rousset & Raymond 1997) and performing a Bonferroni correction. Average pairwise FST values in the metapopulation (nine colonies only) were compared before plague and after recolonization with a paired Wilcoxon test. Estimates of differentiation are confounded by the degree of diversity present; therefore, we verified all inferences of differentiation by calculating standardized estimates of FST (Hedrick 2005) by dividing by the maximum possible values (obtained in the program recodeData, Meirmans 2006) and re-assessed differentiation. All inferences were the same with raw and standardized FST values, except where noted. The degree of isolation by distance was determined before plague and after recolonization by performing Mantel tests in the Vegan package (Oksanen et al. 2010) for R using linearized FST values (Slatkin 1995) and the natural log of geographic distance (Rousset 1997). The degree of isolation by distance was then compared before plague and after recolonization by performing separate regressions in Genepop, estimating the slopes and determining whether the 95% confidence intervals (estimated by bootstrapping over loci) for the slopes overlapped.
We used GeneClass2 (Piry et al. 2004) to assess migration rates among colonies and characterize founding individuals as immigrants or residents. Because there were unsampled colonies in the landscape, our goal was not to assign individuals to actual source colonies, but rather to estimate the number of immigrants in recolonized colonies and estimate the number of putative source populations. Using the recommended frequencies-based method (Paetkau et al. 1995), we inferred immigrant status by choosing individuals that were assigned to their colony of capture with a probability <0.05 (as in Berthier et al. 2006). Individuals were assumed to originate in one of the 15 colonies present before plague (i.e. we did not allow them to be assigned to a postrecolonization colony) or a nearby unsampled colony with a genetic signature similar to the sampled colony. Individuals assigned to particular colonies (especially extirpated colonies) should be interpreted as originating from a population with a genetic signature that matches the source colony, rather than originating necessarily from that colony. Inferences regarding source populations (genetic entities), rather than colonies (spatial entities), are more robust to varied sampling designs.
Analysis of genetic variation within colonies
Genetic variation at the colony scale was assessed within each colony before plague and after recolonization. We tested all colonies for significant departures from Hardy–Weinberg equilibrium both before and after the plague epizootic, controlling for multiple tests with a Bonferroni correction, using Genepop software (Rousset 2008). We estimated allelic richness within colonies before plague and after recolonization using permutation analysis in the program fstat (Goudet 1995) as above and then tested for changes in allelic richness using a paired Wilcoxon test for difference in means, implemented in R. We used a linear model in R to test for a relationship between allelic richness in recolonized colonies and the number of inferred source colonies from the GeneClass2 analysis. We then calculated average relatedness and variance in relatedness among individuals within a colony relative to all individuals in the metapopulation both before plague and after recolonization using permutation analysis in SPAGeDi (Hardy & Vekemans 2002) with the kinship coefficient of Ritland (1996), recommended for microsatellite studies (Vekemans & Hardy 2004). We tested whether there was a change in mean or the variance in relatedness among individuals using unpaired Wilcoxon tests for each colony. Next, we assessed whether the degree of linkage disequilibrium (LD) increased within each colony (indicative of a founder effect) after recolonization by comparing |D′| values estimated in midas software (Gaunt et al. 2006) with unpaired Wilcoxon tests. An unpaired test was used because midas uses information from all allele combinations, some of which were not present either before plague or after recolonization.
We calculated temporal differentiation within colonies by estimating FST in the program fstat and performing 10 000 permutations (Waples 1989; Goudet 1995), which allowed us to assess whether colonists were genetically different from residents present before plague. To confirm that temporal changes in genotypes are not expected in the absence of extirpations, we estimated the degree of temporal differentiation in control colonies and compared this with the degree of differentiation in extirpated colonies with an unpaired Wilcoxon test in R. Finally, we performed a principal component analysis (PCA) in the ade4 package (Chessel et al. 2004; Dray & Dufour 2007) for R on individual genotypes within each colony before plague and after recolonization. Using the ‘between’ function in ade4, we performed 10 000 randomizations of genotypes within a colony across time to assess whether the first two principal components describing genotypes were significantly different before plague and after recolonization.
Analysis of genetic variation within individuals
Genetic variation at the individual scale was examined using observed heterozygosity (Ho) of individuals (calculated as the proportion of loci that were heterozygous) before plague and after recolonization. After determining that heterozygosity values were normally distributed, we used unpaired t-tests to assess the difference in mean observed heterozygosity between (1) individuals from all extirpated colonies before plague and after recolonization, (2) individuals from all control colonies before and after 2006 and (3) inferred immigrants and residents (nonimmigrants). Next, we compared the proportion of individuals with Ho < 0.4 and Ho < 0.2 before plague and after recolonization. To account for among-colony differences in (1) above, we corroborated the t-test by performing a bootstrap without replacement procedure in which we randomly drew values from a vector of preplague observed heterozygosity values across all extirpated colonies. The number of values drawn was equal to the average number of individuals in postrecolonization colonies (N = 43). This sampling procedure was repeated 10 000 times to generate a probability distribution of observing particular average heterozygosity values, and the percentage of times simulated postrecolonization heterozygosity was lower than the average from the preplague distribution was reported as a P-value. For individual colonies, we repeated the bootstrap procedure to test for significant differences in individual heterozygosity by randomly drawing observed heterozygosity values from each preplague colony separately. In this case, the number of values drawn was equal to the actual number of individuals in each postrecolonization colony. This bootstrap procedure was used instead of t-tests because of low samples sizes (e.g. 11 individuals in postrecolonization colony 30A) and was conducted in R (script in supporting online information).
The genotypes of six individuals that survived plague (see 2.1) were compared with those of pre-plague and postrecolonization individuals by performing a permutation test on principal component scores in the ade4 package for R. Next, to compare observed heterozygosity of survivors to a random sample of prairie dogs in the same colonies, we randomly sampled six individuals from those colonies and calculated average heterozygosity. We repeated this sampling 10 000 times and generated a distribution of observed heterozygosity values based on individuals present in the same colonies. The proportion of times survivor heterozygosity was lower than the simulated average heterozygosity was reported as a P-value (script in supporting online information).
Results
During seven field seasons, we captured 1187 prairie dogs from 15 colonies (Figs 1 and 2, Table S1, Supporting information). Rates of population growth after recolonization varied among extirpated colonies, ranging from 11 to 47 individuals captured 2 years after plague (Figs 2 and S1, Table S1, Supporting information). The majority of individuals in recolonized colonies were captured (estimated by visual counts and within-trapping session recapture rates). By retyping a subset of individuals, we estimated our genotyping error rate to be 2.1%; errors were distributed randomly across individuals and loci. Null alleles were not detected at any loci, and departures from neutrality were not detected. Statistical tests supported the pooling of genetic data among years into ‘preplague’ and ‘postrecolonization’ time points: there was no significant differentiation between years within time points in any colony, with the exception of one pairwise comparison in each of two colonies (2007 vs. 2009 in 1A and 12A; in both of these colonies, genotypes in 2007 alone and 2009 alone were also different from those present before plague). Similarly, anovas and a posteriori Wilcoxon tests on differences in allelic richness across years showed significant differences in only one colony (1A) at one time point (2007–2009, P = 0.020; Fig. S2a, Supporting information). Tests for changes in heterozygosity found no significant differences among years (Fig. S2b, Supporting information).
Genetic variation within the regional metapopulation
The degree of allelic richness across all colonies within the metapopulation was not significantly different before plague (6.456) and after recolonization (6.247, Wilcoxon P = 0.529; 90% CI for difference in median −0.3815 to +1.0185), even when including the six colonies present before plague that were not sampled after plague (6.397; P = 0.675; 90% CI: −0.4035 to +0.8340; Table S2, Supporting information). All colonies were significantly differentiated from each other, with the exception of four pairwise comparisons involving 60A; the lack of differentiation was probably an artefact of small sample size (N = 4; Table S3, Supporting information). Average pairwise FST among nine colonies was significantly higher after recolonization (0.1136) than before plague (0.0867, Wilcoxon P = 0.015, 95% CI: −0.04180 to −0.00435). When using standardized FST (Hedrick 2005), differentiation was not significantly higher after recolonization (Wilcoxon P = 0.307, 95% CI: −0.07286 to +0.01917). The isolation-by-distance signal present before plague (Mantel P = 0.029, r = 0.4418) was evident after recolonization (Mantel P = 0.011, r = 0.4856), and the degree of isolation by distance was significantly higher after recolonization (slope and 95% CI of regression after: 0.040 [0.025, 0.080]) than before plague (slope and 95% CI of regression before: 0.016 [−0.002, 0.027]; Table S2, Supporting information).
Following recolonization of six colonies, we identified 94 of 257 founders (36.6%) as immigrants (Table S4, Supporting information). The remaining founders were inferred to originate from a nearby unsampled source colony with a genetic signature indistinguishable from the recolonized colony. Similarly, founders that were assigned to extirpated colonies (e.g. one founder of colony 1A originated in colony 47A, Table 1) were interpreted as either migrants from a nearby unsampled, not extirpated colony with a similar genetic signature, or as a migrant leaving before the colony was extirpated. For example, recolonization of colony 1A began in spring of 2007, so migrants from 47A may have arrived in 2007 before colony 47A was extirpated (Fig. 1); this timing is consistent with the fact that most dispersal occurs in the spring (Garrett & Franklin 1988; Hoogland 1995). Some colonies were repopulated from a large number of source populations (e.g. 19A, 11 sources), while others were repopulated from a small number of sources (e.g. 30A, three sources; Table 1). The average number of inferred source populations was 7, and the proportion of immigrants in each colony ranged from 20.4% (colony 47A) to 46.7% (colony 19A). Our estimate of migration rate is higher than previously observed in prairie dogs in urban landscapes (Magle et al. 2010) but seems consistent with the rapid repopulation of extirpated colonies.
| 1A | 2A | 3A | 5A | 6A | 9A | 12A | 15A | 17A | 19A | 20A | 30A | 47A | 60A | 106A | Other | Ncol | M | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1A | 2 | 2 | 1 | 2 | 1 | 1 | 7 | 0.290 | ||||||||||
| 12A | 1 | 3 | 5 | 9 | 8 | 1 | 7 | 0.491 | ||||||||||
| 17A | 1 | 1 | 1 | 2 | 1 | 2 | 1 | 2 | 9 | 0.297 | ||||||||
| 19A | 1 | 5 | 1 | 1 | 2 | 3 | 3 | 9 | 6 | 1 | 11 | 0.467 | ||||||
| 30A | 1 | 2 | 3 | 0.273 | ||||||||||||||
| 47A | 2 | 6 | 1 | 1 | 5 | 0.204 |
Genetic variation within colonies
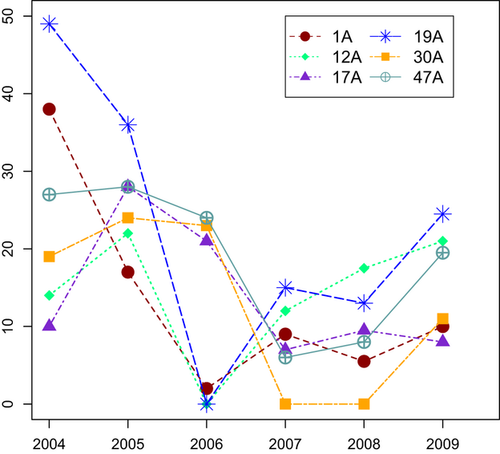
Within colonies, no systematic departures from Hardy–Weinberg equilibrium were observed either before plague or after recolonization. In colonies that experienced plague, changes in allelic richness varied widely when compared with control colonies, which showed no change in richness. There was a decline in richness in three recolonized colonies [1A (Wilcoxon P = 0.164; 95% CI for difference in median: −0.4210 to 1.4885), 30A (P < 0.001, 95% CI: 0.3505–1.2085), and 47A (P ≪ 0.001, 95% CI: 0.5280–1.2415); Table S1, Supporting information], but the other three colonies (12A, 17A and 19A) experienced a slight, but not significant, increase in allelic richness (Fig. 3). Allelic richness did not change systematically throughout recolonization (Fig. S2, Supporting information). Richness in postrecolonization colonies was positively related to the number of inferred source colonies (adjusted R2 = 0.631, P = 0.037; Table S5, Supporting information). Changes in relatedness among individuals in a colony were consistent with the changes in allelic richness (Fig. 3): average relatedness increased in the three colonies that experienced a decline in allelic richness [1A (P ≪ 0.001, 95% CI for difference in mean: −0.1374 to −0.08721), 30A (P = 0.027, 95% CI: −0.04025 to −0.003312), and 47A (P ≪ 0.001, 95% CI: −0.05912 to −0.0370); Table S5, Supporting information]. On the other hand, average relatedness decreased in the three colonies that did not exhibit a significant decrease in allelic richness [12A (P < 0.001, 95% CI: 0.08771–0.1573), 17A (P ≪ 0.001, 95% CI: 0.01730–0.04686), and 19A (P = 0.211, 95% CI: −0.002504 to 0.01296)]. The variance in relatedness among individuals decreased significantly (P < 0.02) in three colonies (12A, 19A and 30A) but was unrelated to the number of individuals sampled or the mean change in relatedness. Linkage disequilibrium (measured by |D′|) increased significantly in the three colonies that lost allelic richness (1A: Wilcoxon P ≪ 0.001, 95% CI: −2.636 × 10−2 to −6.411 × 10−5; 30A: P = 0.004, 95% CI: −2.909 × 10−5 to −1.885 × 10−5; 47A: P = 0.004, 95% CI: −4.887 × 10−5 to −4.816 × 10−5; Table S5, Supporting information), which may be indicative of founder effects. LD did not change in two colonies (17A and 19A, P > 0.1) and decreased in one colony (12A, P ≪ 0.001, 95% CI: 6.383 × 10−5 to 3.191 × 10−5).
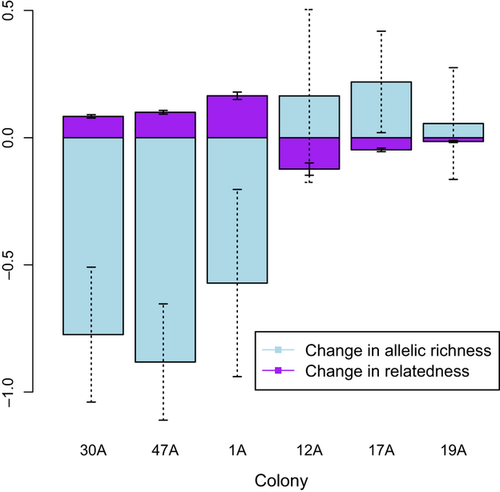
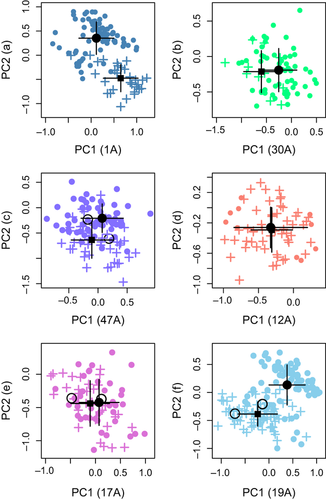
Temporal differentiation before and after 2006 was an order of magnitude higher in colonies affected by plague than in control colonies (FST = 0.0689 (range 0.015–0.113) plague; 0.005 (range −0.006 to 0.006) control; P = 0.009; Fig. 4). The average spatial FST was 0.0867 (Table S2, Supporting information). Principal component analysis of genotypes showed that, in some cases, postrecolonization individuals were significantly different from preplague individuals from the same colony, suggesting that founders originated from other colonies. For instance, founders of colony 19A were more genetically similar to residents of colony 30A than to individuals present in colony 19A before extirpation (Fig. S3, Supporting information). The mean genotype within four colonies (1A, 12A, 19A and 47A) changed over time (1A: P ≪ 0.001, σ = 3.519 × 10−5, 12A: P < 0.05, σ = 0.00013, 19A: P ≪ 0.001, σ = 1.767 × 10−5, 47A: P = 0.013, σ = 3.987 × 10−5; Fig. 5a–f), with the change more pronounced in some colonies (e.g. 1A and 19A, P ≪ 0.001; Fig. 5a, f) than others. One colony with plague survivors, 17A (Fig. 5e), experienced no shift in genotype space (P = 0.265, σ = 0.00013); 17A and 30A were probably recolonized from a source population that was genetically similar to the original colonies.
Genetic variation within individuals
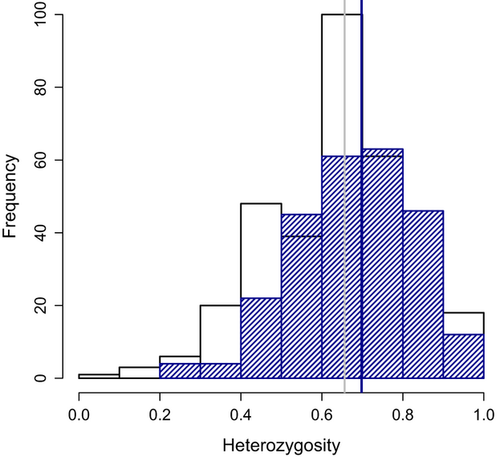
Observed heterozygosity in recolonized colonies ranged from 0.632 (1A; 20 individuals captured in 3 years) to 0.718 (19A; 74 individuals captured in 3 years). Across all recolonized colonies, there was a significant increase in heterozygosity after recolonization (mean Ho in preplague colonies = 0.656; mean Ho in recolonized colonies = 0.698; t = 2.99, P = 0.003, 95% CI for difference in means: −0.06905 to −0.01430). The bootstrap procedure supported this conclusion when the average number of individuals in postrecolonization colonies (N = 43) or a larger number of individuals was sampled (P < 0.05). This increase happened during the first year of recolonization (Fig. S2, Supporting information) and did not change systematically throughout the recolonization process. During the same time period, heterozygosity did not change in colonies that did not experience plague (t = 0.083, P = 0.934, 95% CI: −0.0392 to +0.03606). Individuals inferred by GeneClass2 to be immigrants did not have higher heterozygosity than residents (t = 0.763, P = 0.450, 95% CI: −0.0961 to +0.0433). Recolonized colonies with plague survivors (17A, 19A and 47A;) and 12A exhibited significantly higher heterozygosity than before plague (bootstrap P ≪ 0.001), whereas the other two colonies were not different before plague and after recolonization (bootstrap P > 0.1). Interestingly, the increase in mean heterozygosity among recolonized colonies resulted partly from the loss of almost all individuals in the lowest heterozygosity classes (Fig. 6). Fewer individuals with low (Ho < 0.4) or very low (Ho < 0.2) heterozygosity were present after recolonization [Ho < 0.4: 8 (3.11%); Ho < 0.2: 0 (0%)] than before plague [Ho < 0.4: 30 (8.77%); Ho < 0.2: 3 (0.87%)]. Postrecolonization heterozygosity was approximately equal to that of a hypothetical population created by culling approximately 10% of the most inbred individuals present before plague.
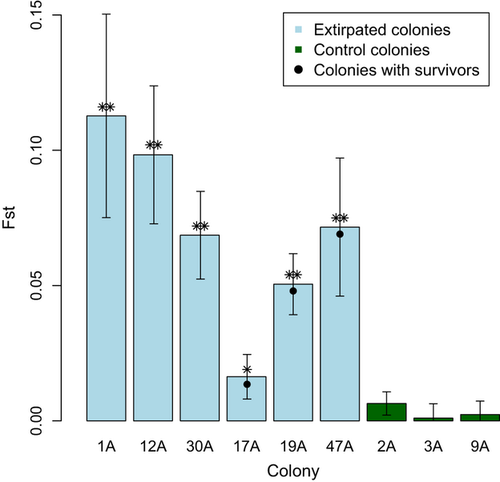
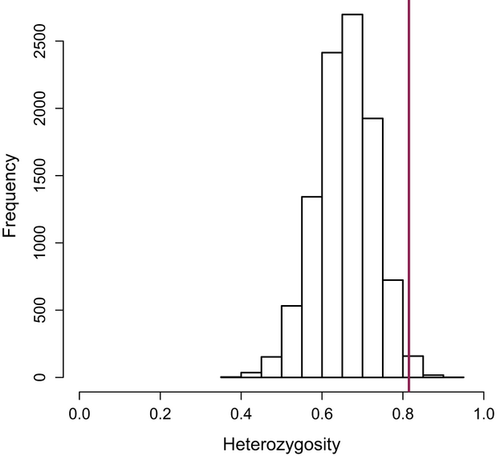
During 2007 and 2008, we confirmed that six individuals survived exposure to plague (assessed by plague antibodies in 1 year and recapture the next); two individuals survived in three colonies (17A, 19A and 47A). Of these six, five were adults, three were males and three were females. Five of these animals also tested positive for antibodies in the second year of capture, indicating that plague antibodies can persist for at least a year or that there was repeated exposure to plague. The PCA indicated that genotypes of survivors were not significantly different from other individuals in their colonies (Fig. 5c, e, f), and in one colony (19A, Fig. 5f) survivors were more genetically similar to colonists (no difference; P = 0.61) than to preplague residents (marginally significant difference, P = 0.10). In the other two colonies (17A and 47A, Fig. 5c, e), survivors were not genotypically distinct from residents before or after plague (P > 0.1). Two of the survivors, both sampled in colony 17A, were inferred immigrants. One was assigned to colony 60A, a historically plague negative colony, and the other was assigned to colony 20A, which was extirpated by plague in 2008 (Fig. 1; data from Boulder Open Space and Mountain Parks). The two survivors from 19A and one survivor from 47A were not excluded from their sampled colony (i.e. not inferred immigrants) but were assigned with a higher probability to colony 20A. The other survivor from 47A was assigned unambiguously to its sampled colony. Heterozygosity of the six plague survivors was significantly higher than expected if survival was random with respect to heterozygosity (mean survivor Ho = 0.815, bootstrap P = 0.006; Fig. 7).
Discussion
The effects of plague extirpation and subsequent recolonization on genetic diversity varied depending on the scale at which diversity was assessed. Within the metapopulation, there was no change in diversity. Within colonies, the magnitude and direction of change varied, with some colonies experiencing a large decline in allelic richness and others exhibiting a slight (nonsignificant) increase in allelic richness. Within individuals, the processes of extirpation and recolonization resulted in individuals with higher heterozygosity. Furthermore, the six survivors had significantly higher heterozygosity than expected by chance. Collectively, our results show that although the effects of extirpations on genetic diversity of colonies varied, plague and recolonization resulted in individuals with higher genetic diversity, with the implication that pathogen-induced extirpations provide a mechanism for the maintenance of genetic variation.
Genetic variation within the metapopulation
In our system, plague extirpations did not depress allelic richness of prairie dogs at the regional scale, a pattern that held even with the inclusion of the six colonies for which we have no postrecolonization data. There was, however, an increase in average pairwise differentiation and isolation by distance, likely due to reductions in population size (Wright 1931; Van Treuren et al. 1991). It is probable that the maintenance of genetic diversity within this metapopulation was due to its urban landscape matrix, which may have slowed the spread of Y. pestis (Collinge et al. 2005) such that source populations remained at all times, allowing recolonization of extirpated colonies (data from Boulder Open Space and Mountain Parks; see Fig. 1). For instance, colony 19A could have been a source colony in 2005 but not 2006, whereas colony 2A could have been a source in 2006. In contrast, a system that experienced simultaneous extirpation of all populations would likely exhibit a loss of genetic diversity (Larson et al. 2002).
Genetic variation within colonies
Three lines of evidence suggest that three colonies (1A, 30A and 47A) experienced founder effects: these colonies experienced (i) a decline in allelic richness, (ii) an increase in average relatedness among individuals and (iii) an increase in LD. Founder effects were not evident in the other three recolonized colonies (12A, 17A and 19A), which experienced a slight increase in allelic richness, a decrease in average relatedness and a decrease or no change in LD (Tables S5–S6, Supporting information). Prairie dogs in the three recolonized sites without founder effects (and in 47A) were clustered in spatially discrete portions of their colonies that were separated by distances requiring dispersal (Sackett, unpublished data; Hoogland 1995), suggesting that they may represent distinct family groups and separate immigration events from different source populations. This inference is supported by the high number of inferred source populations of immigrants in colonies 17A and 19A (Table 1). Coupled with the slight increase in allelic richness and the change in genetic composition before plague and after recolonization, spatial clustering supports the idea that immigrants arrive from multiple source populations with unique genetic signatures.
The reasons for the disparity in patterns of change in diversity are probably largely dependent on population connectivity, which is influenced by the landscape context in which colonies occur (Magle et al. 2010) and the associated relative rates of extirpation and recolonization. We observed that colonies with the largest putative number of sources also contained the highest allelic richness (Table S5, Supporting information), whereas colonies with low migration rates from few source populations experienced a loss of richness. Increased numbers of source populations for recolonization may thus mitigate founder effects (Kolbe et al. 2004). Thus, for the maintenance of genetic variation, the importance of connectivity for recolonization appeared to outweigh the increased risk of extinction (Hess 1996), perhaps because so few colonies escaped exposure to plague (data from Boulder Open Space and Mountain Parks). Quantifying the landscape context in relationship to genetic diversity will be a fruitful avenue for further research.
Temporal genetic differentiation within colonies before plague and after recolonization may occur for three reasons. First, migration routes could vary over time, causing genetic inputs into a colony to also change over time (Mackey et al. 2011). This explanation may be particularly likely in systems where extirpations are spatially proximate, disrupting the typical dispersal corridors used by individuals. For instance, most genetic exchange probably occurs between closely situated populations (e.g. colonies 17A and 47A, Fig. 1), but if plague eliminates both populations simultaneously, then immigrants into each colony will necessarily originate from farther away (e.g. colony 106A). Second, temporal differentiation within colonies before and after plague could arise from stochastic demographic fluctuations. During colony formation, one or two genotype groups could become prevalent due to colonization order or other nonadaptive effects. Finally, only certain genotypes may be able to colonize (as in migratory culling, Bartel et al. 2011) after plague. Successful colonists may need to be excellent dispersers, resistant to plague, or both.
Genetic variation within individuals
At the individual scale, heterozygosity within individuals increased, partially reflecting the loss of individuals with the lowest heterozygosity. This suggests one of two nonmutually exclusive hypotheses: (i) a cost of low heterozygosity to either the ability to successfully colonize a new population or the ability to survive plague, or (ii) increased heterozygosity is a result of admixture of multiple genotype groups founding new populations. Migratory culling (Bartel et al. 2011) may be partially responsible for the loss of low-heterozygosity individuals; however, we found no difference in heterozygosity between inferred immigrants and residents, suggesting instead that the role of heterozygosity is linked to survival from plague. Indeed, genome-wide heterozygosity is often associated with resistance to disease (Hawley et al. 2005; Pearman & Garner 2005; Calleri et al. 2006). The overall increase in heterozygosity within postrecolonization individuals probably reflects both the selective loss of inbred individuals and the direct result of admixture (Table 1). Moreover, because prairie dogs avoid inbreeding (Hoogland 1982), reduced population size followed by admixture may increase the likelihood of mating with unrelated mates, resulting in higher heterozygosity in admixed populations. This increase in heterozygosity suggests that successful immigration increases after extirpations: Prairie dog colonies have high population densities and socially structured groups that are defended territorially (Hoogland 1981); thus, immigrants are likely to experience intense competition with residents. After plague extirpations, this competition is eliminated, and immigrants may be more likely to survive. Thus, extirpation could act to effectively increase the degree of successful immigration because territories are no longer defended. This hypothesis should be further tested in species with territorial behaviour that are subject to periodic population extirpations.
The six individuals that survived plague were not genetically different from other individuals in their colonies for the microsatellite loci examined (Fig. 5); however, survivors exhibited unusually high heterozygosity (Fig. 7). Although this pattern could arise from cryptic within-colony genetic structure, we were unable to detect genetic structure below the colony level (analysed in the program Structure, data not shown), suggesting that this effect is not the sole contributor to the heightened heterozygosity of survivors. Admixture resulting from colonization from multiple sources can lead to higher heterozygosity of residents; however, this phenomenon does not explain the higher heterozygosity of plague survivors (0.815) compared with other postrecolonization founders (0.695).
Microsatellites are neutral markers; however, they provide a proxy for genome-wide heterozygosity (Da Silva et al. 2006), which correlates inversely with inbreeding and sometimes reflects diversity at non-neutral loci (Hansson & Westerberg 2002; Slate et al. 2004) such as genes that provide protection against disease (Penn et al. 2002; Turner et al. 2008; Yang et al. 2011). Heterozygosity-fitness correlations at neutral loci may be due either to LD between one or several neutral markers and functional loci under selection (Hansson & Westerberg 2002) or to negative effects on fitness owing to genome-wide homozygosity (Charlesworth & Charlesworth 1987; Hansson & Westerberg 2002). We observed a particularly strong effect of two loci (D1 and D115, Jones et al. 2005), at which all survivors were heterozygous, which may indicate LD with functionally important loci. However, the pattern persisted when removing these loci from analyses; further, these markers showed no signs of selection with the Fdist approach (see 2), suggesting these genomic regions are not the only drivers of the observed effect. Although it is possible that both mechanisms are contributing to differential survivorship of highly heterozygous prairie dogs, the significant relationship between multilocus heterozygosity and survivorship lends support to the idea that survival is conferred by the collective heterozygosity of many loci across the genome.
Implications of this study for understanding pathogen influences on diversity
The genetic consequences of plague across scales support the hypothesis that extirpation creates a vacant habitat into which colonization can occur from multiple sources, leading to a maintenance of colony genetic diversity and an increase in individual genetic diversity in sufficiently connected populations. The immediate decline in allelic richness in three colonies may be counteracted by immigration in subsequent years (indeed, colony 30A had just begun recolonization at the conclusion of our study and only 11 individuals were captured; Fig. 2). The increase in diversity may occur over multiple generations, and rapid population growth may relax selection, allowing the enhanced survival of new mutations (Carson 1968; Templeton 1980). If founder populations have higher additive genetic variation, they could have a more pronounced response to selection in subsequent generations. Furthermore, the joint processes of extinction/recolonization preferentially eliminated the most inbred individuals. High genome-wide heterozygosity may offer protection against pathogens via overdominance at many loci involved in resistance to pathogens (e.g. MHC, Hughes & Nei 1988; signalling pathways in immune response, Yang et al. 2011). The increase in genetic diversity after extirpation has implications for the way we understand host-pathogen co-evolution (Nuismer & Doebeli 2004), the evolution of virulence (Lenski & May 1994), and evolution of host resistance (Caprio & Tabashnik 1992; Hughes & Boomsma 2007; Rocke et al. 2012).
More research is needed to determine the circumstances under which pathogens facilitate an influx of diversity. First, the spatial arrangement of populations in a landscape can influence extinction probability and recolonization dynamics as discussed earlier. Subdivided populations may be more likely to retain genetic diversity in the face of extinctions than less divided populations (Ray 2001). Second, pathogen virulence is a likely predictor of how genetic diversity is maintained (Tobler & Schmidt 2010). A pathogen of moderate virulence would not eliminate populations (e.g. Breitschwerdt & Kordick 2000) and create a vacant habitat; thus, we would not predict an influx of diversity. Highly virulent pathogens that extirpate populations—provided some populations remain as sources for recolonization—may be the most likely to allow an influx of diversity into a vacant habitat, particularly when coupled with high levels of host movement (Cross et al. 2005). Finally, social structure and life history characteristics of the host also probably play an important role in how extirpations change host genetic diversity. Prairie dogs territorially defend their colonies (Hoogland 1981); thus, immigrants are probably more successful at surviving after a colony has been extirpated. In less aggregated species, however, individuals may have to travel farther to find unrelated mates, and inbreeding may thus increase after extirpations (Lachish et al. 2011).
Overall, our findings suggest that, when population connectivity is sufficient to allow for recolonization from multiple sources, pathogen-induced extirpations serve to maintain genetic variation of populations. Furthermore, the dual processes of extirpation and recolonization eliminated individuals nonrandomly: those with the lowest heterozygosity did not survive. These results challenge the conventional wisdom that pathogens decrease host genetic diversity and suggest instead that virulent pathogens should drive an evolutionary increase in the genetic diversity of their hosts. Further study will help resolve the conditions under which this may occur.
Acknowledgements
We are grateful to numerous assistants on this project, especially Erin Arnold, Jory Brinkerhoff, Ryan Jones and Katherine McClure. We thank Boulder County Parks and Open Space, Boulder Open Space and Mountain Parks and Boulder Parks and Recreation for access to colonies and the Colorado Division of Wildlife for permission to trap prairie dogs. We appreciate analytical discussions with Nic Kooyers and helpful comments on an earlier draft of this manuscript from Alan Templeton, Ken Olsen and the Olsen lab at Washington University in St. Louis. We thank the five anonymous reviewers who provided thoughtful comments on the previous version of this manuscript. This research was funded by the Boulder County Nature Association, the University of Colorado and the National Science Foundation/National Institutes of Health joint program in Ecology of Infectious Diseases (DEB-0224328).
References
L.C.S. was responsible for sampling design during 2007–2009, for study conception and for data analysis and writing. S.K.C. was responsible for sampling design from 2003 to 2006. A.P.M. was responsible for data analysis and writing.
Data accessibility
R script: uploaded as online supporting information.
Sample locations, microsatellite genotypes, and identification of individuals that produced plague antibodies: uploaded as online supporting information: DRYAD http://dx.doi:10.5061/dryad.p2j15.




