Combined analysis of gut microbiota, diet and PNPLA3 polymorphism in biopsy-proven non-alcoholic fatty liver disease
Abstract
Background and aims
Non-alcoholic fatty liver disease (NAFLD) is a global health burden. Risk factors for disease severity include older age, increased body mass index (BMI), diabetes, genetic variants, dietary factors and gut microbiota alterations. However, the interdependence of these factors and their individual impact on disease severity remain unknown.
Methods
In this cross-sectional study, we performed 16S gene sequencing using fecal samples, collected dietary intake, PNPLA3 gene variants and clinical and liver histology parameters in a well-described cohort of 180 NAFLD patients. Principal component analyses were used for dimensionality reduction of dietary and microbiota data. Simple and multiple stepwise ordinal regression analyses were performed.
Results
Complete data were available for 57 NAFLD patients. In the simple regression analysis, features associated with the metabolic syndrome had the highest importance regarding liver disease severity. In the multiple regression analysis, BMI was the most important factor associated with the fibrosis stage (OR per kg/m2: 1.23, 95% CI 1.10-1.37, P < .001). The PNPLA3 risk allele had the strongest association with the histological grade of steatosis (OR 5.32, 95% CI 1.56-18.11, P = .007), followed by specific dietary patterns. Low abundances of Faecalibacterium, Bacteroides and Prevotella and high abundances of Gemmiger were associated with the degree of inflammation, ballooning and stages of fibrosis, even after taking other cofactors into account.
Conclusions
BMI had the strongest association with histological fibrosis, but PNPLA3 gene variants, gut bacterial features and dietary factors were all associated with different histology features, which underscore the multifactorial pathogenesis of NAFLD.
Abbreviations
-
- AIC
-
- Akaike information criterion
-
- AST
-
- aspartate aminotransferase
-
- BMI
-
- body mass index
-
- BMR
-
- basal metabolic rate
-
- EI
-
- energy intake
-
- GGT
-
- gamma-glutamyl-transferase
-
- HbA1c
-
- glycated haemoglobin
-
- IDF
-
- International Diabetes Foundation
-
- INR
-
- international normalized ratio
-
- NAFLD
-
- non-alcoholic fatty liver disease
-
- NASH
-
- non-alcoholic steatohepatitis
-
- OTUs
-
- operational taxonomic units
-
- PNPLA3
-
- patatin like phospholipase domain containing protein 3 (adiponutrin)
Lay summary
Non-alcoholic fatty liver disease (NAFLD) is the most common chronic liver disease globally. In this study, we investigated in one cohort of patient with NAFLD, which of the individual gut microbial, dietary, genetic (PNPLA3 variants) and clinical factors have the most important association with disease severity when taking all factors into consideration. The body mass index had the strongest association with histological fibrosis, but PNPLA3 gene variants, gut bacterial features and dietary factors were all associated with different liver histology features, which underscore the multifactorial pathogenesis of NAFLD.
1 INTRODUCTION
Non-alcoholic fatty liver disease (NAFLD) has evolved as the most common chronic liver disease in the world affecting approximately 25%-30% of the population in Western countries.1 Whereas nonalcoholic fatty liver, the most common type, has a relatively benign prognosis, the risk for disease progression including the development of liver fibrosis, cirrhosis and the development of hepatocellular cancer is increased in individuals with non-alcoholic steatohepatitis (NASH) and progressive fibrosis.2 As approximately 20% of patients with NAFLD will develop progressive disease3 and no treatment options except for lifestyle modifications are available, the early identification of patients at risk is important.
NAFLD represents a multifactorial disease, and several risk factors for disease progression in the setting of NAFLD have been identified. These include older age, increased body weight, the presence of type 2 diabetes, genetic variants such as the common PNPLA3 (encoding patatin like phospholipase domain-containing protein 3) p.I148 M polymorphism and dietary factors.4 In addition, changes in the gut bacterial microbiota composition with an overgrowth of potentially detrimental bacterial species and a deficit of potentially beneficial gut bacteria, as well as a reduced bacterial diversity, have been identified as additional cofactors that might contribute to the deterioration of NAFLD.5-8 Several mechanisms could explain how specific changes in the microbiota composition might modulate NAFLD. Gut barrier dysfunction, which has been described in patients with NAFLD, can lead to the translocation of microbes or microbial metabolites to the liver, where they can induce an inflammatory response and activate profibrotic pathways. In addition, gut bacteria are responsible for the deconjugation of bile acids that are produced by the liver and for the conversion of primary into secondary bile acids. Bile acids act as signalling molecules that bind to host nuclear and G-protein-coupled receptors, which have an impact on several host metabolic functions. Synthesis of short-chain fatty acids by gut bacteria, an increased energy harvest by gut bacteria and endogenous ethanol production represent other potential mechanisms on how the gut bacterial microbiome might affect NAFLD.9
Because most studies investigated associations between these factors and disease severity individually, it is unclear whether the observed gut microbiota changes in more advanced NAFLD are independently associated with the disease or if they are rather secondary to the frequently observed metabolic comorbidities such as type 2 diabetes, obesity and dyslipidemia. Further, the importance of the gut microbiota in relation to clinical factors, diet and the PNPLA3 polymorphism remains unclear.
In this study, we analysed gut microbiota, dietary data, PNPLA3 p.I148M genotypes and various clinical parameters from a very well-characterized NAFLD cohort. We aimed to investigate, which of the individual gut microbial, dietary, genetic and clinical factors have the most important association with NAFLD severity when taking all factors into consideration. For this purpose, we performed simple and multiple stepwise ordinal regression analyses using the liver histology features steatosis, inflammation, ballooning, the NAFLD activity score and the stage of fibrosis, as outcome parameters.
2 MATERIALS AND METHODS
2.1 Patient cohort
A total of 180 NAFLD patients were prospectively enrolled in this cross-sectional observational study between March 2015 and December 2018 in the outpatient liver department of the Clinic for Gastroenterology and Hepatology, University Hospital of Cologne, Germany (Figure 1A). The protocol was approved by the Ethics Commission (reference # 15-056) of Cologne University's Faculty of Medicine, and written informed consent was obtained from each patient. The study was performed in accordance with the Declaration of Helsinki.
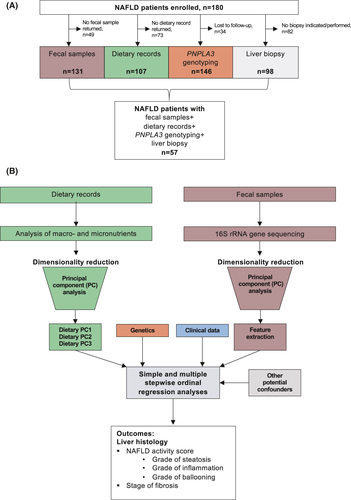
Patients were referred to our tertiary referral centre with elevated liver function tests and/ or liver abnormalities on ultrasound for further diagnostic tests or with already diagnosed NAFLD in order to assess disease activity and severity. If NAFLD diagnosis was made or confirmed, patients were consecutively enrolled in this observational study.
Within the study, a detailed medical history including drug treatment, physical exam, laboratory analysis, anthropometric and blood pressure measurements, ultrasound and/or magnetic resonance imaging (MRI), transient elastography and liver biopsy, if clinically indicated, as per standard of care were performed. NAFLD was diagnosed, if the following conditions were met: hepatic steatosis on liver imaging (ultrasound and/or magnet resonance imaging) and/or the presence of ≥5% fat in histological analysis of liver biopsy; self-reported daily alcohol consumption of less than 10 g in women and less than 20 g in men; absence of steatogenic drugs such as glucocorticoids, methotrexate, amiodarone and tamoxifen; absence of other diseases causing secondary steatosis such as human immunodeficiency virus infection, celiac disease or inflammatory bowel disease; absence of other chronic liver diseases, for example, viral hepatitis, autoimmune hepatitis, toxic liver injury, alcoholic steatohepatitis, cholestatic liver disease, Wilson's disease and hereditary hemochromatosis. Exclusion criteria for all study subjects were oral- or intravenous antibiotic treatment within the last 6 months prior to the study, known malignancy, pregnancy and age <18 years. Further exclusion criteria for NAFLD patients were ongoing successful lifestyle modifications defined as more than 5% loss of body weight within the last 3 months prior to enrolment or current or prior participation in an interventional NASH study.10, 11
Any recommendations or treatment suggestions for study participants did not differ from usual patient care. Thus, NAFLD patients were treated according to the recommendations of the current European guideline.12
Abdominal ultrasound was performed for all patients. All blood samples for laboratory analyses were collected under fasting conditions. Anthropometric measurements were carried out by physicians or trained research assistant nurses.
Type 2 diabetes was defined as glycated haemoglobin (HbA1c) ≥6.5% and/or fasting glucose ≥126 mg/dL and/or use of antidiabetic medications. Metabolic syndrome was defined following the International Diabetes Foundation (IDF) criteria.13 Arterial hypertension was defined as office blood pressure ≥140/90 mmHg on ≥2 measurements during ≥2 occasions or antihypertensive drug treatment. Dyslipidemia was defined as increased plasma cholesterol (>200 mg/dL) and/or triglycerides ≥150 mg/dL and/or low high-density lipoprotein levels (<50 mg/dL for women and <40 mg/dL for men).
2.2 Liver biopsies
Liver biopsy was performed in patients with NAFLD with history of persistently elevated serum alanine aminotransferase (ALT) and/or aspartate aminotransferase (AST) for at least 6 months, to rule out potential other liver diseases than NAFLD and if there was clinical suspicion for advanced liver disease. If liver biopsy was performed, samples were evaluated by an experienced liver pathologist who was blinded for all clinical and laboratory patient data. The NASH clinical research network histological scoring system14 was used to evaluate disease activity and severity. Accordingly, the NAFLD activity score (NAS) was obtained for each biopsy. This score is defined as the unweighted sum of the scores for steatosis (0-3), lobular inflammation (0-3) and ballooning (0-2), thus ranging from 0 to 8.14, 15 Fibrosis was staged from 0 to 4:0 none, 1 perisinusoidal or periportal, 2 perisinusoidal and portal/periportal, 3 bridging fibrosis and 4 cirrhosis. Stages 1a, 1b and 1c were summarized as Stage 1.
2.3 Genotyping of the PNPLA3 variant
Genotyping of the common PNPLA3 variant rs738409 (p.I148M) was performed centrally in the genetic laboratory of the Department of Medicine II (Saarland University Medical Center) by technicians blinded to the phenotypes of patients.16 PNPLA3 genotypes were included in the regression models, using the wild-type genotype (CC) as reference versus heterozygous (CG) and homozygous (GG) carriers, and in addition, we combined the genotypes CG and GG using CC as reference.
2.4 Gut bacterial sequencing
The DNA was isolated using the RNeasy Power Microbiome Kit (Qiagen, Hilden, Germany). Seven of the nine variable bacterial 16S rRNA gene regions (pool 1: V2, V4 and V8; pool 2: V3, V6/7 and V9) were amplified with the Ion 16S Metagenomics Kit (Thermo Fisher Scienctific, Waltham, USA) utilizing two primer pools (an integrated research solution for bacterial identification using 16S rRNA sequencing on the Ion PGM™ System with Ion Reporter™ Software https://www.thermofisher.com/content/ dam/LifeTech/Documents/PDFs/Ion-16S-Metagenomics-Kit-Software-Application-ote.pdf). Amplicons were pooled and cleaned using the NucleoMag NGS Clean-up (Macherey-Nagel, Düren, Germany). The Qubit system was used to determine amplicon concentration; the library was prepared with the Ion Plus Fragment Library Kit (Thermo Fisher Scienctific, Waltham, USA). For the template preparation, amplicon concentration was diluted to 30 ng/mL. The Ion Chef Kit and the Ion Chef system (both, Thermo Fisher Scienctific, Waltham, USA) were used to enrich and prepare the template-positive Ion Sphere Particles (ISPs). Amplicon library was sequenced using the Ion Torrent S5 system (pH-dependent, Thermo Fisher Scienctific, Waltham, USA). The amplicon sequences were clustered into operational taxonomic units (OTUs) before taxonomical alignment with the MicroSEQ 16S-rDNA Reference Library v2013.1 (Thermo Fisher Scienctific, Waltham, USA) and Greengenes v13.5 databases; 97% similarity was used to genus level assignment and 99% similarity for species level assignment. Data files were assigned by the Ion Reporter metagenomics 16S w1.1 workflow (Thermo Fisher Scienctific, Waltham, USA). The raw data were processed using the programming language R version 3.5.1.10, 17 We obtained a total of 231 206 (range 30 397-659558) clean reads per sample on average.
2.5 Accession numbers sequence data
Sequence data were registered at NCBI under BioProject PRJNA540738. BioSample IDs included in this study can be found in Table S1.
2.6 Dietary records
The food intake was recorded using an open, 14-day self-administered food record. Patients were instructed to report each daily portion of consumed food and all beverages in as much detail as possible directly after ingestion, to weigh foods or to estimate doses in gramme and not to change their usual dietary and physical activity habits during the recording period. EBISpro 2016 professional scientific software was used to analyse energy intake, basal metabolic rate and all macro- and micronutrients. The intake of all macro- and micronutrients was divided by the total energy intake to obtain the relative intake of the respective food component.
Energy misreporting is a very frequently observed issue in self-reported dietary assessment and is considered to be unavoidable.18, 19 However, excluding misreporters leads to a loss of statistical power and may bias estimates of associations.19 As an alternative approach, we calculated the ratio between energy intake (EI) and the basal metabolic rate (BMR) (EI:BMR ratio). Overall, 40% of the cohort were definite energy misreporters (EI:BMR below 1). To account for energy misreporting in our study, we included the EI:BMR ratio in our multiple analyses in similarity to previous publications.17, 19, 20
2.7 Statistical analysis
Results are expressed as median and range in parentheses for each continuous outcome and as number and percentage for categorical variables. A two-sided P value less than .05 was considered as statistically significant. Simple and multiple proportional ordinal regression models as implemented in the “Ordinal” package in R,21 using liver histology features that are measured on an ordinal scale as outcome, were used to associate the PNPLA3 polymorphism and various clinical and dietary features with liver disease severity (Figure 1B). We used ordinal regression analyses because dichotomizing variables, originally measured on an ordinal scale, leads to a loss of information, and ordinal regression methods have been proposed in order to reduce sample size and increase statistical power.22, 23 Given an alpha level of 0.05, the sample size of n = 57 provides 80% power to detect odds ratios of 3.8 (NAS), 3.9 (fibrosis), 4.1 (steatosis), 4.1 (ballooning) and 4.2 (inflammation). After performing a simple regression with calculation and comparison of the Akaike information criterion (AIC), we used a forward stepwise approach. The AIC is a measurement of the regression model performance and can be used to compare different models, whereas a lower AIC indicates a better performance. Adding variables to the model was stopped as soon as the model performance could not be further improved by adding other variables, that is, when the AIC values start to pick up if adding one more variable. Outliers were not excluded from the analysis, and the analysed variables included in the regression models were not transformed prior to entering the model.
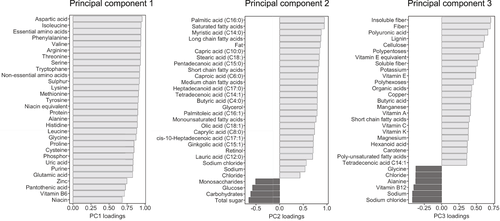
Principal component (PC) analyses were used for dimensionality reduction of the dietary and 16S gene sequencing data. The first three dietary PCs, which covered three main food groups (Figure 2), were included in the regression models. For the 16S gene sequencing data, the major contributing bacterial taxa at genus level were represented in all the first six PCs (Figure S1). In order to improve interpretability of the regression models, we therefore included these specific bacterial taxa individually in the regression models (Figure 1B). Statistical analysis was performed using R statistical software, R version 3.5.1.24
3 RESULTS
A total of 180 NAFLD patients were enrolled, of whom 131 returned fecal samples, and 107 returned the dietary record, PNPLA3 genotypes could be determined in 146 patients, and liver histology data were available for 98 patients (Figure 1A). The final study cohort was composed of 57 patients with NAFLD where all data were available (16S gene sequencing results, dietary record, PNPLA3 genotypes and liver histology, Figure 1). Their median age was 52 years, and 44% were female. The median BMI was 30.0 kg/m2, 21% suffered from type 2 diabetes, 40% carried the heterozygous PNPLA3 genotype, and 18% were homozygous PNPLA3 risk allele carriers (Table 1).
| Variable | Number of patients | ||
|---|---|---|---|
| Demographics | |||
| Age, years | 57 | 51.9 (19.1-76.2) | |
| Gender female, n (%) | 57 | 25 (43.9) | |
| Body mass index, kg/m2 | 57 | 29.7 (22.5-52.9) | |
| Waist circumference (cm) | 50 | 105 (81.0-143.0) | |
| Type 2 diabetes, n (%) | 57 | 12 (21.1) | |
| Metformin use, n (%) | 57 | 8 (14.0) | |
| Arterial hypertension, n (%) | 57 | 32 (56.1) | |
| Dyslipidemia, n (%) | 57 | 33 (57.9) | |
| Statin use, n (%) | 57 | 9 (15.8) | |
| Proton pump inhibitor use, n (%) | 57 | 5 (8.8) | |
| Genetics | |||
| PNPLA3 p.148IM (CG), n (%) | 57 | 23 (40.4) | |
| PNPLA3 p.148MM (GG), n (%) | 57 | 10 (17.5) | |
| AST, U/L | 56 | 32.0 (17.0-143.0) | |
| ALT, U/L | 56 | 47.5 (16.0-232.0) | |
| GGT, U/L | 56 | 71.5 (14.0-660.0) | |
| Alkaline phosphatase, U/L | 56 | 74.0 (43.0-160.0) | |
| Bilirubin, mg/dL | 56 | 0.5 (0.2-2.1) | |
| Albumin, g/L | 56 | 45.0 (38.0-51.0) | |
| Triglycerides, mg/dL | 56 | 159.0 (29.0-1104.0) | |
| Total cholesterol, mg/dL | 56 | 184.0 (86.0-329.0) | |
| HDL cholesterol mg/dL | 54 | 45.5 (21.0-88.0) | |
| LDL cholesterol mg/dL | 52 | 112.5 (15.0-247.0) | |
| Fasting glucose, mg/dL | 56 | 95.5 (63.0-196.0) | |
| HbA1c, % | 52 | 5.4 (4.7-8.0) | |
| Alpha-fetoprotein kU/L | 53 | 6.0 (2.0-12.0) | |
| Creatinine, mg/dL | 56 | 0.9 (0.5-1.4) | |
| Urea, mg/dL | 56 | 29.0 (12.0-47.0) | |
| Uric acid, mg/dL | 56 | 6.1 (1.9-10.8) | |
| Ferritin, µg/L | 56 | 217.5 (17.0-1701.0) | |
| Platelet count, x1E9/L | 56 | 213.5 (60.0-386.0) | |
| INR | 56 | 1.0 (0.9-2.0) | |
| Liver histology data | Scoring | Number (%) | |
| Grade of steatosis, n (%) | 0 | <5% | 0 (0.0) |
| 1 | 5%–33% | 17 (29.8) | |
| 2 | >33%–66% | 24 (42.1) | |
| 3 | >66% | 16 (28.1) | |
| Ballooning, n (%) | 0 | None | 18 (31.6) |
| 1 | Few balloon cells | 25 (43.9) | |
| 2 | Prominent ballooning | 14 (24.6) | |
| Grade of inflammation, n (%) | 0 | No foci | 11 (19.3) |
| 1 | <2 foci | 28 (49.1) | |
| 2 | 2-4 foci | 18 (31.6) | |
| 3 | >4 foci | 0 (0) | |
| NAFLD activity score | 1 | 2 (3.5) | |
| 2 | 8 (14.0) | ||
| 3 | 15 (26.3) | ||
| 4 | 10 (17.5) | ||
| 5 | 9 (15.8) | ||
| 6 | 9 (15.8) | ||
| 7 | 4 (7.0) | ||
| 8 | 0 (0) | ||
| Stage of fibrosis, n (%) | 0 | None | 16 (28.1) |
| 1 | Perisinusoidal or periportal | 18 (31.6) | |
| 2 | Perisinusoidal and portal/periportal | 13 (22.8) | |
| 3 | Bridging fibrosis | 3 (5.3) | |
| 4 | Cirrhosis | 7 (12.3) | |
Note
- Values are presented as median and range in parentheses. The number of patients with available data for this variable is indicated in the second column.
- Abbreviations: ALT, alanine aminotransferase; AST, aspartate aminotransferase; BMI, body mass index; GGT, gamma-glutamyl-transferase; HbA1c, glycated haemoglobin; HDL, high-density lipoprotein; INR, international normalized ratio; LDL, low-density lipoprotein; NAS, NAFLD activity score; PNPLA3, patatin-like phospholipase domain-containing protein 3.
3.1 Multiple ordinal regression analyses reveal associations with liver histology features
First, we performed PC analyses in order to reduce dimensionality of the high-dimensional dietary data and 16S gene sequencing data. The first dietary PC (PC1) was predominantly represented by several amino acids, sulphur, niacin, phosphor, uric acid and purine. PC2 was mainly represented by fat components, sugar and carbohydrates; PC3 was represented by fibre and several vitamins (Figure 2). These three individual PCs were included in the multiple regression models. For the 16S gene sequencing data, the first six PCs that accounted for more than 90% of the variances in the data were all predominantly represented by the bacterial genera Faecalibacterium, Bacteroides, Blautia, Prevotella, Bifidobacterium, Roseburia, Ruminococcus, Eubacterium and Streptococcus (Figure S1). To improve interpretability of the regression models, we included these specific bacterial taxa individually in the models, together with the Shannon diversity index, which was calculated including all detected bacterial taxa. We further included the PNPLA3 variant, age, BMI, gender, type 2 diabetes, dyslipidemia and arterial hypertension as risk factors for NAFLD progression and the EI:BMR ratio as well as proton pump inhibitor, metformin and statin use as potential confounding factors.
We first visualized associations between explanatory variables that were entered in the simple and multiple regression models to obtain an overview about collinearity. As expected, we observed positive associations between clinical variables such as type 2 diabetes and arterial hypertension and the body mass index (BMI). We also observed several correlations between bacteria. A higher BMI was further associated with a lower EI:BMR ratio, indicating higher energy misreporting with increasing BMI as well as a lower intake of fibre and fibre components, the vitamins E-, A-, C- and K, copper, short chain fatty acids and poly-unsaturated fatty acids with increasing obesity (Figure S2). Because collinearity of the explanatory variables was expected, we have chosen a forward selection process in which a choice is made over two variables that are correlated with each other and where the variable with the higher importance over the other variable regarding the respective outcome is included in the final multiple regression model.
3.2 Hepatic steatosis
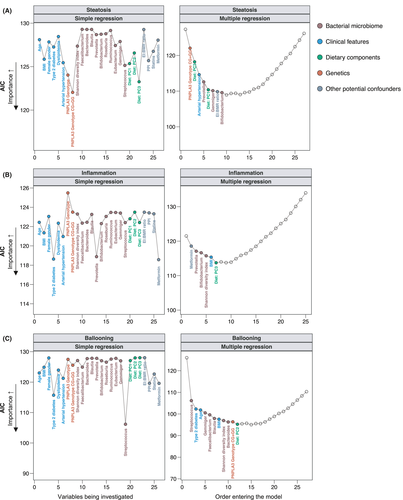
We first looked at histological steatosis, measured in grades G0-G3 (Table 1). In the simple regression analysis, variant PNPLA3 risk allele, dietary factors and clinical features had a higher importance as indicated by lower AIC values than features of the gut bacterial microbiota (Figure 3A, Table S2). In the multiple regression analysis, the presence of PNPLA3 p.148IM or PNPLA3 p.148MM was significantly associated with higher histological grades of hepatic steatosis (P = .007) and had the highest importance in relation to other variables, followed by a significant negative association with the dietary PC3 (P = .001) (Figure 3A, Table 2). Some major components of PC3 were fibre and fibre components, the vitamins E-, A-, C- and K, copper, short chain fatty acids and poly-unsaturated fatty acids (Figure 2). A lower intake of these compounds and a higher intake of sodium were significantly associated with higher degrees of steatosis on liver histology. Similarly, PC1 was significantly (P = .027) positively associated with higher degrees of steatosis (Figure 3A, Table 2), whereas PC1 was composed of several amino acids, sulphur, niacin, phosphor, uric acid and purine (Figure 2).
| Outcome | Steatosis (OR, 95% CI) | Inflammation (OR, 95% CI) | Ballooning (OR, 95% CI) | NAFLD activity score (OR, 95% CI) | Fibrosis (OR, 95% CI) |
|---|---|---|---|---|---|
| Clinical features | |||||
| Age | 1.09 (1.03-1.16), P = .003 | 1.05 (1.01-1.10), P = .021 | 1.06 (1.01-1.11), P = .018 | ||
| BMI | 1.12 (1.00-1.25), P = .041 | 1.16 (1.00-1.34), P = .048 | 1.26 (1.12-1.45), P = .001 | 1.23 (1.10-1.37), P < .001 | |
| Female gender | 0.26 (0.07-0.98), P = .046 | ||||
| Type 2 diabetes | 2.56 (0.28-23.75), P = .407 | 2.11 (0.46-9.81), P = .339 | |||
| Dyslipidemia | |||||
| Arterial hypertension | 10.42 (2.51-43.31), P = .001 | ||||
| Genetics | |||||
| PNPLA3 p.148IM (heterozygous, CG) | |||||
| PNPLA3 p.148MM (homozygous, GG) | |||||
| PNPLA3 CG or GG | 5.32 (1.56-18.11), P = .007 | 3.82 (0.90-16.12), P = .068 | 4.66 (1.45-16.23), P = .016 | ||
| Bacterial microbiome | |||||
| Shannon diversity index | 0.17 (0.03-1.187), P = .073 | 0.16 (0.02-1.12), P = .066 | 0.24 (0.03-1.49), P = .138 | 0.33 (0.07-1.49), P = .149 | |
| Faecalibacterium | 0.72 (0.57-0.91), P = .006 | 0.81 (0.70-0.94), P = .009 | 0.77 (0.65-0.91), P = .002 | ||
| Bacteroides | 1.07 (0.99-1.15), P = .088 | 0.91 (0.86-0.97), P = .002 | |||
| Blautia | 0.88 (0.78-1.00), P = .049 | ||||
| Prevotella | 0.96 (0.91-1.01), P = .096 | 0.93 (0.88-0.98), P = .009 | 0.93 (0.88-0.98), P = .012 | ||
| Bifidobacterium | 1.08 (0.98-1.18), P = .125 | 0.92 (0.85-1.00), P = .051 | 0.86 (0.73-1.01), P = .061 | ||
| Roseburia | 1.22 (1.02-1.45), P = .026 | ||||
| Ruminococcus | |||||
| Eubacterium | 0.83 (0.67-1.04), P = .101 | ||||
| Gemmiger | 1.25 (0.99-1.57), P = .063 | 2.08 (1.38-3.13), P < .001 | 1.45 (1.12-1.87), P = .006 | ||
| Streptococcus | 1.66 (1.08-2.54), P = .021 | 1.07 (0.99-1.18), P = .106 | 0.89 (0.81-1.98), P = .024 | ||
| Dietary components | |||||
| Diet: PC1 | 1.13 (1.01-1.26), P = .027 | 1.12 (1.01-1.24), P = .033 | |||
| Diet: PC2 | 0.87 (0.75-1.02), P = .084 | ||||
| Diet: PC3 | 0.69 (0.55-0.87), P = .001 | 1.19 (0.99-1.43), P = .062 | |||
| Other potential confounders | |||||
| EI:BMR ratio | 5.58 (0.70-44.56), P = .105 | 10.27 (1.08-99.18), P = .048 | |||
| PPI | |||||
| Statins | 17.46 (2.63-116.06), P = .003 | ||||
| Metformin | 4.10 (0.76-22.12), P = .101 | ||||
Note
- Multiple stepwise proportional ordinal regression analyses including clinical features, the PNPLA3 polymorphism, features of the gut bacterial microbiome, dietary components and potential confounding factors; 57 biopsy-proven NAFLD patients were included in the analysis. No missing data present.
- Abbreviations: BMR, basal metabolic rate; CI, confidence interval; EI, energy intake; OR, odds ratio; PC, principal component; PNPLA3, patatin-like phospholipase domain-containing protein 3; PPI, proton pump inhibitor. Bold font indicates significance (P < .05).
3.3 Liver inflammation
The presence of type 2 diabetes, the related metformin use and lower relative abundances of Prevotella were the most important factors associated with higher degrees of hepatic inflammation in simple regression analyses (Figure 3B, Table S2). Low Prevotella abundance (P = .009) and an increased BMI (P = .041) remained significant in the final multiple regression model (Figure 3B, Table S2).
3.4 Hepatocellular ballooning
We next investigated hepatocellular ballooning. In simple regression analyses, besides the relative Streptococcus abundance, several clinical factors (type 2 diabetes, arterial hypertension, age and dyslipidemia) were significantly positively associated with higher degrees of ballooning (Figure 3C). The most important features in the simple regression models that also remained significant in the final model were a higher relative Streptococcus abundance (P = .021), a higher age (P = .003), higher relative abundances of Gemmiger (P < .001) and lower relative abundances of Faecalibacterium and Blautia (Figure 3C, Table 2).
3.5 NAFLD activity score (NAS)
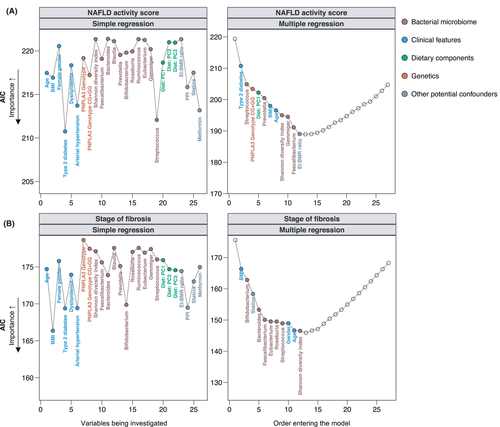
A higher NAS indicates more severe liver damage and has been associated with worse outcome in patients with NAFLD.25 In the simple regression analysis, type 2 diabetes, arterial hypertension and high relative abundances of Streptococcus had the highest association with a higher NAS (Figure 4A, Table S2). In the multiple model however, the most important variables that remained significant in the final model were the PNPLA3 risk genotypes PNPLA3 p.148IM or PNPLA3 p.148MM (P = .016), diet enriched in several amino acids, sulphur, niacin, phosphor, uric acid and purine (P = .033, corresponding to PC1, Figure 2), low relative abundances of Prevotella (P = .012) and a higher BMI (P = .001) (Figure 4A, Table 2).
3.6 Liver fibrosis
Among all histological parameters, the fibrosis stage represents the strongest predictor feature of future liver-related complications in NAFLD patients.26 In the simple regression analysis, clinical factors (BMI, P = .001; type 2 diabetes, P = .005; arterial hypertension, P = .005; female gender, P = .046) were overall the most important features associated with higher fibrosis stages (Figure 4B, Table S2). In the multiple stepwise regression analysis, a higher BMI remained the most important and significant feature associated with higher degrees of fibrosis (P < .001), followed by low relative abundances of Bacteroides (P = .002) and Faecalibacterium (P = .002) (Figure 4B, Table 2).
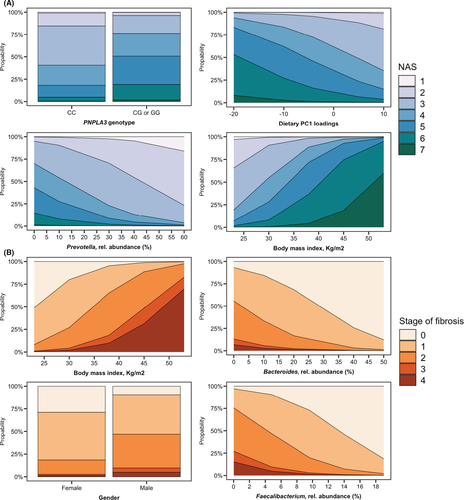
To visualize some key findings of our stepwise models, we plotted the predicted probabilities generated by the final multiple stepwise regression for the individual histological stages of the NAS and fibrosis (Figure 5). These data demonstrate that the probability of having higher degrees of the NAS was low in patients with high abundances of Prevotella, whereas it was high among patients with PNPLA3 risk genotypes PNPLA3 p.148IM or PNPLA3 p.148MM and with increasing BMI (Figure 5A). Patients with NAFLD and obesity, low abundances of Bacteroides or Faecalibacterium and NAFLD patients with a male gender were more likely to have significant or advanced liver fibrosis already (Figure 5B).
4 DISCUSSION
NAFLD is considered a multisystem disease. Due to its close interconnection with other metabolic diseases and shared risk factors with them (eg, obesity and overnutrition), it is inherently difficult to dissect the impact of distinct pathogenic pathways to the overall phenotype. However, in order to therapeutically target the most relevant of these pathways, to adapt targeted interventions to the stage of NAFLD and to reduce the risk of non-efficacious treatment strategies, it is of utmost importance to understand which of the manifold factors promote features of NAFLD progression in the clinical setting of a real-life patient cohort. To our knowledge, this is the first study investigating associations between the gut microbiome, the PNPLA3 polymorphism, dietary factors and clinical features in one well-described NAFLD cohort. We have chosen stepwise multiple ordinal regression analyses as a powerful approach assessing associations across a relevant range of outcomes. Dichotomizing outcomes that are originally measured on an ordinal scale is a common practice but results in loss of information.23
We found that covariates that we investigated were differently associated with specific features of liver histology, which supports the multifactorial pathogenesis of NAFLD severity. Given all investigated variables, the BMI had the strongest association with liver fibrosis and was also associated with inflammation, ballooning and the NAS in the multiple regression analyses. This indicates that a higher BMI is one of the most important modifiers of NAFLD severity and underlines that lifestyle modification to achieve weight loss remains the first-line intervention in patients with NAFLD, as recently emphasized in a clinical practice guideline update of the American Gastroenterological Association.27 The presence of type 2 diabetes was one of the most important factors associated with liver inflammation, ballooning, the NAS and stages of fibrosis in the simple regression analysis but seems to be dependent from other clinical factors such as age, the BMI and the presence of dyslipidemia in the multiple regression analysis. This indicates that those clinical factors that typically co-occur with the presence of type 2 diabetes are potentially moderating associations with liver disease severity.
The carriage of the PNPLA3 p.I148M polymorphism was the most important feature independently associated with higher histological degrees of steatosis and NAS, which is in line with other studies showing an increased risk for disease progression, the occurrence of liver-related events28 and hepatocellular cancer29 in patients with either a heterozygote or homozygote variant. In our study, this association remained significant even after taking additional potential synergistic cofactors such as an increased BMI or dyslipidemia into account, indicating a pivotal role of this variant as endogenous determinant of severe NAFLD.
Whereas several studies investigate associations between the gut microbiota composition and disease severity in NAFLD individually, we further demonstrate in our analysis that specific gut bacterial features are associated with NAFLD severity even after considering other cofactors that might otherwise bias the results. The strongest associations were observed for liver inflammation, hepatocellular ballooning and the fibrosis stage. Translocation of gut bacteria through the portal vein to the liver is one mechanism of how the gut bacterial microbiota might mediate disease. Once having reached the liver, microbial-derived pathogen associated molecular patterns (PAMPs) such as lipopolysaccharide (LPS) are recognized by specific receptors such as toll-like receptors, where their activation can mediate hepatic inflammation. Further, activation of the NOD-like receptor protein 3 (NLRP3) inflammasome has been identified as another trigger for liver inflammation responding to PAMPs in the setting of NAFLD. NLRP3 is significantly upregulated in the liver of patients with NASH compared with simple steatosis, and preclinical data have demonstrated that NLRP3 activation results in the activation of caspase 1 as well as in the production of several inflammatory cytokines, which ultimately results in programmed cell death, inflammation and fibrosis.30 Of the gut microbial taxa at genus level, lower abundances of Faecalibacterium were associated with higher stages of fibrosis, a higher NAS and higher degrees of ballooning in the multiple regression analyses. As a member of the Ruminococcaceae family, Faecealibacterium prausnitzii (F prausnitzii) is the only species within the genus of Faecalibacterium that has been successfully isolated.31 F prausnitzii is an important short-chain fatty acid producer and has anti-inflammatory properties.32 A lower abundance of Faealibacterium in more advanced NAFLD has been described before.33, 34 In a randomized-controlled trial, treatment of obese women with prebiotics led to an increase of F prausnitzii, which was associated with lower circulating levels of LPS.35 Overall, our data indicate that a depletion of intestinal Faecalibacterium might play an important role in NAFLD disease severity.
A higher relative abundance of Gemmiger, on the other hand, was associated with more severe disease in our patients. Gemmiger is a gramme-negative bacterium that also belongs to the Ruminococcaceae family and has been associated with the presence of early hepatocellular carcinoma36 and Crohn's disease.37 We have shown previously that the relative Gemmiger abundance is positively correlated with increased liver enzymes, ferritin and overweight.17 Overall, further mechanistic studies are needed to clarify a potential detrimental role of Gemmiger in the setting of NAFLD. Our study revealed associations between the gut microbiome and NAFLD disease severity that remained significant even after taking other cofactors into account. Microbe-based treatments such as administration of prebiotics, probiotics or a combination of both (synbiotics) might be an attractive treatment approach and should be studied in clinical trials.
Our study has several limitations. Although we found associations between the gut microbiome, the PNPLA3 polymorphism, dietary factors and clinical features, we cannot make causal conclusions due to the cross-sectional study design. Particularly when considering factors with high colinearity such as conditions of the metabolic syndrome, it is important to bear in mind that failing to be included in the multiple regression models does not necessarily indicate that those factors are not of relevance. We have chosen a forward selection process in which a choice is made over two variables that are correlated with each other and where the variable with the higher importance over the other variable regarding the respective outcome is included in the final multiple regression model. Therefore, failure to enter the final model rather indicates that other colinear factors are of higher importance when considering all variables. Overall, longitudinal studies are needed to investigate which of the studied factors are the most relevant in terms of liver-related and overall outcomes.
Focusing only on patients with all data completely available allowed us to analyse a very well-characterized cohort and limited our sample size. In relation to the high number of explanatory variables, this might have led to model instability, and the results need to be confirmed in larger cohorts.
Collecting diet over 14 days provides very detailed information about food choices and has some advantages over other collection methods such as food frequency questionnaires.38, 39 Dietary assessment however is generally very vulnerable to distortion and even if patients were instructed on how to use the dietary report in as much detail as possible, high motivation is needed to follow the requirements, and the analysis relies on honesty and accuracy of the patients’ disclosures. Even if we included energy misreporting in our analyses, miscalculation or underreporting of food portions might have occurred and could have biassed the results. In addition, focusing more than usual on the diet due to the dietary recording can create more awareness and therefore may change dietary habits, even though patients were instructed not to change usual dietary habits over the recording period.
In conclusion, specific gut microbial, dietary, the PNPLA3 polymorphism and clinical features are significantly associated with different histology features, which underscore the multifactorial pathogenesis of NAFLD. A higher BMI, the PNPLA3 risk variant, lower relative abundances of Faecalibacterium or Prevotella, higher relative abundances of Gemmiger and dietary patterns low in fibre and specific vitamins as well as enriched in amino acids, uric acid and purine seem to represent factors of crucial importance for NAFLD disease severity. A better understanding of synergistic effects driving simple steatosis to more advanced disease stages might help to develop improved strategies for the prevention of disease progression and could result in better individualized treatment strategies such as personalized nutrition or microbe-based therapeutics resulting in effective prevention of disease progression and improved treatment of the global burden of NAFLD.
ACKNOWLEDGEMENTS
This study was supported by the ‘Marga und Walter Boll-Stiftung,' project number 210-03-2016, and the ‘Köln Fortune' research pool, Faculty of Medicine, University of Cologne, Germany, project number 160/2014 (to MD). SL was supported by a DFG fellowship (LA 4286/1-1) and the ‘Clinical and Translational Research Fellowship in Liver Disease' by the American Association for the Study of Liver Diseases (AASLD) Foundation. The study was supported by services provided by the San Diego Digestive Diseases Research Center (SDDRC) supported by NIH P30 DK120515 (to BS).
CONFLICT OF INTERESTS
BS has been consulting for Ferring Research Institute, Intercept Pharmaceuticals, HOST Therabiomics, Patara Pharmaceuticals, Mabwell Biotehrapeutics and Takeda. BS's institution UC San Diego has received grant support from BiomX, NGM Biopharmaceuticals, CymaBay Therapeutics, Synlogic Operating Company and Axial Biotherapeutics. MJGT has received research grants from 3M, Astellas Pharma, Biontech, DaVolterra, Evonik, Gilead Sciences, Glycom, Immunic, MaaT Pharma, Merck/MSD, Organobalance, Seres Therapeutics, Takeda Pharmaceutical, has received speaker fees from Astellas Pharma, Basilea, Gilead Sciences, Merck/MSD, Organobalance, Pfizer and has been a consultant to Alb Fils Kliniken GmbH, Arderypharm, Astellas Pharma, Biomérieux, DaVolterra, Farmak International Holding GmbH, Ferring, Immunic AG, MaaT Pharma, Merck/MSD, Roche, SocraTec R&D GmbH.
AUTHORSHIP STATEMENT
S.L. was responsible for collection of samples, statistical analysis, interpretation of data and writing of the manuscript; A.M. was responsible for collection of samples; X.Z. and F.F. provided help with statistical analysis; H.W. was responsible for sequencing of fecal samples; M.J.G.T.V. was responsible for interpretation of data and edited the manuscript; M.K. and F. L. were responsible for the PNPLA3 analysis and edited the manuscript; A.N., A.K. and C.S. were responsible for fecal DNA extraction and sequencing; P.K., T.G., B.S., H-M.S, R.M., F.T. and C.R. were responsible for interpretation of data and edited the manuscript; M.D. was responsible for the study concept and design, collection of samples, data collection, analysis of dietary records, interpretation of data, editing the manuscript, and study supervision. All authors approved the final version of the article.
ETHICS APPROVAL STATEMENT
The protocol was approved by the Ethics Commission (reference # 15-056) of Cologne University's Faculty of Medicine, and written informed consent was obtained from each patient. The study was performed in accordance with the Declaration of Helsinki.