Cryptic diversity of marine gastropod Monodonta labio (Trochidae): did the early Pleistocene glacial isolation and sea surface temperature gradient jointly drive diversification of sister species and/or subspecies in the Northwestern Pacific?
Abstract
Formation of glacial refugia during the Pleistocene climatic oscillations has been put forward to elucidate the diversification of marine organisms in the north-western Pacific. The marine gastropod Monodonta labio is one of the most common species in the Northwestern Pacific and possibly possesses cryptic diversity. Here, we investigate the phylogeographic pattern of this species to test the potential mechanisms driving its diversification in the Northwestern Pacific. Genetic information for two mitochondrial genes (Cytochrome oxidase subunit I and 16S rDNA) and one nuclear gene (internal transcribed spacer 1) was acquired to detect genetic structuring and to reconstruct the gastropod's phylogenetic history. Our results revealed that M. labio is comprised of five main clades, and divergence time estimates place their cladogenesis as corresponding to the initiation (c. 2.5 mya) and intensification (c. 0.9 mya) of large-scale Northern Hemisphere ice sheets. The early and middle Pleistocene divergence times are consistent with the emergence times of the Dongshan land bridge, which would seperate the ancient East China Sea and the ancient South China Sea forming two potential refugia. In addition, the deep trough in the Qiongzhou Strait would possibly act as another potential refugium with the uplift of the Qiongzhou Strait at mid-Pleistocene. This study suggests that the current genetic architecture of M. labio is probably correlated with glacial isolation and sea surface temperature gradient. We also put forward the possibility that these factors were probably an important driver for the diversification of sister species or subspecies of other taxa in the Northwestern Pacific.
1 INTRODUCTION
Mechanisms of population subdivision and speciation in the marine realm have long been little understood topics in evolutionary biology (Bowen, Rocha, Toonen, Karl, & ToBo, 2013; Mayr, 1954; Miglietta, Faucci, & Santini, 2011; Palumbi, 1992). Speciation via allopatry (Dobzhansky & Dobzhansky, 1937; Mayr, 1942) continues to play a dominant role in species diversification. By contrast, due to dispersal potential of marine species' planktonic phase and absence of absolute barriers in the sea, speciation via ecological isolation and natural selection (Bowen et al., 2013; Rundle & Nosil, 2005; Schluter, 2001) has been proposed to decipher the phenomenon of sister species with overlapping distributions. Overall, diversification may proceed along both geographic and ecological partitions in marine systems (Dawson & Hamner, 2008). Nonetheless, a combination of physical and ecological partitions is probably considered to be the most effective avenue to drive speciation (Bowen et al., 2013), or intra-specific subdivision.
Paleoclimate oscillations and plate tectonics during the past 3,000,000 years have repeatedly produced such a combination of physical barriers (such as land bridges exposed during glaciation or oceanic currents) and ecological partitions between adjacent regions. These factors resulted in intermittent isolation of marine organisms and great changes in species' distributions, which may be reflected in their contemporary genetic structures (Hewitt, 2000; Provan & Bennett, 2008). Along the vast coastline of China, isolated sea basins (the semi-closed South China Sea and the enclosed Okinawa Trough) have been put forward to act as historical refugia, driving divergence between corresponding geographic populations in some marine species (Kojima, Segawa, & Hayashi, 1997; Liu, Gao, Wu, & Zhang, 2007; Ni, Li, Kong, & Zheng, 2012). However, this cannot be the explanation to species diversification in this region due to the late formation time of the Okinawa Trough. In fact, speciation duration is about 1–2,000,000 years proceeding unhindered through the Pliocene and Pleistocene (Hewitt, 1996, 2000). Sea level and temperature fluctuations during Plio-Pleistocene epochs have been considered to play the major role in facilitating contemporary species diversity of the Northwestern Pacific (Shen, Jamandre, Hsu, Tzeng, & Durand, 2011). In addition, speciation in Chthamalus in the Western Pacific is thought to have been facilitated by the formation of glacial refugia (Cheang et al., 2012), whereas no potential refugia corresponding to this epoch have been discovered. Overall, information on how the geological history of the marginal seas of China has affected the process of speciation remains scarce.
Understanding the evolutionary history and tectonic structure of China's seas will contribute to unraveling the most important factors governing the formation of species. From the Pliocene to mid Pleistocene, the geographic scope of the ancient marginal seas differed a lot from that of the present day. In ancient times, the South China Sea (SCS), which was opened in the late Oligocene-middle Miocene (c. 30–15 mya; Taylor & Hayes, 1983), reached a range similar to that of the present day. By comparison, large parts of the East China Sea (ECS) were exposed as landmass till the mid-Pleistocene (Letouzey & Kimura, 1985), in addition to the Southern ECS containing the Taiwan Strait (Wan, 2012). Ryukyu arc, connected with the Taiwan Island in the south, formed the eastern margin of the Asian continent (Wang, Li, & Li, 2014). Consequently, the Kuroshio Current flows to the east side of the present Ryukyu Islands in the Pacific. Notably, Lin (1982) discovered the Dongshan land bridge linking the mainland with Taiwan Island, which is about 40 m below the sea surface. Under this scenario, when the sea level was 40 to 70 m below that of the present day during the glacial epoch, the land bridge would emerge and the ECS would become an enclosed shallow sea serving as a potential isolated refugium. Thereafter, with the region around the Qiongzhou Strait rising to 40 m below the present sea level in the late early-Pleistocene (Yao, 1980), the deep trough in the Qiongzhou Strait may have been isolated from the SCS during the ice ages by the emergence of the Qiongzhou Strait landmass. After its reopening and large-scale depression in the early (<1 Ma) and middle Pleistocene (<0.7 Ma), the Okinawa Trough of the ECS rifted and subsided along its full length (Kimura, 1990; Tanaka & Nomura, 2009). The seawater flowed into the northern part of the ECS and the entire Yellow Sea. Meanwhile, together with the tectonic changes in this region, several climatic events occurred that caused environmental changes (Wang et al., 2014), such as the initiation (c. 2.5 mya) and intensification (c. 0.9 mya) of large-scale Northern Hemisphere ice sheets. These major events may have left marks in the population histories of some species.
Monodonta labio is a common gastropod species in inter-tidal shores, widely distributed along the coast of East Asia. It occupies a wide range of inter-tidal habitats, including rocky, cobble and boulder shores as well as mangroves (Chin, 2003). Study of this species' thermal and desiccation tolerance has been conducted. It was revealed that M. labio was able to live in a salinity range from 13‰ to 35‰, and survive for an average of 9 days when exposed to air in a temperature range of −7 to 25°C (Wang & Zeng, 1994). The species' ability to adapt well to eurythermic and euryhaline environments has allowed it to survive the intermittent paleoclimate oscillations. Furthermore, M. labio may be a species complex harboring cryptic species. However, the intra-specific variation in shell morphology adds to the difficulty in defining its systematics. Within this species, two commonly recognized Japanese subspecies, referred to as Monodonta labio labio and Monodonta labio confusa, are documented based on differences in shell morphology (Higo, Callomon, & Gotō, 1999). Donald, Kennedy, and Spencer (2005) found that they are genetically divergent and treated them as separate species, Monodanta labio and Monodonta confusa. In addition, they noticed that individuals obtained from Australia turned out to be genetically distinct from the two Japanese subspecies. Thus, detecting M. labio's genetic relationships among the different geographically distributed populations along the coastline of China seas may facilitate our understanding of how variations in paleoclimate promoted speciation and/or population subdivision. In this study, we obtained sequences from two mitochondrial genes and one nuclear marker to: (i) examine patterns of genetic diversity of M. labio along the Chinese coastline; and (ii) test the mechanisms shaping the genetic structure of this species.
2 MATERIAL AND METHODS
2.1 Sampling
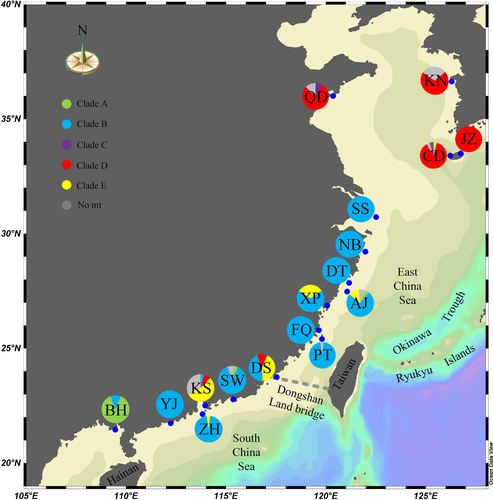
We collected a total of 425 individuals of Monodonta labio from 17 localities distributed across the coast of China and Korea between October 2008 and June 2015. Samples were collected by hand and preserved in 95% ethanol. Figure 1 and Table S1 show the localities sampled in this study.
2.2 DNA extraction, PCR amplification, sequencing and alignments
Genomic DNA was extracted from foot tissue of each specimen with a cetyltrimethylammonium bromide extraction protocol (Schulenburg et al., 2001). Partial sequences of the mitochondrial genes Cytochrome oxidase subunit I (COI), 16S ribsomal DNA (16S) and the nuclear internal transcribed spacer 1 (ITS1), were obtained for subsequent analyses of Monodonta labio. The above COI fragment was amplified using the self-designed primer set (LDAN: 5′-CTTTAACTTTACTCCTGGCATCCT-3′, RDAN: 5′-CGGTGAAATAAGCACGGGT-3′). Universal primer sets (16Sar: 5′-CGCCTGTTTATCAAAAACAT-3′, 16Sbr: 5′-CCGGTCTGAACTCAGATCACGT-3′; Palumbi, 1996) and (ITS1F: 5′-TAACAAGGTTTCCGTAGGTGAA-3′, ITS1R: 5′-GCTGCGTTCTTCATCGATGC-3′; Armbruster, van Moorsel, & Gittenberger, 2000) were used to amplify the target 16S and ITS1 markers separately. PCR was carried out in a 50 μl volume containing 2 U Taq DNA polymerase (Takara Co.), about 100 ng template DNA, 0.25 mm of each primer, 0.2 mm deoxy-ribonucleoside triphosphate, 1× PCR buffer and 2 mm MgCl2. The reaction conditions were as follows: an initial 3-min denaturation at 94°C, 30 cycles of 30 s at 94°C, 30 s at various annealing temperatures (COI and 16S, 48°C; ITS1, 52°C) and 45 s at 72°C, and a final 5-min extension at 72°C. We performed the sequencing reaction on an ABI PRISM 3730 (Applied Biosystems) automatic sequencer, using a BigDye Terminator Cycle Sequencing Kit (v. 3.1, Applied Biosystems).
All mtDNA fragments were sequenced in two directions. To check for heterozygous sequences, the nuclear ITS1 amplicon was initially sequenced using the forward primer. For the samples containing heterozygous insertions/deletions, the reverse-direction sequencing was conducted. Then, phase determination for individuals with 2 sequences of different lengths was performed with CHAMPURU 1.O software (Flot, 2007). Finally, for all other individuals with heterozygous sequences, we cloned the amplicon and obtained three to six copies for each individual by sequencing the clones at random.
Forward and reverse sequences of individual loci were trimmed, assembled and merged into consensus sequences using DNA BASER SEQUENCE ASSEMBLER v. 4 (2013, Heracle BioSoft, www.DnaBaser.com). Alignment was conducted with CLUSTAL_X 1.81 under the default settings (Thompson, Gibson, Plewniak, Jeanmougin, & Higgins, 1997). All sequences were deposited in GenBank with the accession numbers KU848769–849171 (COI), KU849172–849586 (16S) and KU849587–850465 (ITS1).
2.3 Phylogenetic analyses and genealogical network construction
A concatenated data set (COI + 16S) of 389 individuals was produced with the help of SEQUENCEMATRIX (Vaidya, Lohman, & Meier, 2011). Identical haplotypes were removed from the data set, leaving unique haplotypes only. Phylogenetic relationships among the unique haplotypes were inferred, using Bayesian inference and maximum-likelihood (ML) reconstruction methods implemented in MRBAYES 3.2.5 (Ronquist et al., 2012) and RAxML 8.2.4 (Stamatakis, 2014), respectively. Prior to phylogenetic reconstruction, the best-fit model of nucleotide substitution was determined by JMODELTEST v. 2.1.1 (Darriba, Taboada, Doallo, & Posada, 2012) under the Akaike information criterion. General time-reversible model with gamma distribution (GTR+G) was selected as the most appropriate model for subsequent analyses. Branch supports were assessed using 1,000 bootstrap replicates for ML tree construction. In MRBAYES, two independent Markov-chain Monte Carlo searches were performed with one cold chain and three heated chains each. Trees were sampled every 1000 iterations in 20,000,000 generations, with the first 25% of generations discarded as burn-in. To investigate the relationship among closely related haplotypes, a median-joining network was constructed in NETWORK v. 4.6.1.3 (Bandelt, Forster, & Rohl, 1999) for the three markers.
The ITS1 marker of M. labio is short in length and includes very few parsimony informative sites (4/261). While the insertions/deletions found in the ITS1 sequences were recoded utilizing the simple coding method of Simmons and Ochoterena (2000) in FASTCODE v. 1.2 (Borchsenius, 2009), no credible phylogenetic trees were constructed finally. As an alternative, only a haplotype network was constructed to visually resolve the relationships among haplotypes.
2.4 Population genetics analyses
Population genetics analyses were based on the COI data set, because it has extensively been shown in the literature to be the most informative marker for mollusks among the genes used in this study and because of the extensive use of COI marker in species delimitation. Five clades were formed in the phylogenetic analyses. Clade C was excluded from the analyses because it contained only three individuals. To test the genetic divergences among the remaining four clades, Kimura two-parameter (K2P) distances between and within clades were calculated in MEGA 6 (Tamura, Stecher, Peterson, Filipski, & Kumar, 2013). Genetic diversity was analysed in DNASP v. 5 (Librado & Rozas, 2009).
The absence of calibrated molecular clocks for 16S and ITS1 markers precludes the accurate estimation of time since expansion. Thus, historical demographic analysis was performed through two different methods using the COI marker only. Tajima's D and Fu's Fs statistics were calculated in ARLEQUIN 3.5 (Excoffier & Lischer, 2010) to verify neutrality for each clade. The significance levels were evaluated under 10,000 permutations. Mismatch distribution analyses were also conducted to detect whether sudden population expansion occurred in the demographic history. For the clades exhibiting a unimodal distribution, the parameter τ was used to estimate the time elapsed through the equation τ = 2μt, where μ is the mutation rate of the marker (per locus per generation) and t is the number of generations. The divergence rate was set at 2.4% per million years for COI to convert parameters into quantitative estimates of time. The exact hatching period for Monodonta labio has never been studied in detail; therefore, the generation time was assumed to be 1 year deduced from the annual recruitment of this species (Chin, 2003).
2.5 Divergence time estimates
Given the absence of a clear fossil record and molecular clocks of M. labio calibrated for the 16S and ITS1 markers, estimation of the divergence times between the distinct clades defined was conducted in BEAST v. 1.8.2 (Drummond, Suchard, Xie, & Rambaut, 2012) on COI gene data using a calibrated molecular clock method. A GTR+G model was implemented with four rate categories recommended by JMODELTEST. We used the Yule process of speciation and the uncorrelated log-normal relaxed clock model as the tree prior. Two independent runs of 200,000,000 generations were performed with tree sampling every 5,000 generations. The first 10% of generations was discarded as burn-in. Convergence and effective sample size of estimated parameters were checked in TRACER v. 1.6. As geological events can also work as reliable calibration points for molecular clocks (Ho et al., 2015), we utilized the mtCOI molecular clock (2.4% nucleotide divergence per million years) calibrated by two Tegula species separated by the Isthmus of Panama (Hellberg & Vacquier, 1999). Note that sequence divergence rate = substitution rate × 2 (Wilke, Schultheiss, & Albrecht, 2009). Detailed prior settings for the molecular clocks are shown in Table 1.
| Molecular clock | Sequence divergence rates | Substitution rates specified in BEAST |
|---|---|---|
| COI marker | mean = 2.4% SD = 0.5% | mean = 1.2% SD = 0.25% |
- COI = Cytochrome oxidase subunit I; Prior distribution = normal.
3 RESULTS
In this study, 403 individuals for the COI gene and 415 individuals for the 16S rDNA gene were sequenced. In addition, a total of 879 sequences were obtained for the ITS1 marker after discarding identical copies from the same individuals. The lengths of the COI, 16S and ITS1 markers were 555, 570 and 261 bp, respectively.
3.1 Demographic history
Neutrality tests conducted on the COI data set suggest that all clades experienced a demographic expansion as indicated by significant negative values of Tajima's D and Fu's Fs statistics (Table 2). The mismatch distribution was unimodal solely for examined individuals belonging to Clade B, supporting a model of sudden expansion. The expansion time parameter was estimated from mismatch distribution analysis to be 4.557, and the time since expansion was estimated to be 342,000 years ago.
| Clade | n | Tajima's D | Fu's Fs | h | π |
|---|---|---|---|---|---|
| A | 24 | −1.690* | −3.344* | 0.377 | 0.409 |
| B | 238 | −2.193* | −26.449* | 0.874 | 2.594 |
| D | 88 | −1.663* | −22.592* | 0.935 | 3.971 |
| E | 50 | −1.562* | −10.066* | 0.833 | 2.278 |
- n = number of sequences; Tajima's D = Tajima's D statistic; Fu's Fs = Fu's Fs statistic; h = haplotype diversity; π = nucleotide diversity.
3.2 Phylogenetic analyses and gene genealogy
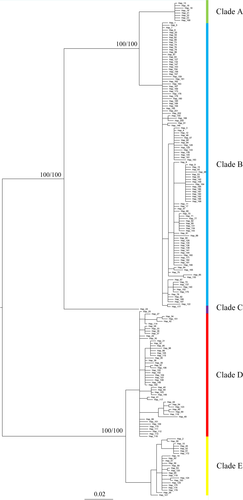
The mitochondrial concatenated data set contains 389 taxa with 202 unique haplotypes, which were used to conduct the phylogenetic analyses. The ML and Bayesian methods yielded identical topologies (Figure 2). Five well-supported clades were formed in the phylogenetic trees. Since the primers of COI marker were self-designed in the present study, no corresponding COI fragments were found in M. labio's closely related species. Phylogenetic trees was constructed without outgroup, and the directionality of the trees was inferred according to the relationships between the clades in the haplotype network. The average pairwise K2P differences between these clades varied from 2.4% to 9.7%, and the intra-clade distances were 0.074%–0.72% (except Clade C; Table 3).
| Clade | A | B | C | D | E |
|---|---|---|---|---|---|
| A | 0.00074 | ||||
| B | 0.024 | 0.00547 | |||
| C | 0.072 | 0.065 | 0 | ||
| D | 0.088 | 0.092 | 0.079 | 0.00724 | |
| E | 0.096 | 0.097 | 0.082 | 0.024 | 0.00405 |
Locality Beihai is the only sampling site situated in the Gulf of Tonkin. However, most of the individuals in BH, together with another three samples, one from each locality of Pingyang, Dongshan and Shanwei cluster into Clade A.
Clade B was dominant in the southern and central part of the vast coastline of China (the South China Sea and the East China Sea), from the southernmost locality of Beihai to the Shengsi Islands. Moreover, all localities situated in this range contained individuals belonging to Clade B, and samples from six (Shengsi, Ningbo, Dongtou, Fuqing, Pingtan, Yangjiang) of the 13 localities solely fell within this clade.
Only three samples collected in Qingdao and Jeju-do (CD) with an identical haplotype formed Clade C. The remaining individuals distributed north of the Yangtze River fell within Clade D. However, Clade D also includes samples in DS and KS south of the Yangtze River.
Clade E is mainly restricted to the subtropical zone, extending from the locality Zhuhai to the locality CD (only one sample).
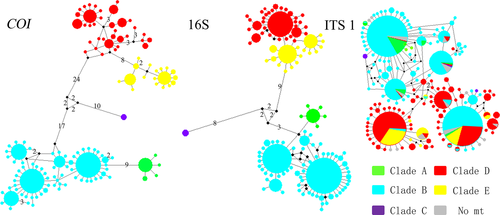
The mitochondrial fragments and ITS1 markers showed different resolutions in the haplotype network (Figure 3). The COI haplotype network exhibited well-resolved and robust relationships corresponding to the phylogenetic tree, and 16S haplotype network exhibited a similar pattern to the COI haplotype network. However, the relationship between clades in the 16S haplotype network was not resolved as well as that in the COI haplotype network (Figure 3). However, no specific pattern was detected in the ITS1 haplotype network associated with the five clades. A complex network with some relatively abundant haplotypes was revealed, and many individuals belonging to different clades shared an identical haplotype. Nonetheless, most individuals within Clade D+E and Clade A+B are rather close to each other.
3.3 Divergence time
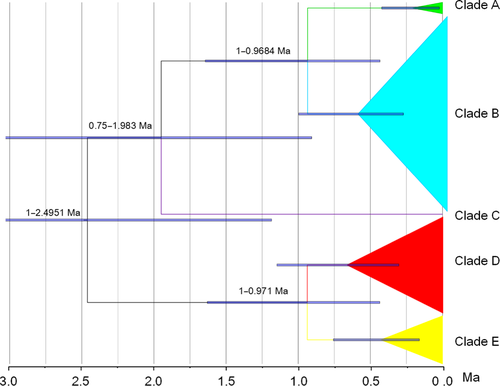
Figure 4 shows the Monodonta labio phylogeny based on mtCOI gene sequence variation with a tectonic event calibration. The topology is congruent with that previously constructed using the mitochondrial concatenated data set. The divergence dating estimates reveal relatively young split times among the main clades. The initial divergence between Clade A+B+C and Clade D+E was approximately 2.4591 mya, corresponding to the early Pleistocene. Within the first main clade (Clade A+B+C), estimated divergence age between Clade A+B and Clade C is 1.983 mya. The divergence of Clade D and Clade E dates back to 0.971 mya, while the youngest split, between Clade A and Clade B, is dated to 0.9684 mya, corresponding to the middle Pleistocene.
4 DISCUSSION
4.1 Divergence among the main mitochondrial clades and diversification mechanisms
Determining divergence times among the main clades is crucial for evaluating potential diversification mechanisms. As the molecular clock used in the analyses was calibrated for the COI of two Tegula species, which belongs to the same family (Trochidae) as Mondonta labio, our molecular clock analysis is very likely to acquire reasonable estimates. The divergence time between Clade A+B+C and Clade D+E dates back to the early Pleistocene (2.4951 mya), a time point that is consistent with the first significant build-up of Northern Hemisphere ice (about 2.57 mya; Jansen & Sjoholm, 1991; Tian, Wang, Cheng, & Li, 2002). The estimated divergence times of two subspecies of oyster (2.0–3.6 mya; Ren et al., 2016) and two mitten crab species (2.24 mya; Wang, Li, & Li, 2008) along the coast of China were similar to ours. Vicariant events induced by the sea-level changes in the early Pleistocene were put forward to elucidate the speciation of three cryptic species in Mugil cephalus (Shen et al., 2011); however, no specific barrier was discovered.
Climate changes occurred on Earth during the early Pleistocene about 2.5 mya when there was a shift from predominantly warm conditions characterized by low amplitude climatic changes to a cooler period characterized by major Northern Hemispheric glaciations and high amplitude climatic cycles. Cronin (1991) found that fluctuating environments characterized the period from 2.5 to 2.3 mya in the Eastern Asia region; global ice volume changes were about 1/2 to 2/3 the scale of the 120 m change that occurred in the late Pleistocene deglaciation and sea level fell during glacial periods by as much as 50–60 m. Under these circumstances, the Dongshan land bridge emerged and acted as a physical barrier for the populations between the ancient SCS and ancient ECS. According to their present-day distributions, members of Clade A+B+C were possibly restricted to the ancient SCS and Clade D+E may have been distributed in the enclosed ancient ECS during that epoch. Notably, the China Sea experienced several large-scale glaciations, especially the Last Glacial Maximum. The repeated range contraction and expansion might have led to a breakdown of the distribution boundary of Clade A+B+C and Clade D+E. Moreover, the record of planktonic foraminifers revealed a climate change from warm to cool during the latest Pliocene to early Pleistocene in the subtropical Western Pacific (Wang, 1994). Evidence indicated that increasing seasonality and steepening of the sea surface temperature (SST) gradient that were most significant between 2.4 and 2.2 mya occurred in the ancient ECS, were located in the subtropics, whereas the ancient SCS located in the tropics showed minor or no cooling. Nonetheless, study of an isopod in North America and of a marine clam in Australia revealed that SST gradients can also be primary drivers of cladogenesis (Eberl, Mateos, Grosberg, Santamaria, & Hurtado, 2013; Li, Foighil, & Park, 2013). The SST gradient between the ECS and the SCS possibly also exerted an effect on the divergence of the clades studied here. Therefore, isolation combined with the SST gradient were probably correlated with the divergence between Clades A+B+C and D+E.
The split between Clade A and Clade B is dated to 0.9684 mya and that between Clade D and Clade E dates back to 0.971 mya. Both of them date to during marine isotope stage (MIS) 22 of the Mid-Pleistocene Climate Transition (MPT). The MPT is the fundamental changes in climate between 1,250,000 and 700,000 years ago, when the earth's dominant periodicity of climate cycles shifted from 41,000 to 100,000 years with no relation to orbital forcing (Pisias & Moore, 1981). During this period, a secular expansion of subarctic water masses lowered the SST in the Pacific during both glacial and inter-glacial epochs, and reached a maximum at nearly 1.0 mya (Head & Gibbard, 2015). Winter SST declined from 24–25 to 17–18°C even in the northern ancient SCS, a region that is close to the core of the West Pacific Warm Pool (Li et al., 2008). As the subtropical region is more sensitive to SST changes than the tropical region, a larger extent of SST decrease may have occurred in the ancient ECS. The average global ice volume increased during the MPT (Clark et al., 2006), and the most intense changes occurred at around MIS 24–22, the so-called 0.9-mya event (McClymont, Sosdian, Rosell-Mele, & Rosenthal, 2013). During the MPT, mean glacial sea level dropped by about 20 m, whereas a 40–50 m decrease was detected from 1 to 0.9 mya (Kitamura, 2016; Sosdian & Rosenthal, 2009). With this latter decrease, the Dongshan land bridge would have emerged again, impeding the connection between the ancient SCS and the ancient ECS, and Clade D and Clade E would probably have been isolated by the land bridge. Additionally, the uplifting of the Qiongzhou Strait to 40 m below the present sea level in the late early-Pleistocene would have made the deep trough in it isolated from the SCS, which thus could have been another refugium for M. labio. Individuals belonging to Clade A possibly originate from this refugium.
4.2 Discrepancy between mitochondrial and nuclear topologies
As Clade C includes only three individuals, we exclude it from our discussion. Intra-specific divergences are usually <2% (Avise, 2000). Mean K2P divergence among the members of Clades A+B and D+E is 0.9% and 1.5%, respectively. Clade A+B is 9.4% divergent from Clade D+E using K2P distance, a divergence that corresponds to species-level differentiation (10%–25%) in marine metazoans (Bucklin, Steinke, & Blanco-Bercial, 2011; Hebert, Ratnasingham, & deWaard, 2003) better than the intra-specific divergences (<2%). However, the level of divergence between Clade A+B and Clade D+E is close to the threshold of the 10× inter-specific to intra-specific rule used in DNA barcoding (Hebert, Cywinska, Ball, & DeWaard, 2003). We hypothesize that Clade A+B and Clade D+E may correspond to the two Japanese subspecies (Monodonta labio labio and Monodonta labio confusa). They probably represent two subspecies and their speciation is possibly still in progress. Although they are monophyletic to each other, the systematics remained unresolved solely revealed by the data for COI marker. Moreover, occurrence of divergent mitochondrial lineages alone would not prove the existence of distinct evolutionary lineages (Makino & Tanabe, 2009). As species boundaries are obscured by hybridization or introgression, supplemental analyses of one or more nuclear genes will be needed to discover the relationship between Clade A+B and Clade D+E (Hebert, Cywinska et al., 2003).
Our nrITS topology was not consistent with the mitochondrial genetic split. A similar phenomenon has been recorded for Atrina pectinata during the corresponding period (5–1.5 mya; Liu, Li, Kong, & Zheng, 2011). The conflict between the mtDNA and nuclear genes seen here may be the result of either incomplete lineage sorting or post-glacial introgression due to secondary contact (Toews & Brelsford, 2012). In fact, both situations can lead to similar patterns of hybridization between divergent clades, and they are difficult to distinguish. Two cases exist that allow the two processes to be discriminated. First, there has been enough time to accumulate genetic differences before hybridization in the case of introgression. However, incomplete lineage sorting requires that the speciation event post-dates the corresponding split in the gene tree (McGuire et al., 2007). It is possible that the ITS genes would still remain homogeneous while mtCOI achieved coalescence, as the mean time for nuclear genes to coalesce is four times longer than that for mitochondrial genes (Avise, 2000). Thus, one should not predict nuclear loci to recover lineage splits if the mitochondrial gene tree shows reciprocal monophyly (Zink & Barrowclough, 2008). This means that mtDNA can be used to detect recent splits for which nuclear loci will not have had enough time to diverge. In the present study, the divergence between Clade A+B+C and Clade D + E was placed in the early Pleistocene, a relatively young split. This scenario seem to be in accordance with the case of the incomplete lineage sorting. However, the post-glacial disruption of isolation could have resulted in a second contact, making it difficult to rule out the possibility of introgression. Second, incomplete lineage sorting is not expected to promote the geographic proximity of inter-specifically shared haplotypes that may be seen under local introgression (Hare & Avise, 1998). In the present examples, samples from different clades, which share identical haplotypes for the nrITS marker, are not concentrated near the Dongshan land bridge, but are rather randomly distributed throughout the ranges of the sampling localities. Therefore, we prefer incomplete lineage sorting as an explanation for the discrepancy between the patterns seen here in the mitochondrial and nuclear genes.
ITS regions tend to evolve by insertion and deletion rather than substitution and often lack variation among closely related species, making phylogenetic reconstruction problematic. Given the possibility of incomplete concerted evolution in the ITS region (Dover, 1982), the results derived from the ITS1 marker here should be interpreted with caution. As the boundaries of our study taxa are obscured by hybridization or introgression, supplemental analyses of other nuclear genes with higher variability, such as microsatellites, will be required to determine whether they are distinct species.
5 CONCLUSIONS
This work covered the large range of occurrence of Monodonta labio in the Northwestern Pacific. Five well-supported clades were detected in the mitochondrial phylogenetic trees. However, the systematic status of Clade A+B+C and Clade D+E remains unresolved owing to the lack of verification of nuclear markers. The present-day genetic divergence revealed by the mitochondrial marker originated from two main vicariance and climate events, the initiation (c. 2.5 mya) and intensification (c. 0.9 mya; MIS 22) of large-scale Northern Hemisphere ice sheets. The enclosed ancient ECS and the semi-closed ancient SCS separated by the emergence of the Dongshan land bridge acted as potential refugia in the late Pliocene (c. 2.5 mya). The emergence of the Qiongzhou Strait and the Dongshan land bridge divided the China Sea into another three refugia during the MPT (c. 0.9 mya; MIS 22). The potential refugia combined with the SST gradient were possibly correlated with the observed genetic structure.
Our study of M. labio stresses the importance of potential refugial areas and a SST gradient in shaping the genetic structure of this species. We also put forward the possibility that these factors were probably important drivers for the diversification of sister species or subspecies of other taxa in the Northwestern Pacific.
ACKNOWLEDGEMENTS
We are grateful to Joong-Ki Park for supplying specimens. This study was supported by research grants from the National Natural Science Foundation of China (41276138), Doctoral Program of Ministry of Education of China (20130132110009) and Fundamental Research Funds for the Central Universities.




