Phylogeographic advances in midwife toads (Alytes) support the existence of a novel taxon endemic to the Central Pyrenees
Contributing author: Axel Hernandez ([email protected])
Abstract
Although necessary to promote conservation, defining evolutionary units and naming biodiversity remain a difficult task, especially in problematic species groups that experienced a dynamic biogeographic history. In this article, we undertake such task for midwife toads of the Alytes obstetricans complex by integrating recent molecular studies altogether—multilocus phylogenies and population genetic barcoding. Despite a partly unresolved phylogeny underlain by deep cyto-nuclear discordances, nuclear and mitochondrial evidence support the validity of six genuine lineages assigned to two different species (A. obstetricans and A. almogavarii), which could be accurately mapped across most of their ranges. In particular, we demonstrate the existence for an overlooked yet genetically distinct lineage previously confounded with A. almogavarii, confined to the northern part of Huesca Province in the Spanish Central Pyrenees. We describe this micro-endemic as the subspecies Alytes almogavarii inigoi ssp. nov., with reports on the mating call and the larvae. Conservation genetics of eight populations of this new taxon revealed two independent conservation units, separated by topographic barriers. In the wait for upcoming genomic analyses to unravel many elusive aspects of the evolution, diversity and systematics of Alytes, the present paper offers an integrative phylogeographic overview to guide future investigations and generally illustrates how multiple lines of molecular evidence can be combined to clarify the confusing taxonomy of complex species groups.
1 INTRODUCTION
The process of describing newly discovered phylogeographic lineages as valid species or subspecies is fraught with many challenges. The most obvious one is our limited confidence in traditional molecular evidence. It has now become clear that mitochondrial DNA (mtDNA) might be a poor predictor of contemporary genetic structure, due to demographic or hybridization events (e.g., Wielstra & Arntzen, 2020). Consequently, mtDNA lineages do not always represent evolutionary entities (e.g., ghost mtDNA; Dufresnes et al., 2020) and should alone not call for taxonomic revisions (Speybroeck et al., 2020).
Genetic variation at commonly used nuclear markers is not straightforward to interpret either: intron genes sometimes display ancestral polymorphism and/or incomplete lineage sorting that may create false impressions of divergence between panmictic populations or conspecific relationships between genuinely diverged populations, respectively. In parallel, population structure inferred from multilocus nuclear datasets such as microsatellites can reflect recent population disconnection rather than long-term genetic independence. In the wait of more decisive data, as obtained from genomic phylogeography (e.g., Dufresnes et al., 2020), multiple lines of traditional molecular evidence are thus needed to gauge the validity of recently diverged taxa. Even when such evidence exists, different concepts and delimitation methodologies are preventing researchers to reach clear-cut conclusions on species versus subspecies ranks (Hillis, 2019, 2020), especially when it comes to cryptic species (Chenuil et al., 2019; Heethoff, 2018), and even more so when phylogenetic relationships are poorly resolved. For these reasons altogether, taxonomic revisions in species groups that have experienced dynamic biogeographic histories—which left complex genetic signatures of structure and diversity—can be a tremendous endeavor, even in well-studied clades (e.g., Dufresnes et al., 2021).
We emphasize such situation for midwife toads of the Alytes obstetricans (Laurenti, 1768) complex, an emblematic group of Mediterranean anurans widely distributed in the Iberian Peninsula and Western Europe. For more than two decades, these have received significantly phylogenetic (e.g., Martínez-Solano et al., 2004), biogeographic (e.g., Rodríguez-Rodríguez et al., 2020), and taxonomic attention (e.g., Arntzen & García-París, 1995; García-París & Martínez-Solano, 2001), notably to clarify phylogenetic ambiguities caused by deep discordances between mitochondrial and nuclear markers (Gonçalves et al., 2007). Much has been achieved since the last attempt to refine taxonomic units in this complex (Maia-Carvalho, Gonçalves, Martínez-Solano, et al., 2014), including higher-resolution phylogenies (Maia-Carvalho, Gonçalves, Ferrand et al., 2014) and fine-scale multilocus population genetic barcoding across most of the range (Dufresnes & Martínez-Solano, 2020; Gonçalves et al., 2015; Maia-Carvalho et al., 2018). Building up on this recent research, here we re-interpreted patterns of diversity from multiple sources to propose an updated taxonomic arrangement for the A. obstetricans complex, with a specific focus on the unnoticed presence of a novel taxon endemic to the Central Pyrenees.
2 METHODS
To comprehensively summarize and discuss patterns of diversity and structure within the A. obstetricans complex, we adapted key figures featured in the original phylogeographic studies and partially re-analyzed some of their published datasets.
First, we mapped the mitochondrial barcoding data (NADH-ubiquinone oxidoreductase chain 4 gene, hereafter ND4) from Gonçalves et al. (2015) and Dufresnes and Martínez-Solano (2020), which altogether cover 125 localities across the entire range. Gonçalves et al. (2015) already incorporated data from previous studies (e.g., Gonçalves et al., 2007; Maia-Carvalho, Gonçalves, Martínez-Solano, et al., 2014; Martínez-Solano et al., 2004).
Second, we displayed the original phylogenetic trees published by Maia-Carvalho, Gonçalves, Ferrand, et al. (2014), which were built from the most extensive mitochondrial (9045 bp) and nuclear alignments (3142 bp from four introns) available to date for this group. Because the two trees are discordant (see also Gonçalves et al., 2007), we refrained to consider phylogenies combining both nuclear and mitochondrial datasets.
Third, we performed additional analyses of the microsatellite dataset of Maia-Carvalho et al. (2018), which includes the genotypes of 965 individuals (142 populations) at 12 loci. We initially re-run the Bayesian clustering algorithm of STRUCTURE (Pritchard et al., 2000) on all samples following the authors' methodology, in order to build color-explicit ancestry barplots and maps and to compute allele-frequency divergences between the inferred clusters. The software Treefit was used to build a neighbor-joining tree of pairwise Fst from the results of Maia-Carvalho et al. (2018). To visualize the genetic variation among individuals, especially among those assigned to different clusters, we performed a principal component analysis (PCA) with the R package adegenet (Jombart, 2008).
Finally, in the framework of the description of a new Pyrenean taxon (see Results and discussion section), we performed STRUCTURE and PCA analyses on the corresponding subset of populations. The specimen chosen as the holotype of this taxon was measured with an analogical caliper (1 mm precision). Mating calls recorded at the type locality were analyzed with Raven Pro 1.61 and the R package seewave.
The genetic data re-analyzed in this paper are available in Table S1.
3 RESULTS AND DISCUSSION
3.1 Phylogeographic structure in the A. obstetricans complex
- an unnamed Pyrenean lineage (color-coded beige), referred to as the Huesca lineage, as per its range in the Huesca Province of Spain;
- an unnamed southwestern lineage (color-coded green), often referred to as “cf. boscai,” as it encompasses the presumed range of subspecies A. o. boscai Lataste, 1879, although this name applies to another lineage (see below);
- a Catalonian lineage (color-coded yellow), corresponding to A. o. almogavarii Arntzen & García-París, 1995 (type locality: “Rasos de Peguera, Berga, Provincia de Barcelona, Spain”);
- a northwestern lineage (color-coded red), corresponding to A. o. boscai sensu stricto (type locality: “Tuy, in the Provincia Pontevedra, Spain,” García-París & Martínez-Solano, 2001);
- a northern lineage (color-coded orange), corresponding to the nominal subspecies A. o. obstetricans (type locality: “France”), which expands from the Spanish Cantabrian coast to northwestern Europe (France, Switzerland, Belgium, Netherland, and Germany);
- a southeastern lineage (color-coded blue), corresponding to A. o. pertinax García-París & Martínez-Solano, 2001 (type locality: “La Tola, Municipio de Casas Ibáñez, Provincia de Albacete, Spain”).
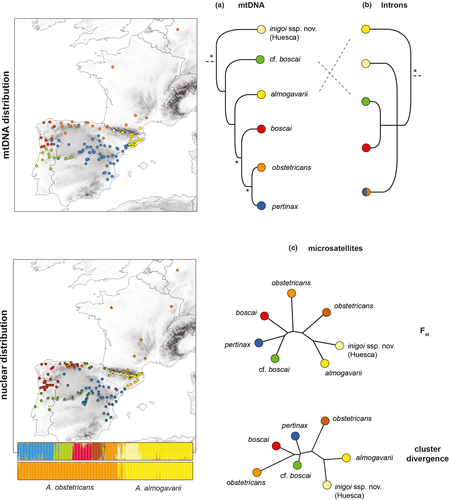
Despite extensive alignments (~9 kb), the position of two deeply diverged mitochondrial lineages (cf. boscai and almogavarii) could not be robustly inferred (Maia-Carvalho, Gonçalves, Ferrand, et al., 2014; Figure 1a).
The mitochondrial phylogeography received fairly convincing nuclear support (Maia-Carvalho, Gonçalves, Ferrand, et al., 2014). The phylogeny based on four introns (~3.4 kb) recovered all mitochondrial splits but the most recent one (obstetricans/pertinax) (Figure 1b). The topology is however conflicting with the mitochondrial tree, especially for two weakly supported nodes: the almogavarii lineage is the most early diverged in the nuclear tree (a pattern previously recovered from allozyme data; Arntzen & García-París, 1995), while the remaining four lineages form an unresolved polytomy. Such discrepancies potentially reflect an intricate history of hybridization, coupled with incomplete lineage sorting at the analyzed nuclear markers: all lineages differ by only few mutational steps (Gonçalves et al., 2007, 2015).
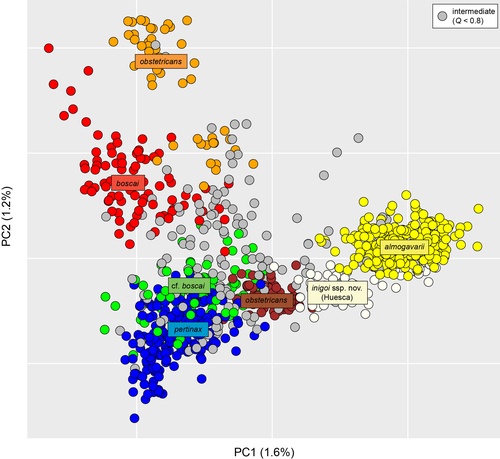
More recently, the fine-scale microsatellite survey by Maia-Carvalho et al. (2018) mapped seven well-defined genetic groups. These nicely matched the distribution of all six mitochondrial lineages and some additional substructure within the widespread A. o. obstetricans (northern Spain vs. northwestern Europe, Figure 1c). Our PCA on this dataset distinguished most of the Bayesian clusters, notably the unnamed Huesca populations (Figure 2).
According to the available molecular evidence, all six mitochondrial lineages thus qualify as valid taxa in the sense that nuclear analyses (intron phylogeny and microsatellite clustering) support their genetic and thus evolutionary independence. It remains unclear whether the microsatellite substructure found in A. o. obstetricans represents a seventh evolutionary unit, since populations from northern Spain do not harbor a specific mtDNA haplogroup nor diagnostic intron alleles (Gonçalves et al., 2015). This structure could therefore stem from recent population fragmentation, potentially linked to glacial or post-glacial distribution shifts (Maia-Carvalho et al., 2018). If we presently retain six taxa in the A. obstetricans complex, two unnamed lineages would thus be candidates for taxonomic descriptions: the Huesca lineage from the Central Pyrenees and the cf. boscai lineage from central Iberia.
3.2 How to revise the A. obstetricans complex?
Any satisfying update must account for the fact that we now know the A. obstetricans complex to comprise at least two distinct species. Recent hybrid zone analyses have suggested genetic incompatibilities between the lineages pertinax and almogavarii in Catalonia, which imply an advanced stage of reproductive isolation and thus incipient speciation (Dufresnes & Martínez-Solano, 2020). The latest species list for the European herpetofauna consequently recognized A. almogavarii and A. obstetricans (Speybroeck et al., 2020). Although the phenotypic variation in Alytes remains vastly understudied, these two resembling species might still feature some external disparities, as suspected from analyses of skull osteology (Martínez-Solano et al., 2004), bioacoustics (Márquez & Bosch, 1995), and coloration. Yet, given the unresolved phylogeny, how should the remaining lineages be treated within a two-species scheme?
If one trusts their weak mitochondrial divergence (Figure 1a), A. o. pertinax and A. o. boscai should both remain subspecies of A. obstetricans. For A. o. pertinax, this is further supported by the intron polymorphism, where alleles are shared with A. o. obstetricans (Figure 1b). The assignment of lineages branching deeper in the phylogeny is less straightforward. From the nuclear tree (Figure 1b), it is parsimonious to also assign the remaining cf. boscai to A. obstetricans: it forms a shallow nuclear clade with A. o. boscai (Figure 1b), clusters together with Spanish A. o. obstetricans and A. o. pertinax in the microsatellite analyses (Figures 1 and 2 and Table 1), and appears to admix over large distances with both A. o. boscai and A. o. pertinax (Figure 1c)—a cue that all three likely belong to the same species. In contrast, the old mitochondrial divergence of the Huesca lineage agrees with an early split from all A. obstetricans taxa. Instead, the Huesca clade bears the lowest microsatellite differentiation with the almogavarii populations (Table 1), unambiguously clusters with the latter in the two-group STRUCTURE analysis (Figure 1c), and both partly overlap on the PCA (Figure 2). The discriminant PCA in Maia-Carvalho et al. (2018) already suggested stronger similarities between the Huesca populations and those assigned to almogavarii (Figure 2c). Accordingly, the Huesca toads are traditionally considered as almogavarii as per their allozyme genotypes (García-París, 1995) and phenotypic resemblance (coloration).
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
|---|---|---|---|---|---|---|---|
| A. almogavarii | |||||||
| 1 – almogavarii | — | 0.115 | 0.228 | 0.191 | 0.169 | 0.201 | 0.150 |
| 2 – inigoi ssp. nov. (Huesca) | 0.479 | — | 0.245 | 0.169 | 0.172 | 0.223 | 0.163 |
| A. obstetricans | |||||||
| 3 — obstetricans (NW-Europe) | 0.800 | 0.851 | — | 0.231 | 0.177 | 0.151 | 0.145 |
| 4 — obstetricans (N-Spain) | 0.721 | 0.650 | 0.809 | — | 0.154 | 0.189 | 0.131 |
| 5 — pertinax | 0.778 | 0.782 | 0.754 | 0.692 | — | 0.090 | 0.062 |
| 6 – boscai | 0.864 | 0.923 | 0.623 | 0.792 | 0.528 | — | 0.077 |
| 7 – cf. boscai | 0.761 | 0.809 | 0.697 | 0.656 | 0.471 | 0.521 | — |
Note
- Below diagonal, pairwise Fst (taken from table 3 of Maia-Carvalho et al., 2018); above diagonal, allele-frequency divergence (hereby computed with STRUCTURE).
Hence, from these observations, the cf. boscai lineage corresponds to an unnamed subspecies of A. obstetricans, while the Huesca lineage corresponds to an unnamed subspecies of A. almogavarii. Both cases imply that the position of the almogavarii matriline in the phylogeny does not reflect the evolution of this taxon, which presumes that it lost its mitochondrial identity by past hybridization with the common ancestor of A. o. obstetricans/boscai/pertinax (Maia-Carvalho, Gonçalves, Ferrand, et al., 2014).
Two sets of future studies are needed to certify this taxonomic scenario. First, a genomic phylogeography of the genus should provide a decisive species tree. If our interpretation is correct, the Huesca and almogavarii lineages should branch together in a robust sister clade, well differentiated from all the remaining mitochondrial lineages (obstetricans, pertinax, boscai, and cf. boscai). Such study would also prove useful to elucidate the nature of the two microsatellite groups of A. o. obstetricans. Second, analyses of secondary contact zones in the Pyrenean region can provide additional insights on the Alytes speciation continuum. Assuming a gradual evolution of reproductive isolation through time (which seems to be the rule in European anurans; Dufresnes, Brelsford, et al., 2021), we predict more extensive introgression between the Huesca and almogavarii lineages (if they are indeed conspecific) in the Central-Eastern Pyrenees than between the Huesca and obstetricans lineages (if they indeed belong to two different species) in Central-Western Pyrenees. The microsatellite phylogeography already located both contact zones (Figure 1c), which remain to be targeted by fine-scale transect sampling for comparing their degree of admixture.
In the wait of such studies, the available data agree on one thing: the Huesca lineage represents a novel taxon endemic to the Central Pyrenees, and it is probably a subspecies of A. almogavarii. In the following, we provide a formal taxonomic description for this lineage, as the subspecies Alytes almogavarii inigoi ssp. nov.
3.3 Description of Alytes almogavarii inigoi ssp. nov. Dufresnes & Hernandez, 2021
3.3.1 ZooBank registration
This paper is registered to Zoobank. Link: LSID: http://zoobank.org/urn:lsid:zoobank.org:pub:18747A8C-6B72-4288-98DB-B57AFECA857B
3.3.2 Identity and diagnosis
A small, stocky built toad reaching about 5 cm of body length that is characterized by large bulging eyes with a vertical pupil, thin parotids, a visible tympanum, a slightly warty skin, three rounded metacarpal tubercles, and one small olive-shaped metatarsal tubercle. Dorsal coloration is whitish to pale gray, broadly covered by irregular greenish blotches reaching down the flanks. Dorsolateral warts can show pale orange spots that extend outside the warts, especially on the parotids. Ventral coloration is white with darker spots on the throat and chest. Sexes are presumably indistinct, although only males carry clutches on their back. Most of these characteristics are shared by other members of the Alytes obstetricans complex, with which the new taxon was previously confounded. In particular, the green coloration on the flanks and orange spots extending outside the warts are presumably typical of A. almogavarii. As thorough phenotypic comparisons remain to be established, the identity of the new taxon so far appears essentially genetic. Specifically, the new taxon features substantial divergence at mitochondrial genes (≥3.3% at ND4) and slow-evolving nuclear introns, that is, private haplotypes at three out of the four genes analyzed, with at least three and two mutational steps from other lineages for the commonly used beta-fibrinogen intron 7 and cellular myelocytomatosis oncogene genes, respectively (Gonçalves et al., 2015). It is also distinguished by unique microsatellite profiles that reflect strong nuclear differentiation from other lineages of the A. obstetricans complex (Maia-Carvalho et al., 2018; Figures 1 and 2 and Table 1): the corresponding STRUCTURE cluster features an allele frequency divergence of 0.11 with the almogavarii cluster and 0.16–0.25 with the clusters assigned to A. obstetricans (average 0.19). For comparison, the divergences found between the A. obstetricans subspecies were 0.06–0.23 (average 0.14). As discussed previously, and although Alytes lacks a decisive phylogeny, these relative genetic distances suggest to preliminary consider the new taxon as a subspecies of A. almogavarii, which is also consistent with their similarities in coloration.
3.3.3 Holotype
MNCN 50762, adult male collected on August 3, 2021, near a small reservoir along road Hf-0104AA, 4.5 km from Escuaín, Huesca province, Spain (42.573°N, 0.139°E, 1260 m a.s.l.; beige star in Figure 3); curated at the herpetological collection of the Museo Nacional de Ciencias Naturales (MNCN); and depicted in Figure 4.


3.3.4 Description of the holotype
Small-sized body (snout-vent length: 38 mm) with a broad oval-shaped head (16 mm wide); blunt, rounded snout, and with nostrils small and round; eyes large, interorbital space flat, and much wider (10 mm) than the internarial distance (4 mm); tympanum well defined, round, and with a diameter (3 mm) half of the eye diameter (6 mm); tympanum annulus is distinct and thick at the upper margin; vomerine teeth present, disposed at a 150° angle, but not in contact; three prominent metacarpal tubercles, the middle one slightly smaller; four fingers with a relative length of II < II = IV < III (9 mm for the longest) and a single subarticular tubercle on III; hind limbs relatively short, tibia slightly longer (16 mm) than the thigh (14 mm) and the tarsus (12 mm); five toes moderately long with rounded tips and relative length as V < IV < I < III < II (14 mm for the longest); webbing almost absent, with webbing formula as I 3–3 II 2–3 III 3–3 IV 3–3 V; and small inner metatarsal tubercle (2 × 1 mm). Dorsum and ventrum slightly granulated; thin parotids, almost indistinct; dorsum, flanks, and upper side of limbs colored with numerous greenish, brownish, or olive blotches οn a pale gray background, with irregular orange warts along the dorsolateral ridges; ventral surface whitish, slightly translucent, with darker pectoral and abdominal areas.
3.3.5 Etymology
No midwife toad has ever been described within the range of the Huesca lineage, so no taxonomic name is presently available. We thus coin a new nomen, Alytes almogavarii inigoi, as a tribute to Dr. Íñigo Martínez-Solano, a distinguished specialist of the Iberian herpetofauna and the leading researcher on Alytes phylogeography and systematics for over two decades. Íñigo was the first to report the Huesca lineage (Martínez-Solano et al., 2004). Moreover, the holotype was coincidently collected on his birthday.
3.3.6 Larvae
Similar to the other taxa of the A. obstetricans complex. According to our observations of c.a. 20 individuals nearby the type locality, tadpoles can reach sizes of ~10 cm with a robust tail flanked by relatively reduced fins. The spiracle is mid-ventral; nostrils are closer to the snout tip than the eyes; dental formula is 2/3 (2 upper and 3 lower dental rows); pale coloration with brown speckles on the tail muscle and marbled spots on the belly; depicted in Figure 5.

3.3.7 Advertisement call
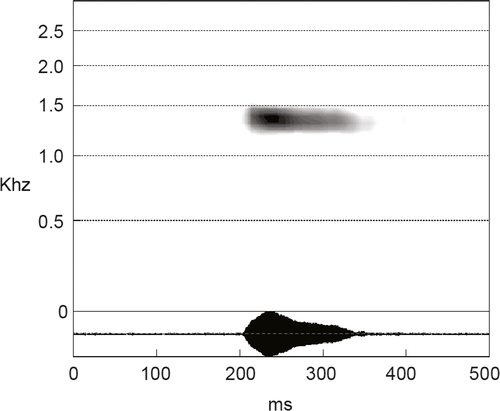
Presumably similar to the other taxa of the A. obstetricans complex, especially A. a. almogavarii, which may feature longer notes than A. obstetricans. Bioacoustic analyses of two males recorded at the type locality indicated an average fundamental frequency of 1.15 Khz, an average dominant frequency of 1.34 Khz, an average note duration of 142 ms (181 pulses, with a rising time of 21 ms). A spectro-oscillogram of the call is shown in Figure 6.
3.3.8 Natural history
Not explicitly documented and presumably similar to other taxa of the A. obstetricans complex. The new taxon was genetically confirmed from 700–1400 m a.s.l, although it probably reaches higher altitudes in suitable habitats. Typical environments include limestone rocky slopes featuring large pools or streams surrounded by semi-xeric shrubland or mesothermal forests, as dominated by pine (Pinus spp.) and oak trees (Quercus spp.). Beside stream puddles (where they coexist with Calotriton asper), tadpoles were also observed in fairly large numbers in washtubs and reservoirs, even in the presence of fish. Like other Alytes, adults are exclusively crepuscular/nocturnal and live a secretive terrestrial life near water bodies, often hidden in rock cracks and under stones from which they call. Species distribution modelling of A. o. inigoi ssp. nov. (as cluster F1n) flagged the bioclimatic variable bio15 (precipitation seasonality) as the most determinant factor of its climatic envelope (Maia-Carvalho et al., 2018). Environmental overlap with the presumed sister taxon A. a. almogavarii (as cluster F2n) was low (Schoener's D = 0.28) but supported neither niche conservatism nor divergence, which suggests broadly similar ecologies.
3.3.9 Distribution
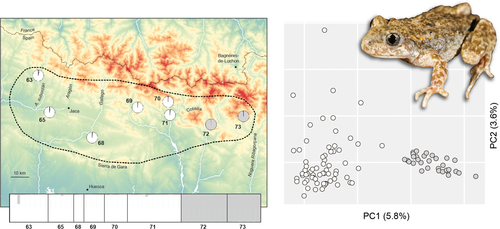
From current knowledge, Alytes almogavarii inigoi ssp. nov. is confined to a narrow ~100 km mountainous band on the southern slope of the Central Pyrenees, all in the highlands of Huesca Province—broadly from −0.9° to +0.7° in longitude and 42.4° to 42.6° in latitude, for a total area of ~3000 km2 (Figure 7). The taxon presumably reaches its western limit in the Noguera Ribagorçana valley (at the border with Leida Province) and its southern limit along the Sierra de Gara, both in parapatry with A. a. almogavarii. The western limit is probably located near the border with Navarre Province. In the north, it is unclear whether the new taxon reaches France in the Parc Nationale des Pyrénnées. Most Pyrenean passes are below the altitudinal limits of Alytes (up to 2500 m), and A. a. inigoi is actually contacted by A. o. obstetricans in the upper parts of several southern valleys adjacent to the French-Spanish border (e.g., Aragón, Aragón Subordán, and Rio Gállego).
3.3.10 Conservation and threats
The new subspecies is potentially threatened given its narrow distribution range and presumed sensitivity to emerging pathogens (Ranavirus, chytrid fungus), which are known to decimate Alytes in northern Spain (Bates et al., 2018; Bosch et al., 2021). Global warming could also have a negative long-term impact, by reducing its range through altitudinal shifts. Genetic diversity of populations is within levels documented for the rest of the A. obstetricans complex (expected heterozygosity He =0.48–0.66, average 0.57). Moreover, Iñigo's midwife toad is genetically structured: we recovered two isolated sets of populations separated by the Cotiella massif, which should be considered as independent conservation units (Figure 7). Nevertheless, A. a. inigoi does not seem under a direct risk of extinction given its local abundance and the remoteness and wilderness of its range, so far spared by urbanization and potentially overlapping three natural parks (Parque National de Ordesa y Monte Perdido; Parque Natural de los Valles Occidentales; and Parque Natural de la Sierra y los Cañones de Guara). Conservation efforts should focus on delimiting the actual boundaries of its range by additional DNA barcoding of populations, informing on the threat of pathogens, and limiting the ecological impact of tourism development in the Central Pyrenees. These would also benefit to other endemic Pyrenean amphibians that presumably share habitats with A. a. inigoi (e.g., Rana pyrenaica and Calotriton asper).
4 CONCLUSIONS
We have updated the incomplete taxonomy of midwife toads of the A. obstetricans complex, by reviewing, re-interpreting, and summarizing the rich phylogeographic literature published in recent years. Several lines of molecular evidence support the validity of six distinct evolutionary lineages, assigned to two different species: (1) A. obstetricans, which includes A. o. obstetricans, A. o. pertinax, A. o. boscai, and an unnamed taxon in southwestern Iberian (A. o. cf. boscai); (2) A. almogavarii, which includes A. a. almogavarii and a novel taxon that we hereby described as A. a. inigoi. The taxonomic description of this genuine micro-endemic should hopefully promote its conservation and call for similar assessments in other candidate Pyrenean lineages of amphibians (e.g., Rana temporaria; Vences et al., 2017). While many aspects of the evolution, diversity, and systematics of Alytes remain to be addressed (e.g., the description of the cf. boscai lineage, a resolved species tree shedding light on the deep cyto-nuclear discordances), the present contribution serves as a checkpoint to guide future investigations, among which phylogenomics, next-generation phylogeography, and hybrid zone genomics hold great promises.
ACKNOWLEDGEMENTS
We are grateful to I. Martínez-Solano for his long-lasting support, our fruitful collaborations, and more specifically here for sharing some of the published data re-analyzed in this article. Thanks must also go to J. Ambu for her assistance with the morphometric and bioacoustic measurements of the new taxon.




