Dietary analysis of harbour seals (Phoca vitulina) from faecal samples and overlap with fisheries in Erimo, Japan
Abstract
The number of harbour seals in Japan has been rebounding since protection began in the mid-1980s. With the increase in seal numbers, increased conflict with fisheries has occurred through depredation and the belief that seals compete with fisheries for prey. However, competition can only be determined if the prey species and quantities seals consume over time are known. We studied the diet of harbour seals in Erimo, site of the largest population of harbour seals in Japan, from 2011 to 2012 and assessed the degree of prey overlap with local fisheries. We used both hard parts and DNA techniques to identify prey items in seal scats, and compared these results to local fisheries data. A total of 46 prey occurrences was detected by both methods, of which 17 matched between techniques at least to the family level. Hard parts methods identified five incidences of prey undetected by DNA methods in five scats (one incidence per scat). DNA methods identified 24 additional prey occurrences in 13 scats, for which no hard part evidence for that prey had been found. This more than doubled the total number of prey occurrences across the 15 scats compared. Overall, the most frequently occurring harbour seal prey were walleye pollock (Theragra chalcogramma), sculpins and snailfishes. In contrast, the top three groups targeted by fisheries were codfishes, salmon and invertebrates. Many species common in the harbour seal's diet such as snailfishes and blennies were not targeted by fisheries. Fishes such as greenlings, sculpins, rockfishes and Japanese anchovy (Engraulis japonicus) were common in the diet of harbour seals, but made up a very small proportion of fisheries catches in Erimo. The importance of other prey species varied between seasons. Sculpins, greenlings and forage fishes were the top three prey groups for harbour seals in spring by percent modified frequency of occurrence, whereas the most important target groups by percentage mass caught by fisheries were codfishes, cephalopods and other invertebrates. Sculpins and rockfishes remained as important prey items for harbour seals in summer and autumn, in addition to codfishes, snailfishes and blennies. In contrast, the main groups caught by fisheries in summer were other fishes, particularly Japanese amberjack (Seriola quinqueradiata) and various species of sharks and eels, and invertebrates. By autumn, Erimo fisheries had focussed on catching salmon. Salmon were taken by harbour seals as well during this period, but at a relatively lower frequency compared to other prey groups. The results of our study show that although harbour seals consumed several of the prey species targeted by fisheries, the relative importance of these species to seals and fisheries and the seasons in which they were targeted were different.
1 INTRODUCTION
In many parts of the world, pinniped populations have increased after protection from over-exploitation for fur and meat (Jeffries, Huber, Calambokidis, & Laake, 2003; Olesiuk, Bigg, & Ellis, 1990; Olesiuk, Bigg, Ellis, Crockford, & Wigen, 1990). With this increase comes an increase in the potential conflict between fisheries and pinnipeds. Competition between seals and man for valuable fish resources is a long-standing contentious issue and of concern with fish stocks in global decline. Any mention of competition between marine mammals and fisheries tends to arouse controversy because of the complex mix of biological, economic, social, political and moral factors involved (Olesiuk, Bigg, & Ellis, 1990; Olesiuk, Bigg, Ellis, Crockford, & Wigen, 1990). Some fishermen believe the presence of marine mammals on their fishing grounds will result in a reduction in potential catch, whereas others see the presence or expansion of fisheries as a threat to the recovery of marine mammal populations from previous heavy exploitation (Harwood & Croxall, 1988). Understanding the mechanisms and the extent to which fisheries are competing with marine mammals would facilitate management decisions regarding conservation of marine ecosystems and protection of endangered marine mammal populations. Consequently, determining the foraging areas and diet of marine apex predators such as marine mammals has attracted much attention for many years (Croxall & Wood, 2002; Guinet et al., 2001; Klomp & Schultz, 2000; Riet-Sapriza et al., 2012; Ryan, Petersen, Peters, & Gremillet, 2004).
The harbour seal is a generalist and opportunistic predator, feeding on many species of fish, crustaceans and cephalopods, including many targeted by fisheries (Bowen & Harrison, 1996; Härkönen, 1987; Payne & Selzer, 1989; Tollit et al., 1998; Tollit, Greenstreet, & Thompson, 1997; Tollit et al., 1997). Their propensity for cod, herring, salmon and other commercially important species has resulted in long-standing conflicts with fishermen in many areas, who believe they are a threat to fish stocks and fisheries. This had led to calls for population control. However, harbour seals tend to feed on locally abundant and easily available prey species, with diets varying by season and region (Iverson, Frost, & Lowry, 1997; Olesiuk, 1993; Tollit et al., 1998). Predation pressure on local fish stocks by harbour seals is therefore likely to be diffuse and reflect the abundances of available prey species, rather than a depletion of local fish stocks. They also tend to prey on smaller fish than those typically taken by fisheries (Trites & Pauly, 1997). Moreover, in any study of potential competition for resources between humans and wildlife, we first need to determine what prey species, how much, what size ranges, where these prey species are taken from and when by both humans and the wildlife species in question. Such information is crucial for minimizing negative direct and indirect interactions between marine apex predators and fisheries. This is becoming of mounting importance as the worldwide exploitation of fisheries continues to increase while at the same time numerous marine apex predator species are in decline or have yet to recover from past exploitation (Costa, Weise, & Arnould, 2006). This information is currently very limited for harbour seals in Japan, owing in part to the remote and insular areas they inhabit, making data collection difficult.
In Japan, the distribution of the harbour seal is restricted to the coast of Eastern Hokkaido. The harbour seal is the only pinniped that lives and breeds in Japan throughout the year (Itoo & Shukunobe, 1986). There are nine rookeries in total. From north to south, they are Yururi and Moyururi islands in Nemuro, two sites in Hamanaka, Daikoku island, three sites in Akkeshi, and Cape Erimo (Figure 1). Of these, Cape Erimo has the largest population of harbour seals in Japan, and also the southernmost in the Western Pacific, with numbers there increasing from about 150 seals since they were protected in the mid-1980s (Fujii, Suzuki, Era, Kobayashi, & Ohtaishi, 2006) to ~700 at present (Kobayashi et al., 2014).The harbour seal was an endangered species in Japan (Nakaoka, 2004). Its status has since been downgraded to “near threatened” in 2015 (Japanese Ministry of the Environment Agency 2015). Since protection began in the mid-1980s, the total number of harbour seals in Japan has been rebounding and is currently estimated at over 1,000 individuals (Kobayashi et al., 2007, 2014). With the increase in seal numbers, increased conflict with fisheries (especially salmon fixed net fisheries) has occurred through depredation and the belief that seals compete with fisheries for prey (Nakaoka, 2004). There is considerable debate over the impact of predation by pinnipeds on salmon populations, many of which are in decline in Japan. The protected status of harbour seals in Japan results in potentially conflicting requirements for government agencies to conserve and manage both pinniped predators and their salmonid prey. Managers must therefore find a compromise between the competing requirements for the conservation and management of seal and salmon populations and their associated economic activities, primarily salmon fisheries and ecotourism.

Accurate information about the diet of pinnipeds is challenging to obtain, yet vital for assessing the impacts of pinnipeds on prey populations and pinniped interactions with fisheries. Scats are abundant, easy to collect, minimally invasive, non-destructive and have proven to be a reliable method for assessing harbour seal diets (Browne et al., 2002; Harwood & Croxall, 1988; Pitcher, 1980; Thomas, Lance, Jeffries, Miner, & Acevedo-Gutierrez, 2011; Tollit, Wong, Winship, Rosen, & Trites, 2003). It is becoming more common to collect undigested hard parts of prey, primarily fish otoliths and cephalopod beaks, from faeces, and their distinctive morphology used to identify taxa of prey consumed by pinnipeds (e.g. Call & Ream, 2012; Thomas et al., 2011; Thompson et al., 1996). The frequencies at which particular prey species occur amongst collections of scats are easily compiled to describe the average diet and can be used to compare diets between and within geographical regions and across years and seasons (Trites & Joy, 2005).
However, there are inherent biases associated with using hard parts from scats to assess the diets of pinnipeds (Bigg & Fawcett, 1985; Bowen, 2000; Pierce & Boyle, 1991), including differential digestion amongst prey species (Harvey, 1989; Tollit et al., 1997; Tollit et al., 1997), varying passage rates owing to differences in prey size (Gales, Pemberton, Lu, & Clarke, 1993), selective consumption of body parts (Hauser, Allen, Rich, & Quinn, 2008) and bias towards nearshore prey (Call & Ream, 2012). As diet assessments based on hard parts analysis alone may be biased, we investigated diet composition using DNA analysis as well. By targeting prey DNA segments that have sequences that are highly variable and in principle species-specific, organisms can be identified by their “DNA bar code” (Moritz & Cicero, 2004). DNA techniques have the potential to identify prey species that leave few or no hard parts owing to digestibility, but as development for these techniques is fairly recent, they have yet to become a mainstream method for diet analysis. Diet studies can be significantly enhanced through incorporation of DNA technologies (Höss, Kohn, Pääbo, Knauer, & Schröder, 1992; King, Read, Traugott, & Symondson, 2008), with obvious benefits to marine wildlife and fisheries managers. DNA analysis has been used to determine the presence and absence of prey in pinniped diets (Deagle & Tollit, 2007; Deagle, Jarman, Pemberton, & Gales, 2005; Deagle, Tollit et al., 2005; King et al., 2008). Recent advances in molecular technologies have already proven useful in a number of marine mammal dietary studies (Casper, Jarman, Gales, & Hindell, 2007; Ford & Ellis, 2006; Jarman, Gales, Tierney, Gill, & Elliott, 2002; Purcell, Mackey, Lahood, Huber, & Park, 2004; Reed, Tollit, Thompson, & Amos, 1997), notably by increasing taxon-level detection rates and improving species resolution. Our study applied PCR and DNA sequencing methodology to describe the diet of the harbour seal in conjunction with diet estimated using conventional morphological identification of diagnostic prey skeletal remains and other hard parts recovered in scat samples.
The magnitude of competition between predators and fisheries will depend partly on the degree of dietary overlap and what prey species are preferred by the competitors. Understanding the extent of the potential resource competition between harbour seals and fisheries is necessary for effective harbour seal conservation and fisheries management. We therefore studied the diet of harbour seals in Erimo and integrated these data with information on fisheries catch to quantify the resource overlap between harbour seals and fisheries. Our specific objectives for this study were to determine (i) the diet and relative quantity of prey consumed by harbour seals using hard parts and DNA analyses of harbour seal scats, and (ii) overlap in prey species between harbour seals and fisheries.
2 MATERIAL AND METHODS
2.1 Study location
Erimo (41°55′28′′N, 143°14′42′′E) is located in Southeastern Hokkaido (Figure 1). It is known for its harbour seal population (~700 seals), the largest and southernmost in Japan. This important rookery and haulout site consists of several rocky outcrops in the sea that stretch for about 2 km from the coast. The pupping season in Erimo is from May to June and pups are weaned at about 1 month old. Erimo's main industries are fishing, kelp harvesting and tourism. Salmon fixed net fishing is carried out from the end of May to mid-November every year.
2.2 Scat collection and hard parts analysis
Harbour seal scats were collected for 1 week/month from July to October 2011 and April 2012 in order to understand seasonal changes in the harbour seal's diet. The target sample size was at least 60 scats per season (spring: March–May, summer: June–August, autumn: September–November) for sufficient statistical power (Trites & Joy, 2005). Scats were collected in plastic zip lock bags and frozen until processing. Subsamples of scat for DNA analysis were obtained by gently pressing ~2 ml homogenized scat slurry through a 0.5 mm sieve using a spatula, scraping the sieved scat slurry from the underside and refreezing it in glass vials. The rest of the scat material was then washed through 0.5 mm sieves and prey were identified to the lowest possible taxon from hard parts (e.g. otoliths, dentaries, cephalopod beaks, crustacean shells) retrieved using a dissecting microscope, reference bones and beaks from the Hokkaido National Fisheries Research Institute, bones extracted from fish caught at the study site and published fish bone, otolith and cephalopod beak keys (Clarke, 1986; Panfili, Pontual, Troadec, & Wright, 2002; Wolff, 1982).
Only scats containing identifiable prey remains were used to calculate the frequency of occurrence (FO). Each scat was treated as an independent sample and the importance of each prey group or species was based on the relative frequency it occurred in the scats. To account for differences in sample sizes between seasons, we calculated the percent FO of prey by dividing the number of scats in which a prey species occurred by the total number of scats collected in the respective seasons (Call & Ream, 2012; Wright, 2010).The number of prey groups or species per scat was used as a measure of prey diversity or richness. To estimate the number of prey items in each scat, we attempted to match right and left otoliths, and upper and lower cephalopod beaks. When the right and left otoliths, or upper and lower beaks could not be distinguished, we estimated the minimum number of prey items by dividing the total number of otoliths, beaks or eye lenses by 2. Unmatched otoliths, beaks and lenses were counted as one prey item.
2.3 DNA diet analysis
We also used DNA techniques to identify fish prey species in harbour seal scats, following an adaptation of the method developed by Tollit, Schulze, and Trites, (2009), Deagle, Jarman, Pemberton, and Gales, (2005) and Deagle, Tollitet al. (2005) for Steller sea lion scats. We randomly selected five scats with identifiable hard parts from each season (15 total) and analysed their subsamples for fish prey DNA. Total genomic DNA was extracted from 180 to 220 mg of each subsample using a QIAmp DNA stool mini kit (Qiagen), following the “Isolation of DNA from stool for human DNA analysis” protocol. The amount and quality of DNA present in each extraction were determined using a Nanodrop (ND-1000) spectrophotometer. The universal primers 16SF1 and 16SallR (Tollit et al., 2009; Table 1) were used in a primary PCR to amplify the extracted prey DNA from scats. A fish-specific semi-nested set of secondary primers, 16SfishF (8 bp internal to 16SF1 and less conserved region of the 16S gene) and 16SallR (Tollit et al., 2009, Table 1), was then used to further amplify the minute quantities of prey DNA while eliminating the amplification of DNA from the host harbour seal and other non-fish species (Jarman, Deagle, & Gales, 2004) under the conditions listed in Table 2. The PCR mixture contained 10 μl PCR Master Mix (Promega) containing Taq DNA polymerase, dNTPs (deoxynucleotide triphosphates), MgCl2 and reaction buffers, 1 μl of each primer, 2 μl template DNA and diluted to a total reaction mixture volume of 20 μl with 6 μl sterile distilled water. Semi-nested PCRs were performed using 2 μl of the primary PCR reaction mixture as template. The primary and semi-nested PCR products were electrophoresed through agarose gels to confirm amplification. The PCR reactions were purified using a GENECLEAN turbo kit. Amplicons from the semi-nested PCRs were cloned using the pGEM-T Easy Vector System (Promega). Colony PCR was performed followed by purification using the Wizard Plus SV Minipreps DNA Purification System (Promega).
| Primers | Sequence (5′–3′) |
|---|---|
| General fish primers mt 16S | |
| 16SF1 | GGACGAGAAGACCCT |
| 16SallR | CGCTGTTATCCCTAGGGTAACT |
| 16SfishF | AGACCCTATGGAGCTTTAGAC |
- These primers were previously developed by Tollit et al. (2009).
| Primer combinations | Amplicon size (bp) | PCR conditions |
|---|---|---|
| General and fish-specific mt 16S gene | ||
| Primary | ||
| 16SF1 × 16SallR | 290–308 | 95°C for 10 min |
| 95°C for 30 s, 56°C for 30 s, 72°C for 1 min (35 cycles) | ||
| 72°C for 7 min | ||
| emi-nested | ||
| 16SfishF × 16SallR | 282–300 | 95°C for 10 min |
| 95°C for 30 s, 58°C for 30 s, 72°C for 1 min (35 cycles) | ||
| 72°C for 7 min | ||
Standard cycle sequencing reactions were conducted with a BigDye Terminator Cycle Sequencing FS Ready Reaction Kit (Applied Biosystems) and nucleotide sequences from prey DNA were determined using an automated DNA sequencer (ABI3700, Applied Biosystems). The obtained DNA sequences were compared to prey sequences at GenBank using BLAST ( http://blast.ncbi.nlm.nih.gov/Blast.cgi). A consensus sequence was assigned to a taxon when the sequence exclusively matched (<1% mismatch) members of that taxon in the database to the exclusion of all other taxa.
The contribution of each prey species to the harbour seal's diet was determined by tallying the number of occurrences of each prey species in all scats. This was counted separately for prey identified using hard parts analysis and DNA techniques. We compared harbour seal diet composition determined using morphological hard parts identification and prey DNA identified within the soft part matrix of the same scats to determine how often species occurrences matched and to what extent the inclusion of prey DNA data increased (i) species richness in scats (i.e. additional prey species incidences for which hard part identification had found no evidence) and (ii) species resolution (i.e. improved prey species identification, typically in cases in which hard parts were identified with certainty down to the family level and DNA identification methods subsequently resolved identification to the species level).
2.4 Overlap between harbour seals and fisheries
Data on the locations of fish nets, the species caught, their respective weights and when each species is caught were obtained from 2011 to 2012 from the Erimo fisheries cooperative and Marinenet Hokkaido ( www.fishexp.hro.or.jp). The species and relative quantities caught by fisheries were compared to those consumed by harbour seals during the same period. The percent modified FO (frequency of occurrence down-weighted so that the sum of all prey frequencies total 100% (Bigg & Perez, 1985)) of harbour seal prey was used in this calculation. Percent modified FO is calculated by dividing each prey species’ percent FO by the sum of all the species’ percent FOs. The frequencies of occurrence of each prey species in the 15 scats analysed using hard parts analysis and DNA techniques were combined for this calculation by adding any new prey occurrences identified using DNA techniques to the scats involved. These measured variables indicate which prey species were most likely to be competed for (if at all), the types of fisheries that are most likely to be affected by harbour seal depredation, which foraging areas and when they are important to harbour seals.
3 RESULTS
3.1 Hard parts analysis
We collected a total of 56 scats from Erimo, of which 43 scats had identifiable prey remains (spring: n = 7, summer: n = 14, autumn: n = 22; Table 3). The average minimum number of prey species per scat was 1.40 ± 0.12 and the average minimum number of prey items was 2.88 ± 0.50. The maximum number of prey species identified in a scat was four and the maximum number of prey items in a scat was 13. Overall harbour seal diet included at least 11 species from 10 families. Hard parts analysis also identified crabs as prey items in the harbour seal's diet, a first in Japan. Overall, the most frequently occurring prey species and groups were walleye pollock (Theragra chalcogramma), sculpins and snailfishes. The importance of other prey species varied amongst seasons (Table 3). Blennies were common prey items in spring, but not in other seasons. Snailfishes and flounders were important in summer and autumn, but not in spring (Table 3). Greenlings were detected only in autumn. Japanese anchovy (Engraulis japonicus) and North Pacific giant octopus (Enteroctopus dofleini) were identified in scats collected in spring and summer, but not in autumn. Squids were detected in summer and autumn, but not in spring. The minimum number of species recorded from scats collected in summer and autumn (eight and seven species, respectively) was higher than in spring (five species; Table 3).
| Phylum | Family | Group or species | Common name | Spring (n = 7) | Summer (n = 14) | Autumn (n = 22) | Total (n = 43) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | FO (%) | No. | FO (%) | No. | FO (%) | No. | FO (%) | ||||
| Chordata | Gadidae | Theragra chalcogramma | Walleye pollock | 1 | 14.3 | 3 | 21.4 | 5 | 22.7 | 9 | 21.4 |
| Gadus macrocephalus | Pacific cod | 0 | 0 | 0 | 0 | 1 | 4.5 | 1 | 2.4 | ||
| Unidentified gadids | Codfishes | 0 | 0 | 1 | 7.1 | 0 | 0 | 1 | 2.4 | ||
| Hexagrammidae | Unidentified hexagrammids | Greenlings | 0 | 0 | 0 | 0 | 1 | 4.5 | 1 | 2.4 | |
| Cottidae | Hemilepidotus jordani | Yellow Irish lord | 1 | 14.3 | 1 | 7.1 | 4 | 18.2 | 6 | 14.3 | |
| Unidentified cottids | Sculpins | 2 | 28.6 | 3 | 21.4 | 3 | 13.6 | 8 | 19.0 | ||
| Pleuronectidae | Glyptocephalus stelleri | Blackfin flounder | 0 | 0 | 0 | 0 | 1 | 4.5 | 1 | 2.4 | |
| Unidentified pleuronectids | Flounders | 0 | 0 | 2 | 14.3 | 2 | 9.1 | 4 | 9.5 | ||
| Liparidae | Unidentified liparids | Snailfishes | 0 | 0 | 3 | 21.4 | 4 | 18.2 | 7 | 16.7 | |
| Engraulidae | Engraulis japonicus | Japanese anchovy | 1 | 14.3 | 1 | 7.1 | 0 | 0 | 2 | 4.8 | |
| Blenniidae | Unidentified blenniids | Blennies | 1 | 14.3 | 0 | 0 | 0 | 0 | 1 | 2.4 | |
| Mollusca | Octopodidae | Enteroctopus dofleini | North Pacific giant octopus | 1 | 14.3 | 1 | 7.1 | 1 | 4.5 | 3 | 7.0 |
| Gonatidae | Unidentified gonatids | Armhook squids | 0 | 0 | 1 | 7.1 | 1 | 4.5 | 2 | 4.8 | |
| Arthropoda | Unidentified Brachyura | Crabs | 0 | 0 | 1 | 7.1 | 0 | 0 | 1 | 2.4 | |
3.2 Comparison of hard parts and DNA techniques
Fish prey DNA remains were isolated from all 15 scats analysed. All obtained DNA sequences displayed ≥99% forward and reverse primer matches with the gene sequences in GenBank. The DNA technique was able to detect more species and families than the hard parts method for the 15 scats analysed. A total of at least 17 fish prey species from 11 families was identified using hard parts and/or DNA techniques (Table 4). The DNA of 16 species of fish prey from 10 families, of which nine species were detected only by DNA methods, were identified from the 15 scats (Table 4). The average number of prey species detected by DNA methods per scat was 2.67 ± 0.40. The average number of prey species per scat detected by hard parts methods for these same 15 scats was 1.40 ± 0.19. The number of prey species detected by DNA methods per scat ranged from one to six, while for hard parts, this ranged from one to three. A total of 46 prey occurrences was detected by both methods, of which 17 matched between techniques at least to the family level (Table 4). Hard parts methods identified five incidences of prey undetected by DNA methods in five scats. DNA methods identified 24 additional prey occurrences in 13 scats, for which no hard part evidence for that prey had been found. This more than doubled the total number of prey occurrences across the 15 scats compared. In addition, DNA techniques identified three prey species previously unreported in harbour seal studies conducted in Japan: great sculpin (Myoxocephalus polyacanthocephalus), ribbed sculpin (Triglops pingelii) and halfbarred pout (Gymnelus hemifasciatus).
| Phylum | Family | Group or species | Common name | Hard parts only | DNA only | Hard parts and DNA |
|---|---|---|---|---|---|---|
| Chordata | Gadidae | Theragra chalcogramma | Walleye pollock | 2 | 2 [1] | |
| Gadus macrocephalus | Pacific cod | 2 [1] | ||||
| Hexagrammidae | Pleurogrammus azonus | Arabesque greenling | 1 | |||
| Hexagrammos lagocephalus | Rock greenling | 1 | 1 [1] | |||
| Hexagrammos stelleri | Whitespotted greenling | 3 | ||||
| Cottidae | Hemilepidotus jordani | Yellow Irish lord | 1 | 4 | ||
| Myoxocephalus polyacanthocephalus | Great sculpin | 1 | 6 [6] | |||
| Triglops pingelii | Ribbed sculpin | 1 | ||||
| Unidentified cottids | Sculpins | 1 | ||||
| Sebastidae | Sebastes taczanowskii | White-edged rockfish | 1 | |||
| Pleuronectidae | Unidentified pleuronectids | Flounders | 1 | |||
| Liparidae | Liparis agassizii | Agassiz's snailfish | 3 | |||
| Unidentified liparids | Snailfishes | 1 | ||||
| Zoarcidae | Gymnelus hemifasciatus | Halfbarred pout | 1 | |||
| Engraulidae | Engraulis japonicus | Japanese anchovy | 3 | 1 | ||
| Trichodontidae | Arctoscopus japonicus | Japanese sandfish | 1 | |||
| Blenniidae | Chirolophis japonicus | Fringed blenny | 1 | 1 [1] | ||
| Salmonidae | Oncorhynchus masou | Cherry salmon | 3 | |||
| Oncorhynchus keta | Chum salmon | 3 |
- Square brackets indicate the number of prey occurrences in which identification was improved from the family to species level by DNA techniques.
Species from the families Sebastidae, Zoarcidae, Trichodontidae and Salmonidae were detected by DNA techniques in the 15 scats analysed, but not by the hard parts method (Table 4). A total of six occurrences of salmon, three of chum and three of cherry salmon, in five scats was detected. The number of prey occurrences in other families identified by the hard parts method also increased with the use of DNA identification techniques (Table 4). In 10 of the 17 prey occurrences shared between hard parts and DNA methods, DNA techniques were able to refine the level of identification from family to species (Table 4). These were mostly of great sculpin (Cottidae). However, DNA techniques failed to detect any species of Pleuronectidae (Table 4). Other families that the DNA method missed in certain scats included members of the families Gadidae, Cottidae and Liparidae (Table 4).
3.3 Overlap between harbour seals and fisheries
Erimo's main fisheries by percentage of the total mass caught in summer and autumn 2011, and spring 2012 were the salmon and invertebrate fisheries (Figure 2). Salmon, particularly the chum salmon (Oncorhynchus keta), was caught almost exclusively in autumn (Figure 2). Invertebrates such as sea cucumbers, urchins, clams and whelks were caught mainly in spring and summer (Figure 2). Codfishes (Gadidae), consisting of walleye pollock and Pacific cod (Gadus macrocephalus), were also important target species and were mostly caught in spring (Figure 2).
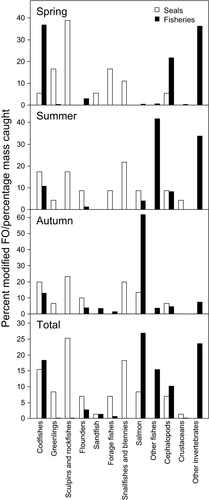
Comparing harbour seal diet with fisheries catches, several species consumed by seals were also caught by fisheries. Codfishes and cephalopods such as the North Pacific giant octopus were all significant harbour seal prey items and also frequently caught by fisheries in all seasons (Figure 2). However, many species common in the harbour seal's diet were not targeted by fisheries. Snailfishes and blennies were not caught at all by fisheries. Fishes such as greenlings, sculpins, rockfishes and forage fishes such as Japanese anchovy (E. japonicus) were common in the diet of harbour seals, but made up a very small proportion of fisheries catches in Erimo (Figure 2).
There were seasonal differences in the importance of harbour seal prey items and fisheries target groups as well. Sculpins, greenlings and forage fishes were the top three prey groups for harbour seals in spring by percent modified FO, whereas the most important target groups by percentage mass caught by fisheries were codfishes, cephalopods and other invertebrates (Figure 2). Sculpins and rockfishes remained as important prey items for harbour seals in summer and autumn, in addition to codfishes, snailfishes and blennies (Figure 2). In contrast, the main groups caught by fisheries in summer were other fishes, particularly Japanese amberjack (Seriola quinqueradiata) and various species of sharks and eels, and invertebrates (Figure 2). By autumn, Erimo fisheries had focussed on catching salmon. Salmon were taken by harbour seals as well during this period, but at a relatively lower frequency compared to other prey groups (Figure 2).
4 DISCUSSION
4.1 Variation in harbour seal diets
We found little evidence to support Erimo fishermen's concern that their target species form a large proportion of the harbour seal's diet and seals are therefore a cause of recent declines in fish catches. The results of our study show that although harbour seals consumed several of the prey species targeted by fisheries, the relative importance of these species to seals and fisheries and the seasons in which they were targeted were different. The main prey groups consumed by harbour seals in our study consisted of many benthic species such as codfishes, greenlings, sculpins, rockfishes, flounders, snailfishes, blennies and cephalopods. In total, we identified at least 20 different species using hard parts and/or DNA techniques from all the collected scats. Fujii et al. (2006) found that harbour seals from Nemuro and Erimo fed on sculpins, blennies and cephalopods that inhabit nearshore waters. Other studies in Erimo found that salmon, North Pacific giant octopus and codfishes formed the major part of the harbour seal's diet in autumn (Nakagawa, 2005; Nakaoka, Hamanaka, Wada, & Tanahashi, 1986). In addition to these species, we found that snailfishes were also important prey in all seasons studied. The sedentary nature of these fishes may make them easy prey for harbour seals, especially pups. Likewise, this may be why we found crab remains in one of the scats, the first record for harbour seals in Japan. Harbour seal pups occasionally feed on shrimp and other crustaceans (Boulva & McLaren, 1979; Bowen & Harrison, 1996; Muelbert & Bowen, 1993; Pitcher, 1980), possibly because of their relative ease of capture. Our findings thus confirm the prevalence of previously identified prey species in harbour seal diets, but other species also contributed significantly.
Amongst pinnipeds, it is common for more than 35 different species of prey to be identified (Olesiuk, Bigg, & Ellis, 1990; Olesiuk, Bigg, Ellis, Crockford, & Wigen, 1990; Sinclair & Zeppelin, 2002; Tollit & Thompson, 1996; Tollit et al., 1997; Tollit et al., 1997). The diet richness recorded here was probably influenced by the high diversity of species around Erimo and the opportunistic foraging habits of harbour seals. All of the top prey taxa described in this study, as well as many others that were consumed less frequently, commonly occur in the waters around Erimo. Geographical, seasonal and inter-annual variations in the diets of harbour seals and other pinnipeds are often attributed to changes in the abundance and distribution of available prey (Härkönen, 1987; Iverson et al., 1997; Olesiuk, 1993; Olesiuk, Bigg, & Ellis, 1990; Olesiuk, Bigg, Ellis, Crockford, & Wigen, 1990; Payne & Selzer, 1989). Species that are dominant in the diets of harbour seals tend to be abundant in the regions in which they are found (Scott & Scott, 1988).
Variation in harbour seal diets can be related to differences in habitat. Other studies have found that on sandy bottoms, sand lance dominated the diet, whereas on rocky bottoms, surrounded by deeper, colder water, rockfish, flounders, salmon, herring and codfishes were more commonly preyed on (Bromaghin et al., 2013; Härkönen, 1987; Payne & Selzer, 1989). The bottom substrate around Erimo is mostly rocky, giving rise to the abundance of benthic taxa such as sculpins, flounders and codfishes in the habitat and consequently in the harbour seal's diet.
Although salmon were taken by both harbour seals and fisheries, their importance was far greater to fisheries, especially in autumn. Salmon smolt move offshore in spring, and adults return to nearshore areas to spawn in autumn. As a result, harbour seal predation on salmon increases in the summer and autumn, when large numbers of these fish pass through the region on the way to their natal streams. Harbour seal populations around the world take advantage of this seasonal abundance in salmon and it becomes an energetically important prey source in autumn (Howard, Lance, Jeffries, & Acevedo-Gutierrez, 2013; Olesiuk, Bigg, & Ellis, 1990; Olesiuk, Bigg, Ellis, Crockford, & Wigen, 1990). At Erimo, although salmon occurred frequently in harbour seal scat samples in summer and autumn, codfishes, sculpins, snailfishes and blennies also formed significant proportions of their diet. There were also taxa such as greenlings and Japanese anchovy that were common only in the harbour seal's diet and were not targeted specifically by any fisheries. This is probably more representative of their diet during the rest of the year when salmon are absent. In marine environments, Olesiuk (1993), Orr, Banks, Mellman, Huber, and Delong, (2004) and Wright, Riemer, Brown, Ougzin, and Bucklin, (2007) calculated that salmonids typically contribute <10% of the harbour seal's diet throughout the year estimated on the basis of scats.
4.2 Comparison between DNA and hard parts diet analysis
The target species of most concern in Erimo is the chum salmon and although hard parts analysis did not reveal the presence of any salmon species in the scats analysed, five out of 15 scats analysed using DNA techniques were found to contain salmon species. This may be because salmon otoliths, although distinguishable between species and one of the best hard parts to use for prey identification, are relatively small and easily digestible (Jobling & Breiby, 1986). In addition, harbour seals may only partially ingest large prey (Boyle, 1990; Tollit et al., 1997; Tollit et al., 1997) or selectively consume body parts (Hauser et al., 2008). If only the fleshy parts are consumed (e.g. the belly: T. C. Y. Hui, personal observations 2012), there may be no hard parts to retrieve for identification. DNA methods were able to discern different species of salmon within the same scat. DNA analysis of salmon occurrence provided species-level taxonomic resolution that could not be obtained by morphological identification and showed that harbour seals were primarily consuming cherry and chum salmon.
We successfully applied group-specific nested primers and BLAST program sequence matching for recovering and analysing prey DNA from scat material collected from harbour seals. The DNA of 16 species of fish prey, of which nine species were detected only by DNA methods, were identified from 15 scats. DNA techniques increased the number of occurrences and taxonomic resolution of several families compared to hard parts identification. This resulted in increased species richness in nearly all the scats compared. DNA techniques were able to identify several fish groups with highly digestible bones such as snailfishes and forage fishes. In many cases, DNA techniques enabled us to identify prey down to the species level, compared to hard parts analysis, which only enabled us to identify prey down to the family level in many cases. This is a significant improvement over using hard parts analysis alone.
However, DNA techniques were unable to detect any members of the Pleuronectidae family whose presence was confirmed by hard parts analysis in the same scats. DNA techniques also missed occurrences of Gadidae, Cottidae and Liparidae in certain scats. This could be a result of incomplete homogenization of the scat slurry before subsampling or improper primer design. The DNA method is heavily dependent on the design of primers used in PCR reactions and only species included in the target area of the primer will be amplified (Tollit et al., 2009). This is why we used a group-specific method in which evolutionary groups, e.g. fish, are amplified using PCR while simultaneously excluding the predator's DNA from amplification. For greater species resolution and breadth, we recommend amplifying samples with different sets of primers targeting more distinct groups or individual species such as that described in Tollit et al. (2009). Another drawback of the DNA method is that neither the number of prey items nor their sizes can be determined, so it cannot provide information on the total amount of food consumed. It can determine the proportion of prey consumed if real-time PCR methods are used (Bowles, Schulte, Tollit, Deagle, & Trites, 2011). The optimal dietary analysis for pinnipeds should therefore use a combination of molecular and hard parts techniques to determine species, composition and size.
4.3 Study limitations
At the population level, diet composition is affected by the number of scats collected (Trites & Joy, 2005). Scats from harbour seals are often tidally washed away each day (Bigg, Ellis, Cottrell, & Milette, 1990) and this is likely to have occurred at Erimo where several of the rocky outcrops become inundated at high tide. Therefore it was not possible to collect the recommended minimum number of scats required (Trites & Joy, 2005). However, we pooled samples from all seasons to give a more robust estimate of the harbour seal's diet. As the seasonal sample sizes are small, our findings should be taken as an indication of the harbour seal's diet to be confirmed by subsequent years of study. Problems associated with the complete digestion of bones can be reduced by using all-structure identification techniques to determine the presence of prey species such as we did in this study (Browne et al., 2002; Cottrell & Trites, 2002; Olesiuk, Bigg, & Ellis, 1990; Olesiuk, Bigg, Ellis, Crockford, & Wigen, 1990; Tollit et al., 2003).
Percent FO is a simple, useful metric for determining common and rare prey taxa, but it offers no indication of the quantity of prey consumed. Enumeration and measurement of hard parts were not possible in this study as no length correction factors were available. We believe percent FO was appropriate for this initial description of harbour seal diet in Erimo. However, to draw any conclusions about the impact of harbour seals on prey populations, future studies should attempt to estimate biomass consumed, particularly for prey species of interest.
4.4 Conflict between harbour seals and fisheries
Overlap between harbour seal prey and fisheries target species does not imply direct competition. However, it is reasonable to infer competitive effects when key prey become scarce and remain subject to heavy fishing pressure (Trites & Pauly, 1997). It is unlikely that harbour seal predation has been a major cause of declines in salmon stocks, but predation by locally abundant pinnipeds has the potential to affect the recovery of many threatened and endangered stocks through both direct mortality and sublethal effects such as injury-related delayed mortality. Although the assessment of dietary overlap is a simple and preliminary approach to understanding complex food web dynamics, alternative and more sophisticated modelling approaches are prone to bias and heavily reliant upon appropriate expertise as well as large reliable data sets (Plaganyi & Butterworth, 2005), which are not yet available in Hokkaido.
The concurrent increase in harbour seals and decrease in fish catches has resulted in an annual cull to remove “problem” seals that enter fishing nets. Culling to reduce the overall numbers of seals has also being proposed. In Erimo, 2%–30% of salmon caught in fixed nets are damaged on average every year (Fujii et al., 2009; Erimo Fisheries Association, personal communication 2012). Although this may seem like a small proportion, the losses incurred are felt more heavily in years when fisheries catches are low, as is the case in recent times. There is resentment towards harbour seals amongst fishermen as they believe that seals target fish caught in nets and that their target species form a large proportion of the harbour seal's diet. The support for culling comes from a persistent notion that there is a mass-for-mass equivalence in the prey of marine mammals and the yields available to fishers. Many fisheries stakeholders believe that seals have major deleterious effects on salmon fisheries and that population control is therefore justified, whereas science suggests that at the salmon population level, seal predation is unlikely to have a significant effect relative to other causes of mortality (Middlemas, Armstrong, & Thompson, 2003). Furthermore, the complexity of ecosystems could well be such that the response to a marine mammal cull could be highly diffused through the food web, involving many other species (Yodzis, 2000). There are concerns over how effective this approach is at targeting those seals that eat salmon and over the negative impact this may have on local seal populations. The harbour seal has already been decimated before by hunting because of depredation and also as a post-war food source (Japanese Ministry of the Environment Agency 2002; Nakaoka, 2004). A cull could revert harbour seal populations to historical lows of the 1960s, when the total population of harbour seals in Japan was under 200 (Nakaoka, 2004).
Our study showed that harbour seals feed on a variety of locally available fish and invertebrates. This enables them to switch between prey species depending on which are the most abundant and readily available. Their diet may thus reflect the local abundance of prey, prey pulses (Thomas et al., 2011) or foraging habitat type (Tollit et al., 1998). We are therefore unable to determine whether competition between harbour seals and fisheries exists, even though several of their prey species overlap. Salmon contributed a component of the harbour seal's diet, but population reduction may not result in a direct compensatory increase in stocks and catches of salmon.
ACKNOWLEDGEMENTS
We thank K. Yanagida for assistance with scat collection, Professors A. Hara and N. Hiramatsu and their students for assistance with DNA sequencing and analysis, and for allowing us to use their laboratory. Drs K. Hattori and Y. Goto provided initial training in DNA techniques. We are also grateful to Erimo Fisheries Cooperative Associations (FCAs) staff and Erimo Seal Club for their support during fieldwork. Our research was financially supported by a Sasakawa Scientific Research Grant from The Japan Science Society, The Explorers Club Exploration Fund and a Grant-In-Aid of Research from the National Academy of Sciences, administered by Sigma Xi, The Scientific Research Society. Financial support for T.C.Y.H. came from a Japanese Ministry of Education, Culture, Sports, Science and Technology (Monbukagakusho) Graduate Student Scholarship and the Japan Asian Youth Fellowship Program.




