Geological and paleoclimatic events reflected in phylogeographic patterns of intertidal arthropods (Acari, Oribatida, Selenoribatidae) from southern Japanese islands
Contributing authors: Maximilian Wagner ([email protected]), Shimpei F. Hiruta ([email protected]), Iris Bardel-Kahr ([email protected]), Wataru Hagino ([email protected]), Satoshi Shimano ([email protected])
Zoobank Link: LSID: http://zoobank.org/urn:lsid:zoobank.org:pub:69E97C95-338A-450C-99D5-FA628D2E5AD9
Online ISSN: 1439-0469
Abstract
enA comprehensive study of the intertidal oribatid mite fauna of southern Japanese islands revealed the presence of the selenoribatid Arotrobates granulatus Luxton, 1992 and two yet undescribed species. The latter are herein described as Indopacifica taiyo n. sp., occurring from the Southern to the Central Ryukyus, and Indopacifica tyida n. sp., which was only found on the most western island of the Ryukyus, namely Yonaguni. A concomitant molecular genetic study using mitochondrial COI and 18S rRNA gene sequences, demonstrated that the phylogeographic pattern of I. taiyo n. sp. reflects recent expansion on the Southern and Central Ryukyus, probably due to existing land bridges during the late Pleistocene. Arotrobates granulatus, on the other hand, shows three distinct lineages, one on Japanese mainland, another on the island of Amami, and the third on part of the Central and Southern Ryukyus. These lineages are most likely the result of the break-up of a large peninsula reaching from China to the Northern Ryukyus about 1.2–1.7 million years ago. Despite emerging land bridges in the late Pleistocene, this species was not able to expand its range again which indicates very low dispersal abilities. Morphometric data of I. taiyo n. sp. show considerable intraspecific variation between island populations correlating with geography. This found variation is suggested to be a result of phenotypic plasticity caused by diverging local environmental factors. From an ecological perspective, all three found species are classified as intertidal rock-dwellers feeding on diverse algae, whereas I. taiyo n. sp. and Arotrobates granulatus occasionally occur in mangrove habitats.
Zusammenfassung
deEine umfangreiche Studie der litoralen Hornmilbenfauna der südlichen japanischen Inseln zeigte das Vorkommen von Arotrobates granulatus und zweier bisher unbeschriebener Arten. Diese werden hier beschrieben als Indopacifica tayio n. sp., welche von den südlichen bis zu den zentralen Ryukyus vorkommt, und als Indopacifica tyida n. sp., welche nur auf der Insel Yonaguni gefunden wurde. Eine begleitende molekulargenetische Studie der mitochondrialen COI und 18S rRNA Gensequenzen zeigte, dass das phylogeographische Muster von Indopacifica taiyo n. sp. eine rezente Ausbreitung auf den südlichen und zentralen Ryukyus widerspiegelt, die vermutlich durch, während des späten Pleistozäns bestehender, Landbrücken möglich war. Arotrobates granulatus hingegen zeigt drei klar getrennte Linien, eine auf dem japanischen Festland, eine andere auf der Insel Amami, und eine dritte auf Teilen der zentralen und südlichen Ryukyus. Diese Linien sind vermutlich ein Resultat des Zerfalls einer großen Halbinsel, die vor ca. 1.2-1.7 Millionen Jahren von China bis zu den nördlichen Ryukyus reichte. Trotz der vorübergehenden Landbrücken im späten Pleistozän, war es dieser Art nicht möglich sich wieder auszubreiten, was auf ein geringes Verbreitungspotential hinweist. Morphometrische Daten von Indopacifica taiyo n. sp. zeigen erhebliche intraspezifische Variation zwischen den Populationen verschiedener Inseln, welche stark mit der Geographie korreliert. Vermutlich ist diese Variation eine Folge phänotypischer Plastizität, verursacht durch sich unterscheidende lokale Umweltfaktoren. Vom ökologischen Standpunkt aus, können alle drei gefundenen Arten als litorale Felsbewohner eingestuft werden, die verschiedene Algen fressen, wobei Indopacifica taiyo n. sp. und Arotrobates granulatus gelegentlich auch in Mangroven vorkommen können.
1 INTRODUCTION
The Selenoribatidae represent, together with Ameronothridae, Fortuyniidae, and Podacaridae, an exceptional oribatid mite group as they have adapted to a marine-associated lifestyle instead of living a typical terrestrial life as soil decomposers (e.g., Pfingstl, 2017). While Ameronothridae and Podacaridae are distributed predominantly on polar and temperate coasts, Fortuyniidae and Selenoribatidae exclusively occur on subtropical and tropical shorelines (e.g., Pfingstl & Schuster, 2014; Schuster, 1989). These mites can tolerate daily tidal flooding by using plastron respiration (Pfingstl & Krisper, 2014; Pugh et al., 1990) and mainly use intertidal algae as substrate and food source (e.g., Pfingstl, 2017). The Selenoribatidae are the most diverse group among these marine-associated oribatid mites, with nine genera and 32 species worldwide. Most members of this family were reported from the Indo-Pacific area (Pfingstl & Schuster, 2014; Procheş & Marshall, 2001); records from neighboring regions, as for example the Japanese Islands, are relatively scarce. Four species, Arotrobates granulatus Luxton, 1992, Schusteria nagisa Karasawa & Aoki, 2005, Schusteria saxea Karasawa & Aoki, 2005 and Rhizophobates shimojanai Karasawa & Aoki, 2005 were found on the southern Ryukyu Islands (Karasawa & Aoki, 2005); the latter three species are only known from this area and so far considered as endemic species for the Japanese Islands. There are yet no records of selenoribatid mites from the close continental coast of China, except for the reports of Arotrobates reticulatus Luxton, 1992, A. granulatus and Psednobates uncunguis Luxton, 1992 from the shore of Hong Kong (Luxton, 1992) and the report of an undetermined species from the littoral of Xiamen (Pfingstl & Schuster, 2014). Therefore, biogeographic connections between those neighboring regions remained mostly unknown.
The Ryukyu Islands represent a chain of more than 200 smaller landmasses stretching for about 1200 km from 24° to 31° latitude between Taiwan and Japanese mainland (Kimura, 2002). These islands are divided into three parts, the Northern, Central, and Southern Ryukyus. The Northern and Central parts are separated by the Tokara gap, the Central, and Southern Ryukyus are divided by the Kerama gap (Kojima et al., 2003). These two gaps were formed during the Pliocene (Ota, 1998) and are suggested to be zoogeographical boundaries that might have played important roles in establishing the biogeographical characteristics of the Ryukyus during the Cenozoic (e.g., Ota, 1998). There is a high number of endemic species across invertebrate and vertebrate taxa, and the fauna is largely divided among the above-mentioned three geographic areas which is supposed to be partly caused by these geographic gaps (Muraji et al., 2012).
A recent phylogeographic study on intertidal mites from the family Fortuyniidae (Pfingstl, Wagner, et al., 2019) showed a slightly divergent picture. There were no endemic species and while the Tokara gap was strongly reflected in the morphological and molecular genetic sequence data, indications of the Kerama gap acting as an effective barrier to dispersal were completely lacking. This pattern was suggested to be the result of low sea level during the last Pleistocene (Ujiié et al., 2003) which allowed recent expansion and gene flow between several island populations of fortuyniid mites (Pfingstl, Wagner, et al., 2019). Apparently, these tiny wingless organisms show better dispersal abilities than partially volant terrestrial animals, such as insects and birds, and it was suggested that their ability to drift on water may be a key factor (Pfingstl, Wagner, et al., 2019).
During the field work for this latter study, we were able to additionally collect numerous populations of selenoribatid mites from various Japanese Islands, which gave us the opportunity to compare their phylogeographic patterns with those of the above-mentioned closely related Fortuyniidae. Therefore, aims of the present study were (a) to determine collected taxa and describe possible new species, (b) to infer phylogeographic scenarios for each found selenoribatid group and compare these to patterns of fortuyniid mites, (c) to interpret the found similarities or divergences, and (d) to update biogeographic information for Japanese intertidal mites.
2 MATERIAL AND METHODS
For the present molecular genetic study, 121 selenoribatid specimens were analyzed and for the morphometric study 75 specimens were used. Additionally, we included sequences (COI, 18S) from GenBank from two Indopacifica pantai specimens (accession nrs: MH285652, MH285654, MH285691, MH285692), from one Indopacifica parva specimen (MH285671, MH285690) and from one Thalassozetes barbara specimen (MW289085, MW298484). For more details see Appendix 1.
2.1 Sample locations
Samples of littoral algae were scraped off rocks, concrete walls, and mangrove roots with a small shovel and then put in Berlese-Tullgren funnels for 12–24 h to extract mites. Afterward, specimens were fixed in ethanol (100%) for morphological and molecular genetic investigation. The majority of samples was taken by Shimano, Hiruta, and Pfingstl and if not, the name of the collector is provided.
Each sample location was given a code; these codes are presented in parentheses and will be used throughout the manuscript to allow easier linking of geographic information. The suffixes “-jima/-shima” and “-gawa” are Romanized Japanese meaning “island” and “river,” respectively; in the graphs and tables showing results, these suffixes are not given due to a shortage of space.
Japanese mainland
- Omi-shima, pref. Ehime: (JP_33) black intertidal algae on mussels and barnacles on concrete wall; 34°13′26.60″N 132°59′0.68″E; 17 Sept. 2018.
- Shikoku, pref. Ehime: Shiraishi-no-hana, Wake, Matsuyama City (JP_20) short green algae growing on barnacles and mussels on rock; 33°52′34.97″N 132°42′12.93″E; 16 Sep. 2018.
-
Kyushu, pref. Kagoshima: Iriki, Fukiage-cho, Hioki City (JP_86) algae on mussels and barnacles on concrete seawall near the mouth of the river; 31°30′14.70″N 130°19′10.56″E; 10 Jun. 2018.
Southern Ryukyus
- Iriomote-jima, pref. Okinawa: Estuary of Goyoshi-gawa (JP_38) brown filamentous algae on mangrove floor, 24°19′17.43″N 123°54′38.87″E; 16 Mar. 2019. Ohara (JP_45) diverse algae growing on concrete pier; 24°16′32.38″N 123°52′58.87″E; 16 Mar. 2019. South shore (statue of the teacher) (JP_46) Bostrychia and other algae from large rocks; 24°16′0.02″N 123°50′45.71″E; 16 Mar. 2019. Nadara-gawa estuary (JP_47) Bostrychia and other algae from boulder; 24°23′52.93″N 123°49′42.72″E; 16 Mar. 2019. Uehara (JP_50) Bostrychia and other algae on artificial stone wall; 24°23′49.18″N 123°49′19.59″E; 17 Mar. 2019.
- Yonaguni-jima, pref. Okinawa: Kataburu Beach (JP_56) Bostrychia on cliffs; 24°26′19.18″N 122°58′21.81″E; 18 Mar. 2019. Kataburu Beach (JP_57) Bostrychia and smaller plants on small rocks; 24°26′25.78″N 122°58′27.44″E; 18 Mar. 2019. Sonai (JP_59) dark black filamentous algae on rocks; 24°28′15.69″N 122°59′55.38″E; 19 Mar. 2019.
-
Ishigaki-jima, pref. Okinawa: Ohama Beach (JP_36) Bostrychia on rocks on sandy beach; 24°20′40.61″N 124°12′5.58″E; 15 Mar. 2019. Kabira Bay (JP_62) Bostrychia on large rocks on sandy beach; 24°27′49.14″N 124° 8′39.09″E; 20 Mar. 2019. Yamabare (JP_65) thick cushions of Bostrychia on large rocks; 24°26′56.11″N 124°10′46.41″E; 20 Mar. 2019. Fukido-gawa estuary (JP_66) diverse algae growing on mangrove roots; 24°29′10.48″N 124°13′48.29″E; 20 Mar. 2019. Kabira (JP_90) unknown algae on rocks; 24°28′25.7″N 124°08′10.5″E; 31 Mar. 2019; leg. H. Uchida.
Central Ryukyus
- Okinawa-jima, pref. Okinawa: Sesoko island, beach next to Marine Biological Station (JP_68) Bostrychia on rocks; 26°38′8.72″N 127°51′52.84″E; 21 Mar. 2019. Oku (JP_71) Bostrychia growing in crevices of rocky shore; 26°50′44.60″N 128°17′22.00″E; 22 Mar. 2019. Sosu (JP_72) Bostrychia in large crack on huge single rock; 26°48′30.23″N 128°19′6.00″E; 22 Mar. 2019. Northern west coast (JP_73) Bostrychia on rock; 26°49′50.91″N 128°14′43.35″E; 22 Mar. 2019. Sosu, Ie no hama (JP_76) Bostrychia on rocks; 26°47′40.24″N 128°19′7.65″E; 22 Mar. 2019.
- Amami-oshima, pref. Kagoshima: Cape Ayamaru (JP_78) Bostrychia from crevices on rocks (rocky shore); 28°28′23.36″N 129°43′8.03″E; 23 Mar. 2019. Isu Bay (JP_81) Bostrychia on large boulder; 28°10′12.79″N 129°21′34.81″E; 24 Mar. 2019. Koniya Port (JP_82) Bostrychia from small crevices on rough boulder; 28°8′51.19″N 129°18′18.99″E; 24 Mar. 2019. Kuji Bay (JP_84) Bostrychia in crevices on rocks; 28°13′27.19″N 129°12′49.47″E; 24 Mar. 2019. Nagara Bay (JP_85) black bushy algae on concrete cay wall; 28°15′21.20″N 129°12′7.56″E; 24 Mar. 2019.
2.2 Molecular genetic analyses
For this study, 121 specimens of the family Selenoribatidae (67 of genus Indopacifica, 53 of the genus Arotrobates and one outgroup specimen of T. barbara) were analyzed. Whole genomic DNA was extracted from ethanol-fixed individuals using Chelex resin according to the adjusted protocols in Pfingstl, Lienhard, et al. (2019). We amplified a partial sequence of the mitochondrial DNA cytochrome c oxidase subunit I (COI) gene (600 bp amplicon length) and the complete 18S ribosomal RNA gene (ca. 1900 bp amplicon length) of major mitochondrial lineages, following the protocols of Pfingstl, Lienhard, et al. (2019). PCR conditions and primers as well as sources are summarized for each locus in Table S1. Subsequent DNA purification steps included enzymatic ExoSAPIT (Affymetrix) and Sephadex G-50 resin (GE Healthcare). Cycle sequencing, using BigDye Sequence Terminator v3.1 kit (Applied Biosystems), was conducted according to Schäffer et al. (2008). Automatic capillary sequencing and sequence visualization was operated on an ABI3500XL (Applied Biosystems) device. Furthermore, already published sequences of I. parva (18S rRNA: MH285690 and COI: MH285671) and I. pantai (18S rRNA: MH285691, MH285692 and COI: MH285654, MH285652) (Pfingstl, Lienhard, et al., 2019) were downloaded from GenBank and included in the alignment. MUSCLE (Edgar, 2004), integrated in the software MEGA 7.0 (Kumar et al., 2016), was used to align sequences. Concatenation of the single locus datasets and indel coding of the 18S alignment was performed in the perl-based program 2matrix (Salinas & Little, 2014). Finally, three alignments were created and analyzed (a) COI (564 bp), (b) 18S (1964 bp; incl. indels and binary coded gaps), and (c) COI + 18S (2528 bp; incl. indels and binary coded gaps). For more detailed information, see alignments in Data S1S–3. All generated sequences have been deposited at GenBank and are accessible at following numbers, MW289085–MW289203 (COI) and MW298484–MW298513 (18S).
The best fitting model of molecular evolution was determined based on Bayesian Information Criterion (BIC) in PartitionFinder v2.1.1 (Lanfear et al., 2017). Additionally, for the COI dataset, based on all three codon positions as starting scheme, a “greedy” partition search (Lanfear et al., 2012) in PartitionFinder v2.1.1 was conducted. The results of these analyses are summarized in Table S1. Phylogenetic inference was conducted for each single locus and the concatenated dataset in RaxML HPC v.8.0. (Stamatakis, 2014) and MrBayes v3.2 (Ronquist et al., 2012). Node support for the maximum likelihood framework was assessed using 10.000 bootstrap replicates and GTR-GAMMA model of evolution. Posterior probabilities for the Bayesian inference were obtained from Metropolis-coupled Markov chain Monte Carlo (MCMC) simulations using two independent runs, eight chains, a sampling frequency of 10,000 and the respective model of evolution. The analyses were run for 20e−6, 15e−6, and 10e−6 generations for the concatenated, COI and 18S dataset, respectively. To ensure parameter convergence and stationarity of chains, split deviation frequencies lower than 0.01 were assured and effective sample sizes (ESS) were investigated in Tracer 1.6 (Rambaut et al., 2014) (available at http://beast.bio.ed.ac.uk/Tracer). The first 25% of obtained trees of the MrBayes run were discarded and the rest summarized in terms of a 50% majority rule consensus tree. All trees were visualized in FigTree v1.4.2 (Rambaut, 2014) (available at http://tree.bio.ed.ac.uk/software/figtree). Net interspecific p-distances between groups, using 1000 bootstraps replications, were calculated in MEGA 7.0. Minimum interspecific and the maximum intraspecific divergence (in %) were calculated in R vs. 3.6.1 (R Core Team, 2017) using the functions “nonConDist” and “maxInDist” from the R-package SPIDER v.1.5 (Brown et al., 2012). Raw intraspecific genetic distances were obtained for both genera by the function “dist.dna” implemented in the R-package APE v.5.4 (Paradis et al., 2004) and visualized using the package PHEATMAP v.1.0.8 (Kolde & Kolde, 2015). Furthermore, to infer the phylogeographic relationships among genera, TCS networks (Clement et al., 2002) were calculated in PopART (Leigh & Bryant, 2015; available at http://popart.otago.ac.nz).
2.3 Morphometric analyses
Specimens were embedded in lactic acid for temporary slides, and measurements were made using a compound light microscope (Olympus BH-2) and ocular micrometer. A set of 15 continuous variables (bl—body length, dPtI—distance between pedotecta I, dbi—distance between inner borders of bothridia, dbo—distance between outer borders of bothridia, nwda—notogastral width on level of seta da, nwdm—notogastral width on level of seta dm, nwdp—notogastral width on level of seta dp; cl—camerostome length, cw—camerostome width, dcg—distance between camerostome and genital orifice, dac3—distance between acetabula 3, gl—genital orifice length, gw—genital orifice width, al—anal opening length, aw—anal opening width, see Figure S1) were measured in 75 Indopacifica specimens from six different populations originating from four different Ryukyu islands (Yonaguni-jima, Iriomote-jima, Ishigaki-jima, and Okinawa-jima). For species discrimination, all populations from a species were pooled; for the comparison of populations in the wider distributed Indopacifica taiyo n. sp., 61 specimens from five different locations from above given four islands were used for analysis. Specimens used for morphometric comparison were not the same as used for molecular genetic analyses, but they belonged to the exact same populations (same sample, approx. 10 cm2 patch of alga). Specimens were sexed based on internal structures (spermatopositor, ovipositor), and sexes were separated for morphometric analyses.
For univariate statistics of Indopacifica, minimum, maximum, mean, and standard deviation for each variable were calculated. Multivariate analyses investigating differences between putative Indopacifica species included a Principal Component Analysis (PCA; using a variance-covariance matrix) and a Non-metric Multidimensional Scaling (NMDS; based on Euclidian distances, two-dimensional); both analyses were performed on log10-transformed size-corrected data. No rotation was applied to the multivariate data. Size correction was done by dividing each variable through the geometric mean of the respective specimen.
For the investigation of divergence among different populations of Indopacifica, a Kruskal–Wallis and a Mann–Whitney U-test (Bonferroni-corrected p-values) were used for comparing the means of variables for pairwise comparisons to clarify whether single variables differ significantly between the populations. Additionally, a Discriminant Analysis (LDA) was performed on log10-transformed raw data to show size and shape differences between populations. All analyses were performed with PAST version 3.11 (Hammer et al., 2001).
Due to the relatively low number of A. granulatus specimens available, we were not able to perform a morphometric analysis of this species.
2.4 Drawings and photographs
Preserved specimens were embedded in Berlese mountant for microscopic analysis in transmitted light. Drawings were performed with an Olympus BH-2 Microscope equipped with a drawing attachment, and then they were scanned and afterwards processed and digitized with the free and open-source vector graphics editor Inkscape (https://inkscape.org).
For photographic documentation, specimens were air-dried and photographed with a Keyence VHX-5000 digital microscope.
Morphological terminology used in this paper follows that of Grandjean (1953) and Norton and Behan-Pelletier (2009).
3 RESULTS
3.1 Description of new taxa
Family Selenoribatidae Schuster, 1963
Genus Indopacifica Pfingstl, Shimano & Hiruta
3.1.1 Indopacifica taiyo Pfingstl, Shimano & Hiruta n. sp.
LSID: urn:lsid:zoobank.org:act:A71290A4-6B56-473F-9ABF-4C02992AF37B
Type material. Holotype: female, Japan, Ryukyus, Iriomote-jima, pref. Okinawa, Nadara-gawa estuary (JP_47) Bostrychia and other algae from boulder; 16 Mar. 2019. Four paratypes: two males and two females, same location and date as holotype. All deposited at the Collection of Arachnida, Department of Zoology, National Museum of Nature and Science, Tokyo (NMST). Two additional paratypes from Yonaguni-jima, pref. Okinawa, Kataburu Beach (JP_56); 18 Mar. 2019, deposited in the collection of the Naturhistorisches Museum Wien/NHM Vienna.
Etymology. The sound of specific epithet “taiyo” has the meaning of both “big ocean” and “the sun” in the general Japanese and “big ocean” refers to the wider coastal distribution from the Southern to the Central Ryukyus. It is given as noun in apposition.
Diagnosis. Prodorsal setae short; bothridial seta clavate and barbed; gastronotic region round in dorsal view; transversal depression on anterior part of notogaster; median longitudinal, hourglass-shaped depression on epimeron I; two pairs of adanal setae; proximoventral tooth on claws present.
Description of adult. Measurements. Females (n = 26), length: 289–325 µm (mean 304 µm), width: 185–203 µm (mean 192 µm); males (n = 33), length: 277–319 µm (mean 296 µm), width: 175–197 µm (mean 189 µm).
Integument. Color brown. Cerotegument of prodorsum, ventral region and legs granular. Notogastral cerotegument densely granulated with larger granules irregularly surrounded by smaller granules.
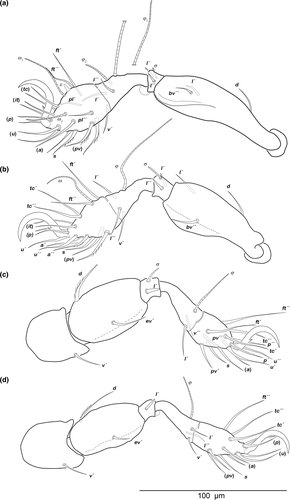
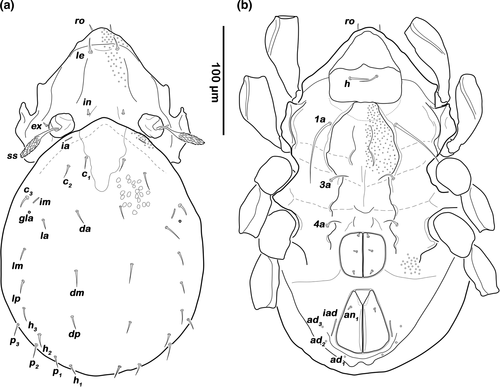
Prodorsum (Figure 1a). Rostrum rounded in dorsal view, clearly demarcated from remainder of prodorsum by transverse ridge. Pair of anteriorly converging faint prodorsal ridges, difficult to observe. Rostral (ro) and lamellar seta (le) simple and short. Interlamellar seta (in) minute, exobothridial seta (ex) minute. Bothridium large cup with lateral incision. Bothridial seta clavate, distally barbed.

Gnathosoma. Chelicera chelate, with two teeth on each digit. Setae cha and chb of approximately same length, both dorsally slightly pectinate. Palp setal formula 0–2–1–3–8 (+solenidion ω). Distal part of rutellum developed as thin, triangular, slightly inwardly curved membrane with longitudinal incision. Setae a and m long, smooth. Mentum regular, finely granular, seta h simple, long.
Notogastral region (Figure 1a). Notogaster rounded, nearly circular in dorsal view. Pair of indistinct humeral projection. A median transversal depression present between lyrifissure ia. Fourteen pairs of thin, short setiform notogastral setae (length 10–16 µm), c1, c3, da, dm, dp, la, lm, lp, h1–3, p1–3; c3 absent. Orifice of opisthonotal gland gla anterior to seta la. Lyrifissure im between setae la and lm.
Lateral aspect (Figure S2). A broad lateral furrow reaching from dorsal to ventral sejugal scissure, caudally delimited by conspicuous ridge. Pedotectum I present, round, small. Lateral enantiophysis consisting of two opposite projections; the anterior rounded, the posterior pointed. Discidium di developed as prominent conical bulge.
Podosoma and venter (Figure 1b). Median longitudinal, hourglass-shaped depression on epimere I covered with granules and inconspicuous semicircular depression on posterior border of epimere III. Three pairs of short, fine genital setae present. Preanal organ triangular in ventral view, interior part anchor-shaped. Two pairs of short adanal setae ad1–2 present. Lyrifissure iad slightly oblique, next to anterior border of anal opening.
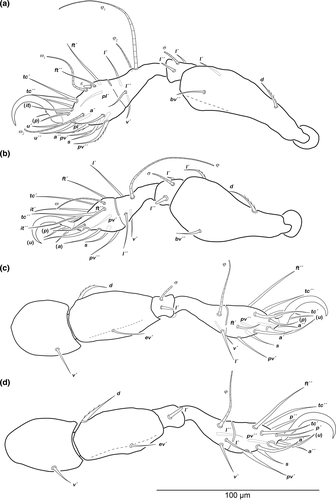
Legs (Figure 2). Long hook-like claw with one small proximoventral tooth. Cerotegument granular. No porose areas detectable. Setation and solenidia: Leg I (0–3–2–3–18) (1–2–2), leg II (0–3–2–3–15) (1–1–1), leg III (1–2–1–2–13) (1–1–0), leg IV (1–2–1–3–12) (0–1–0) (for details see Table 1).
| Trochanter | Femur | Genu | Tibia | Tarsus | |
|---|---|---|---|---|---|
| Leg I | – | d, bv″, l′ | (l), σ | (l), v′, φ1, φ2 | (pl), (pv), s, (a), (u), (p), (it), (tc), (ft), ε, ω1, ω2 |
| Leg II | – | d, bv″, l′ | (l), σ | (l), v′, φ | (pv), s, (a), (u), (p), (it), (tc), (ft), ω |
| Leg III | v′ | d, ev′ | l′, σ | l′, v″, φ | (pv), s, (a), (u), (p), (tc), (ft) |
| Leg IV | v′ | d, ev′ | l′ | (l), v′, φ | (pv), s, (a), (u), (p), (tc), ft″ |
Note
- Parentheses indicate pairs of setae; Greek letters refer to solenidia.
Distribution. The distribution of this species ranges from the Southern Ryukyus (Yonaguni-jima, Iriomote-jima, Ishigaki-jima) to the Central Ryukyus, whereas Okinawa-jima represents its northern limit (Figure 3).

Remarks. Indopacifica taiyo n. sp. can be distinguished from Indopacifica iohanna Resch & Pfingstl, 2019 by the differing notogastral depressions (one transverse vs. two oblique) and the lack of a proximoventral tooth on the claws in the latter. Indopacifica mauritiana Pfingstl & Baumann, 2019 and I. parva Pfingstl, Shimano & Lienhard, 2019 show a circular median depression on epimeron I instead of an hourglass-shaped depression in the present species, and I. pantai Pfingstl, Shimano & Lienhard, 2019 exhibits three pairs of adanal setae instead of only two and is significantly larger than I. taiyo n. sp.
3.1.2 Indopacifica tyida Pfingstl, Shimano & Hiruta n. sp.
LSID urn:lsid:zoobank.org:act:11CAAFEE-1B1C-44D0-B1A9-832439D1F01F
Type material. Holotype: female, Japan, Ryukyus, Yonaguni-jima, pref. Okinawa, Sonai (JP_59) dark black filamentous algae on rocks; 19 Mar. 2019. Four paratypes: two males and two females, same location and date as holotype. All deposited at the Collection of Arachnida, Department of Zoology, National Museum of Nature and Science, Tokyo (NMST). Two additional paratypes from the same location deposited in the collection of the Naturhistorisches Museum Wien/NHM Vienna.
Etymology. The specific epithet “tyida” refers to the Okinawan dialect word for “the sun” and is given as noun in apposition. We choose this term because the type locality Yonaguni-jima is the westernmost point of Japan where one can observe the most beautiful sunsets.
Diagnosis. Prodorsal setae short; pair of anteriorly converging prodorsal ridges; bothridial seta lanceolate and barbed; gastronotic region round in dorsal view; irregular oblique depressions in humeral areas of notogaster; median longitudinal, hourglass-shaped depression on epimeron I; two pairs of adanal setae; proximoventral tooth on claws present.
Description of adult. Measurements. Females (n = 9), length: 308–319 µm (mean 312 µm), width: 188–197 µm (mean 193 µm); males (n = 7), length: 277–313 µm (mean 296 µm), width: 179–191 µm (mean 183 µm).
Integument. Color brown. Cerotegument of prodorsum, ventral region and legs granular. Notogastral cerotegument densely granulated with larger granules irregularly surrounded by smaller granules, resulting in a weak reticulate pattern.
Prodorsum (Figure 1c). Rostrum rounded in dorsal view, clearly demarcated by from remainder of prodorsum by transversal ridge. Pair of anteriorly converging prodorsal ridges. Rostral (ro) and lamellar seta (le) simple and short. Interlamellar seta (in) minute, exobothridial seta (ex) minute. Converging bothridium large cup with lateral incision. Bothridial seta lanceolate, distally barbed.
Gnathosoma. Chelicera chelate, with two teeth on each digit. Setae cha and chb of approximately same length, both dorsally slightly pectinate. Palp setal formula 0–2–1–3–8 (+solenidion ω). Distal part of rutellum developed as thin, triangular, slightly inwardly curved membrane with longitudinal incision. Setae a and m long, smooth. Mentum regular, finely granular, seta h simple, long.
Notogastral region (Figure 1c). Notogaster rounded, oval in dorsal view. Oblique irregular depression in humeral areas. Fourteen pairs of thin, short setiform notogastral setae (length 10–15 µm), c1, c3, da, dm, dp, la, lm, lp, h1–3, p1–3; c3 absent. Orifice of opisthonotal gland gla anterior to seta la. Lyrifissure im between setae la and lm.
Lateral aspect (Figure S2). A broad lateral furrow reaching from dorsal to ventral sejugal scissure, caudally delimited by conspicuous ridge. Pedotectum I present, round, small. Lateral enantiophysis consisting of two opposite projections, both pointed. Discidium di developed as prominent conical bulge.
Podosoma and venter (Figure 1d). Median longitudinal, hourglass-shaped depression on epimeron I covered with granules and inconspicuous semicircular depression on posterior border of epimeron III. Three pairs of short, fine genital setae. Preanal organ triangular in ventral view, interior part anchor-shaped. Two pairs of short adanal setae ad1–2. Lyrifissure iad slightly oblique, next to anterior border of anal orifice.
Legs (Figure 4). Long hook-like claw with one small proximoventral tooth. Cerotegument granular. No porose areas detectable. Setation and solenidia: Leg I (0–3–2–3–18) (1–2–2), leg II (0–3–2–3–15) (1–1–1), leg III (1–2–1–2–13) (1–1–0), leg IV (1–2–1–3–12) (0–1–0) (for details see Table 1).
Distribution. This species was only found in a single location on Yonaguni the south westernmost island of Japan (Figure 3).
Remarks. Indopacifica tyida n. sp. can be distinguished from I. taiyo n. sp. by its faint prodorsal ridges (vs. absent in the latter), the differing notogastral depressions (two oblique vs. one transversal) and a significantly larger distance between camerostome and genital opening (as demonstrated later in the morphometry section). Indopacifica iohanna lacks the proximoventral tooth on the claws, and I. parva and I. mauritiana show a circular median depression on epimeron I (instead of an hourglass-shaped depression). Indopacifica pantai is considerably larger and shows three pairs of adanal setae instead of only two.
3.2 Remarks on the morphology of Arotrobates granulatus Luxton, 1992
The populations of A. granulatus from Japanese mainland (Figure 5) and the Central and Southern Ryukyus show conformity in their morphology and no distinct differences could be detected (Table 2). A slight deviation is shown only in the specimens from Kagoshima (JP_86) which possess very fine adanal setae instead of all being reduced to their alveoli. The specimens from Hong Kong described by Luxton (1992) differ from the herein investigated Japanese individuals by lacking lamellar setae (le) and one pair of notogastral seta and by having only alveolar interlamellar (in) and epimeral setae (3a, 4a). The specimens from the Japanese Ryukyus described by Karasawa and Aoki (2005) also lack the lamellar setae (le) and show only alveolar interlamellar setae (in), but they lack two pairs of notogastral setae and show variable adanal setation (Table 2). However, most of these differences are subtle and may be results of either intraspecific variation or inaccurate observation. In the case of adanal setae, Karasawa and Aoki (2005) already mentioned that they can be present or absent and this variation was also found in the present study which confirms intraspecific variation at least in some aspects. The lamellar setae which were not detected by the other authors (Karasawa & Aoki, 2005; Luxton, 1992) are very delicate structures and partly covered by cerotegument in most of the presently investigated populations and we also had initial difficulties to find them. Consequently, it is possible that these setae were present in the other studies as well but were overlooked due to their very fine nature. Moreover, the interlamellar setae in our specimens are very short and may also be easily mistaken for simple alveoli. The differences in notogastral setation, on the other hand, may represent real divergences though there are certain discrepancies. Seta c3 of the herein studied specimens is located in a very lateral position so that it is difficult to detect from a dorsal view. However, Karasawa and Aoki (2005) depicted a seta in the exact same position but labeled it as la. So, their lack of a seta of the c-row as well as the different position of the lyrifissure im could be explained by a mislabeling, but it still does not explain the lack of notogastral seta h1. Unfortunately, Luxton (1992) did not label any setae in the figure or the text; therefore, it is impossible to make clear assessments.
| Authors | Luxton (1992) | Karasawa and Aoki (2005) | Present study | |||
|---|---|---|---|---|---|---|
| Origin | Hong Kong | Iriomote/Okinawa | Omishima (JP_33) | Kagoshima (JP_86) | Amami (JP_81) | Ishigaki (JP_90) |
| Body length | 320–330 µm | 315–335 µm | 302 µm (N = 2) | 308 µm (N = 2) | 325 µm (N = 2) | 308–325 µm (N = 4) |
| Lamellar seta le | Not seen | Not seen | Normal | Normaldto | Normaldto | Normaldto |
| Interlamellar seta in | Alveolar | Alveolar | Minute | Minute | Minute | Minute |
| Notogastral setae | 14 (c3 absent?) | 13 (c2, h1 absent) | 15 (c3 present) | 15 (c3 present) | 15 (c3 present) | 15 (c3 present) |
| Position lyrifissure im | la/lm? | la/lm | c3/la | c3/la | c3/la | c3/la |
| Epimeral setae 3a, 4a | Alveolar | Short | Short | Short | Short | Short |
| Adanal setae ad1–3 | Alveolar | Fine or alveolar | Alveolar | Fine | Alveolar | Alveolar |
Note
- Japanese mainland: Omishima, Kagoshima; Central Ryukyus: Amami, Okinawa; Southern Ryukyus: Ishigaki and Iriomote. dto = difficult to observe; ? = information not given clearly in literature; c3/la = between seta c3 and la; la/lm = between setae la and lm.
To conclude, the diagnostic value of the existing morphological differences between specimens from the different studies is not clear and needs further investigation.
3.3 Genetic data
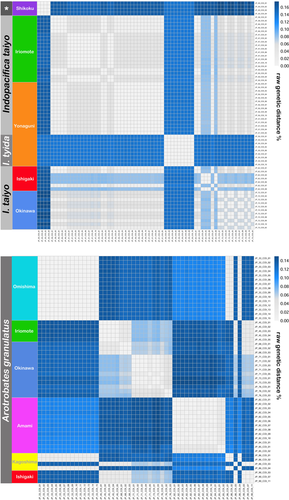
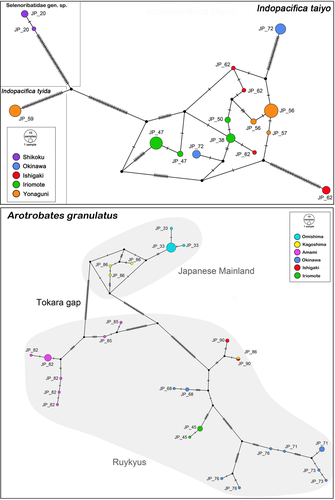
The topology of the Bayesian Inference (BI) tree based on concatenated data (COI and 18S) (Figure 6) is well resolved with each species showing a distinct clade. Despite differing in certain aspects of the topology, the single gene trees are basically consistent in exhibiting the same distinct clades for each species (Figures S3–S7). Although A. granulatus is represented as a monophyletic clade in the BI tree of the concatenated data, it shows a distinct deep structure with more or less three obvious groups. One group consists of specimens from Japanese mainland (JP_33), another group consists only of specimens from Amami-oshima (JP_82) and the third group is a mixture of specimens from Japanese mainland (JP_86), from the Southern Ryukyus (JP_45) and the Central Ryukyus (JP_68, JP_71) (Figure 6). The new Japanese Indopacifica species are well separated, and the Thai I. parva is placed as the sister group to I. taiyo n. sp. The latter consists of two distinct groupings, one including specimens from Iriomote-jima (JP_46, JP_50) and Yonaguni-jima (JP_56) and the other includes only specimens from Iriomote-jima (JP_47) (Figure 6). Indopacifica tyida n. sp., on the other hand, is represented by a homogeneous single clade. Another distinct clade is represented by specimens from Shikoku (JP_20, Japanese mainland) whereas these are placed clearly outside the Indopacifica group and thus they are given as unidentified selenoribatid genus (no specimens or useable remains of these were available for morphological identification).

Maximum intraspecific distances in the COI gene sequence are 0% in I. tyida n. sp. (single haplotype), 8.5% in I. taiyo n. sp., and 14.8% in A. granulatus (Figure 7). Mean net distances between species range from 9.8% to 14.4%, with a net divergence of 12.7% between the two Japanese Indopacifica species (Table 3).
| Indopacifica taiyo | Indopacifica tyida | Selenoribatidae gen. sp. | Arotrobates granulatus | |
|---|---|---|---|---|
| Indopacifica taiyo | 0.013 | 0.013 | 0.010 | |
| Indopacifica tyida | 0.127 | 0.014 | 0.011 | |
| Selenoribatidae gen. sp. | 0.144 | 0.143 | 0.012 | |
| Arotrobates granulatus | 0.098 | 0.116 | 0.124 |
Note
- Net average uncorrected p-distance between species are shown (below diagonal). Standard error estimate(s) are given in grey above the diagonal.
TCS analysis resulted in a network of 13 haplotypes for I. taiyo n. sp. and only one haplotype for I. tyida n. sp. The I. taiyo n. sp. (Figure 8) haplotypes show no specific geographic pattern, that is, haplotypes from the Southern and Central Ryukyus are closely related. There are only two haplotypes, one from Okinawa-jima and one from Ishigaki-jima, that are separated from the others by a large number of mutations. The TCS network for A. granulatus, on the other hand, resulted in 26 different haplotypes showing a distinct geographic pattern (Figure 8). Haplotypes from Omishima and Kagoshima (Japanese mainland) are more or less closely related and form a distinct cluster. Seven haplotypes from Amami-oshima also form a distinct cluster. A third cluster is formed by different haplotypes from the Southern Ryukuys (Ishigaki-jima, Iriomote-jima) and from Okinawa-jima, whereas haplotypes from different sampling locations are separated by a high number of mutations. In the latter cluster, haplotype is shared between Ishigaki-jima (Southern Ryukyus) and Kagoshima (Japanese mainland).
3.4 Morphometric data
3.4.1 Species delimitation/Indopacifica
Univariate data (Table 4) exhibit that size ranges of all measured continuous variables overlap between the two species, except for the distance between camerostome and genital orifice (dcg) which thus may be used as additional diagnostic character. Mann–Whitney U-test, on the other hand, shows that the majority of variables are significantly different between the species, except for the variables nwda, cl, dac3, gl and al. PCA based on size-corrected data results in a clear separation of both species (Figure 9) with the first three components accounting for 72.3% of total variation (PC1 36.2, PC2 23.9, PC3 12.2). Highest loadings on PC1 are shown for the variables dcg and gl (genital orifice length), whereas strong variation in the latter is caused by the sexual dimorphism present in this character, on PC2 again dcg, gl but also gw (genital orifice width) show the highest loadings and on PC3 highest loadings are shown by nwda and nwdp, both characters reflecting body width.
| Indopacifica taiyo n. sp. | Indopacifica tyida n. sp. | |
|---|---|---|
| bl | 277–325 (300 ± 10.2) | 277–319 (305 ± 11.6) |
| dPtI | 132–146 (140 ± 2.9) | 132–139 (136 ± 2.5) |
| dbi | 55–65 (60 ± 2.8) | 52–62 (56 ± 3.1) |
| dbo | 102–114 (110 ± 2.8) | 102–111 (106 ± 2.4) |
| nwda | 163–197 (179 ± 8.3) | 154–194 (174 ± 11.1) |
| nwdm | 175–203 (190 ± 5.6) | 154–194 (176 ± 13) |
| nwdp | 139–182 (157 ± 10) | 151–172 (162 ± 6.3) |
| cl | 77–89 (84 ± 2.6) | 80–86 (84 ± 2.2) |
| cw | 59–68 (63 ± 1.6) | 55–62 (58 ± 2.3) |
| dcg | 63–74 (70 ± 2.8) | 77–89 (84 ± 3.5) |
| dac3 | 99–114 (105 ± 3.4) | 99–108 (104 ± 2.7) |
| gl | 40–52 (44 ± 4) | 37–49 (43 ± 4.3) |
| gw | 43–59 (48 ± 3.6) | 43–55 (48 ± 4.1) |
| al | 62–74 (66±2.9) | 59–71 (65 ± 3.5) |
| aw | 52–59 (54 ± 2.4) | 49–55 (53 ± 1.6) |
Note
- Minimum–maximum (mean ± standard deviation) of each measured variable given in µm. Non-overlapping values given in bold.
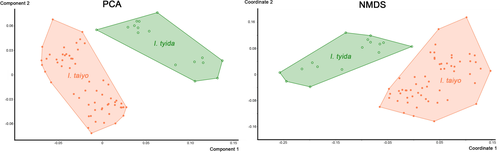
Non-metric Multidimensional Scaling on size-corrected data (Figure 9) shows a clear morphometric distinction between the two species, with a stress of 0.1778. Each species forms a separate cluster without any overlap. PERMANOVA with 10,000 permutations reveals highly significant (p < 0.001) differences between the two species.
3.4.2 Intraspecific variation/Indopacifica taiyo n. sp.
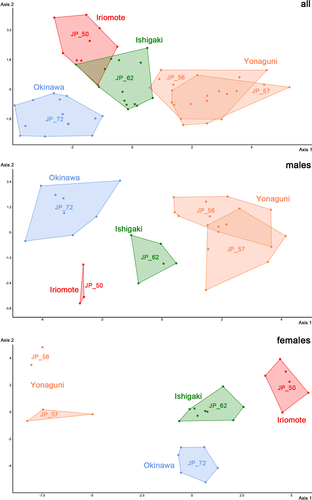
Linear LDA on raw data of I. taiyo n. sp. Results in a more or less clear clustering of the island populations (Figure 10). The Okinawa-jima population is distinctly separated from all other populations, Iriomote-jima and Ishigaki-jima show some overlapping area and only one individual of the Yonaguni-jima population overlaps with the Ishigaki-jima cluster. Variables with the highest loadings are dbi and dbo (distances between bothridia) as well as nwda, nwdm and nwdp (all characters of notogastral width) and 88.14% of the specimens can be correctly classified (Jackknifed 50.85%). MANOVA reveals no significant differences between any of the populations. Kruskal–Wallis test shows significant differences in half of the variables between several populations and Mann–Whitney U-test shows most divergence between the populations from Iriomote-jima (JP_50) and Yonaguni-jima (JP_57) (Table 5).
|
Iriomote-jima JP_50 |
Yonaguni-jima JP_56 |
Yonaguni-jima JP_57 |
Ishigaki-jima JP_62 |
Okinawa-jima JP_72 |
KW | MWU | |
|---|---|---|---|---|---|---|---|
| bl | 277–305 (292 ± 8.5) | 286–319 (301 ± 9.6) | 292–325 (307 ± 11) | 280–313 (300 ± 9.8) | 277–313 (298 ± 9) | – | – |
| dPtI | 135–142 (138 ± 2.2) | 135–146 (141 ± 3) | 139–142 (141 ± 1.3) | 135–145 (141 ± 2.5) | 132–142 (137 ± 3) | *** | b, d, g, i, j |
| dbi | 59–65 (61 ± 2) | 55–65 (58 ± 3.4) | 55–62 (59 ± 2.6) | 55–65 (61 ± 2.4) | 59–65 (62 ± 1.9) | ** | g |
| dbo | 107–111 (110 ± 1.7) | 102–114 (110 ± 3.7) | 108–114 (112 ± 2.4) | 105–114 (111 ± 2.8) | 105–112 (109 ± 2.2) | – | – |
| nwda | 166–182 (174 ± 5.4) | 175–194 (184 ± 7) | 175–197 (186 ± 6.5) | 163–188 (178 ± 8.7) | 166–194 (177 ± 8.2) | ** | b |
| nwdm | 175–191 (186 ± 4.7) | 182–203 (192 ± 6.6) | 187–200 (194 ± 3.7) | 185–200 (192 ± 4) | 179–197 (188 ± 5.9) | * | b |
| nwdp | 142–160 (152 ± 5.9) | 148–172 (156 ± 6.8) | 139–175 (159 ± 11) | 142–182 (161 ± 13.7) | 139–169 (156 ± 8.7) | – | – |
| cl | 77–86 (82 ± 3) | 80–89 (85 ± 2.7) | 83–89 (85 ± 2.4) | 80–86 (82 ± 1.7) | 83–89 (85 ± 1.9) | ** | h, j |
| cw | 62–65 (63 ± 1.3) | 62–65 (63 ± 1.4) | 62–68 (64 ± 2) | 62–65 (63 ± 1.5) | 59–65 (63 ± 1.7) | – | – |
| dcg | 65–71 (68 ± 2.4) | 65–71 (69 ± 2.4) | 65–74 (69 ± 3.1) | 63–74 (70 ± 3.2) | 68–74 (70 ± 2.4) | – | – |
| dac3 | 102–114 (108 ± 4.6) | 102–111 (105 ± 2.7) | 102–108 (105 ± 2.2) | 102–108 (105 ± 2.5) | 99–108 (102 ± 2.6) | ** | d |
| gl | 40–49 (44 ± 3.5) | 40–52 (43 ± 4.3) | 40–52 (45 ± 4.3) | 40–49 (46 ± 3.9) | 40–49 (44 ± 3.9) | – | – |
| gw | 43–52 (48 ± 4) | 43–52 (46 ± 2.7) | 43–52 (47 ± 3.2) | 43–59 (50 ± 4.1) | 43–52 (48 ± 3.5) | – | – |
| al | 62–68 (64 ± 2) | 65–74 (68 ± 2.6) | 62–71 (67 ± 3.1) | 62–71 (65±3.5) | 62–68 (66 ± 1.9) | * | a |
| aw | 52–55 (52±1) | 52–59 (54 ± 2.7) | 52–59 (56 ± 2.3) | 52–59 (54 ± 1.9) | 52–59 (54 ± 2.5) | ** | b |
Note
- Minimum–maximum (mean ± standard deviation) of each measured variable given in µm. Letter indicates significant difference, a = JP_50 vs. JP_56, b = JP_50 vs. JP_57, d = JP_50 vs. JP_72, g = JP_56 vs. JP_72, h = JP_57 vs. JP_62, i = JP_57 vs. JP_72, j = JP_62 vs. JP_72.
- Abbreviations: –, no significant difference; KW, Kruskal–Wallis test; MWU, Mann–Whitney U-test.
- * 0.01 < p < 0.05
- ** 0.001 < p < 0.01
- *** p < 0.001.
LDA on raw data of only male I. taiyo n. sp. shows the same pattern as with all individuals but with clearer separation of each island (Figure 10). Highest loadings are shown by variables dbi, nwda, and al, and 90.1% can be correctly classified (27.3% Jackknifed).
LDA on raw data of only female I. taiyo n. sp. conforms to the pattern found in males (Figure 10). Variables with highest loadings are dbi, dac3, and al and 100% of the specimens can be correctly classified (57.7% Jackknifed).
4 DISCUSSION
4.1 Taxonomy and systematics
The genus Indopacifica was only recently erected (Pfingstl, Lienhard, et al., 2019), and with the discovery of the two Japanese species, it already comprises six different species. The morphology of this genus is quite homogeneous with very few characters allowing to distinguish between species. A similar phenomenon was reported in the intertidal genus Fortuynia where it was suggested to be a result of limited habitat preferences (Pfingstl, 2015) and the same may apply to Indopacifica. In such cases, morphometric data can be a useful tool and provide additional diagnostic characters. In the present study, multivariate analyses clearly separate the two new Indopacifica species and reveal the distance between camerostome and genital orifice to be an additional distinctive feature. Apart from morphology, molecular genetic data also clearly separate the two Japanese species and indicate that they are most likely not closest relatives within the genus. The Thai I. parva is placed as sister species to the Japanese I. taiyo n. sp. whereas I. tyida n. sp. and the Malaysian I. pantai are given as sister groups to the former two. Unfortunately, there are no molecular genetic data available for the Philippine I. iohanna and the Mauritian I. mauritiana allowing us to infer a complete phylogeny. However, morphology as well as molecular genetics depict Indopacifica as a monophyletic homogeneous group with a possible ancestry in the South Eastern Asian area.
Arotrobates granulatus was originally found at the coast of Hong Kong (Luxton, 1992) and later it was reported from the Southern and Central Ryukyus (Karasawa & Aoki, 2005). In this study, we could confirm the distribution on the Ryukyus and we could demonstrate additional occurrences on the Japanese mainland. As outlined earlier, there are slight morphological differences between the specimens from each study whereas most of these very few divergences may actually be the result of inaccurate observations or intraspecific variation. Accordingly, it is not possible to tell with confidence if specimens from the different studies belong to the exact same species. However, it would be highly unlikely that the specimens taken from the same Ryukyuan islands by Karasawa and Aoki (2005) and later by us represent different taxa. Unfortunately, there are no molecular genetic data available from the earlier studies which would allow to definitively solve this issue. However, DNA sequence data of the present study revealed three distinct genetic lineages in A. granulatus, one from Japanese mainland, one from Amami-ohshima, and one from Okinawa-jima and the Southern Ryukyus, whereas all lineages show complete morphological conformity. Intraspecific COI sequence divergences between the geographic lineages range from 11 to nearly 15% which is quite high considering that some authors (Cosgrove et al., 2016) suggested that a 10% divergence in COI exceeds typical species delimitation threshold in arthropods. So, from a genetic point of view, one might consider these lineages already as distinct species though they are morphologically identical. Nevertheless, even if we would be inclined to make the lineages distinct species, we would not be able to because, without genetic data from the type locality in Hong Kong, we cannot exclude that one of the lineages represents the original A. granulatus. Consequently, A. granulatus remains a widely distributed species showing an unusual cryptic genetic diversity until further genetic data are available.
Considering the phylogeny of studied selenoribatid taxa, A. granulatus is placed on a long branch in the 18S trees, distant from the other groups, the outgroup included. There are two possible explanations for this excessively long branch: (a) the 18S gene evolves at a distinctly higher rate in A. granulatus, albeit this is rather unlikely because the rate must be three times higher than in the other groups; (b) the Caribbean T. barbara, which was chosen as outgroup due to its far distant geographic occurrence, is closer related to Indopacifica than Arotrobates is. This is visible in Figures S5–S7 where midpoint-rooted trees (both 18S as well as COI) are shown. Indeed, morphology supports such a relationship because Arotrobates differs more from Indopacifica than Thalassozetes does. However, the phylogenetic position of Arotrobates within the family Selenoribatidae has not been investigated yet, neither on a morphological base nor on a molecular genetic base and we are the first to put this taxon into a phylogenetic context. Nevertheless, for a comprehensive phylogenetic study of Selenoribatidae more taxa from all geographic regions are needed and therefore phylogenetic relationships within this family remain yet uncertain.
4.2 Phylogeography
The high genetic diversity of Japanese A. granulatus coincides with the geological history of this region. The Japanese mainland populations are clearly separated from all Ryukyuan populations which corresponds to the Tokara gap. This gap, which was presumably formed during the late Pliocene, is located on the Tokara tectonic strait and is suggested to have served since then as a barrier for the dispersal of amphibians and reptiles (Ota, 1998). An earlier study on oribatid mites from Japan (Pfingstl, Wagner, et al., 2019) also demonstrated that the Tokara gap is clearly reflected in the genetic structure of the intertidal Fortuynia shibai Aoki, 1974, and the present data confirm this strait as an effective dispersal barrier for small intertidal mites.
The haplotype network of A. granulatus further shows a distinct Amami-ohshima clade which is clearly separated from the other Ryukyuan island populations, including Okinawa-jima. The latter island is, together with Amami-ohshima, part of the Central Ryukyus which is a chain of several neighboring islands separated by smaller stretches of open ocean. In contrast to A. granulatus, the intertidal F. shibai as well as Fortuynia churaumi Pfingstl, Shimano & Hiruta, 2019 appear to be closely related and even share haplotypes between both islands (Pfingstl, Wagner, et al., 2019), which was suggested to be a result of emerging land bridges during the late Pleistocene (Lin et al., 2002). Apparently, A. granulatus has not been able to cross these alleged land bridges probably due to a very low dispersal ability. Furthermore, the results of the haplotype network indicate hardly any closely related haplotypes on the same island or even at the same locality. This implies extremely low gene flow even between geographically close populations and could explain why A. granulatus was not able to cross the oceanic stretch between Amami-ohsima and Okinawa-jima. The same barrier for gene flow was found in certain insects, for example the wood-feeding cockroach Salganea taiwanensis ryukyuanus Asahina, 1988 (Maekawa et al., 1999) or the firefly Curtos costipennis (Gorham, 1880) (Muraji et al., 2012). The channel between the Amami and the Okinawa region opened sequentially around 1.3–1 million years ago resulting in isolation between the islands, and despite emerging super islands and land bridges in the late Pleistocene (Muraji et al., 2012), it apparently remained an insuperable barrier for the above-mentioned taxa. An Atlantic Fortuynia species was observed to show a specific floating behavior when washed into open water which could facilitate dispersal via ocean currents (Pfingstl, 2013). Given that the Japanese Fortuynia species show the same behavior, this could explain why they were able to cross the Kerama gap while A. granulatus was not. But specific behavioral experiments would be necessary to confirm such a hypothesis.
The phylogenetic tree and the haplotype network highlight a third distinct A. granulatus clade, including populations from the Southern Ryukyus and from Okinawa-jima, although no clear substructure can be detected within this cluster. Kimura (2002) postulated a large peninsula reaching from the Chinese continent to the Northern Ryukyus about 1.2–1.7 million years ago and other authors (Yamamoto et al., 2006) suggested that this peninsula began to break 1.6 million years ago. Based on this scenario and the given distribution from Hong Kong (Luxton, 1992) to Japanese mainland, it is possible that an ancestral A. granulatus population was distributed on this peninsula and that the break-up of this landmass induced a sequential vicariant process reflecting the found pattern. The later emergence of several large paleo-islands connecting neighboring landmasses of the Southern and Central Ryukyus in the late Pleistocene (Lin et al., 2002) seemingly did not result in a recent expansion of these segregated A. granulatus populations, which again points to a very low dispersal potential of this species. There is a single-shared haplotype between Kagoshima (Japanese mainland) and Ishigaki-jima (Southern Ryukyus), but this may either be an artifact or recent accidental dispersal via anthropogenic activities.
Molecular genetic sequence data of I. taiyo n. sp. show a fairly different picture. Sequences of this group have not strongly diverged among islands of the Southern and Central Ryukyus suggesting that expansion might have occurred recently. This pattern is well consistent with the above-mentioned existence of late Pleistocene paleo-islands and land bridges connecting the formerly long isolated Ryukyuan islands (Lin et al., 2002). In contrast to A. granulatus, I. taiyo n. sp. was apparently able to use these late Pleistocene low sea level stands to disperse across the Southern and Central Ryukyus. This assumption is further corroborated by the fact that two other Ryukyuan intertidal mites, F. churaumi and Alismobates reticulatus Luxton, 1992, show comparable genetic patterns which were also suggested to be a result of the same evolutionary scenario (Pfingstl, Wagner, et al., 2019).
Apart from all the closely related island populations, there a two genetically distinct haplotypes in I. taiyo n. sp., one from Okinawa-jima and one from Ishigaki-jima. A similar distinct clade from Okinawa-jima was found in F. churaumi, whereas it was proposed that it either represents a relic of more ancient populations or the result of a recent colonization event from more isolated areas of this region (Pfingstl, Wagner, et al., 2019). As two independent colonization events from far isolated areas are less likely, we think that the distinct haplotypes herein rather represent remnants of ancient populations that have subsisted since the late Pleistocene expansion, but certainly more data are necessary to reliably answer this question.
The Tokara gap as well as a barrier between the Amami and Okinawa region are clearly reflected in the sequence data of A. granulatus. However, the Kerama gap, located between the Central and Southern Ryukyus, is not detected in the phylogeographic pattern of any herein investigated species, despite its pivotal role as distribution barrier for insects (Tojo et al., 2017), amphibians, reptiles (Ota, 2000), birds (Tokuda, 1969), and mammals (Motokawa, 2000). Pfingstl, Wagner, et al. (2019) already indicated that the Kerama gap is apparently not an effective zoogeographical barrier for intertidal mites and the present data clearly support this hypothesis.
The current distribution ranges indicate that A. granulatus showed a wide ancestral distribution along the Asian coastline before the opening of the Tokara strait in late Pliocene (Ota, 1998) and the aperture of the Okinawa Trough, approx. 2 million years ago, when the Ryukyu arc was still a continental margin (Osozawa et al., 2012). Indopacifica taiyo n. sp. probably evolved somewhere south of the Tokara gap and was never able to cross this barrier. However, known distributions are likely based on incomplete sampling and hence may differ considerably from real distributions. Despite comprehensive sampling activities, we were only able to find two of the four selenoribatid species reported from the Ryukyus by Karasawa and Aoki (2005), namely A. granulatus and S. saxea, whereas we only collected four specimens of the latter species, two from Amami-ohshima and two from Okinawa-jima.
4.3 Morphological variation between populations
Morphometric data demonstrate that there is a considerable intraspecific variation between island populations of I. taiyo n. sp. and this variation corresponds to geography. The two populations of Yonaguni-jima show a nearly complete overlap in the graph, the populations from Ishigaki-jima and Iriomote-jima cluster very closely and show partial overlap whereas the population from Okinawa-jima forms a distinct cluster. The southern Ryukyu islands Yonaguni-jima, Ishigaki-jima, and Iriomote-jima are part of the Yaeyamas, a group of closely located neighbor islands, and Okinawa-jima is part of the Central Ryukyus which are separated from the former by the Kerama Gap and this geographic pattern seems to be reflected in the morphometric data (i.e., geographic distance correlates with morphological divergence). A similar result was demonstrated in a study on intertidal mites from the Galapagos archipelago (Pfingstl & Baumann, 2017), where closer populations showed less divergence than populations from distant islands. Interestingly, molecular genetic data of the exact same populations of the present study do not correspond to actual geography. Given this discrepancy, it is likely that the morphological variation found is partly a result of phenotypic plasticity caused by diverging local environmental factors. Nevertheless, more comprehensive investigations are necessary to confirm such a correlation.
Another interesting fact is that separating the sexes in the present morphometric analyses leads to a clearer separation of the single island populations. Pfingstl, Wagner, et al. (2019) already mentioned that a weak sexual dimorphism, which is present in most oribatid mite species, can slightly blur the results and that there is a yet not understood sex-related overall variation with males diverging more than females or vice versa. In the present case, females apparently show more variation among the different islands than males, a similar result was found in the fortuyniid Alismobates galapagoensis Pfingstl & Schatz, 2017 occurring on several islands of the Galapagos archipelago (Pfingstl & Baumann, 2017). However, the reason for this variation remains unknown.
4.4 Ecology
Arotrobates granulatus was first found in oyster shells on a rocky shore (Luxton, 1992), later it was also reported from bark of the mangrove Bruguiera gymnorrhiza and from algae on a mangrove forest floor (Karasawa & Aoki, 2005). In the present study, A. granulatus was found in different kinds of algae and once on barnacles and mussels which indicates a wider ecological range for this species. Based on results from the present study, A. granulatus seems to feed on a variety of algae but prefers rocky substrates (though it occasionally may occur in mangrove habitats).
Species of the genus Indopacifica were recently discovered for the first time (Pfingstl, Lienhard, et al., 2019) and therefore knowledge about their ecology is largely incomplete. One of the few existing studies stated that they seem to inhabit and feed on diverse kinds of algae growing on intertidal rocks (Resch et al., 2019) and the present study basically confirms this statement. Indopacifica tyida n. sp. was only found in a single location which was a typical rocky shore and I. taiyo n. sp. occurred mainly in algae growing on rocky substrate, and only in three out of 21 samples they were found in mangrove associated habitats with very low specimen numbers. Thus, similar to A. granulatus, both species apparently prefer rocky intertidal environments but may occasionally be found in other habitats too.
In eight locations I. taiyo n. sp. and A. granulatus were found syntopically but based on the present findings we are not able to tell whether they use different niches within the microhabitat, for example, feeding on different parts of the algae or showing a vertical zonation. Both Indopacifica species, on the other hand, never occurred syntopically and microscopic investigations revealed a blue shimmering gut content in nearly all I. tyida n. sp. specimens, indicating a different diet of this species. The found blue shimmering gut content may indicate that I. tyida n. sp. may use cellular blue-green algae (cyanobacteria) as food source while I. taiyo n. sp. may feed on large multicellular algae and this could explain why they were never found together. However, further gut content analyses and food preference tests are necessary to verify such a statement.
ACKNOWLEDGEMENTS
Thanks to Mr. Koji Uchida for providing additional samples from Ishigaki-jima. We are also grateful to Mr. Yuji Yamakara (Yanbaru-Manabino-mori) and Mr. Hutoshi Taira (Ashi-muri-no-sato) for helping and supporting us during our fieldtrip on Okinawa-jima, and to Dr. Takuma Fujii and Dr. Mariko Suzuki (International Center for Island Studies Amami Station, Kagoshima University) for helping and supporting us during our fieldtrip on Amami-oshima. Special thanks to Prof. emeritus Minoru Shiba (Matsuyama Shinonome Junior College, Kuwabara, Matsuyama) for his help and hospitality during our stay on Shikoku. We would like to thank Dr. Ken-ichi Okumura (National Museum of Nature and Science, Tokyo) for the deposition of type specimens. This investigation was funded by the Austrian Science Fund (FWF): [I 3815] for TP and by bilateral Programs, Joint Research Projects (JSPS) and JSPS KAKENHI Grant Numbers 17K07271, 18K06392 for SS and SFH and Tokyo Metropolitan University (TMU) Fund for Strategic Research (Leader: Prof. Noriaki Murakami; FY2020-FY2022).
APPENDIX 1
Sample IDs, geographic origin, GenBank accession numbers for COI, and 18S sequences comprising all specimens included in genetic investigations. All specimens deposited in the collection of the Institute of Biology, University of Graz (IBUG)
| ID | Location | Taxon | COI | 18S | Source |
|---|---|---|---|---|---|
| JP_20_Selen_01 | Shikoku | Selenoribatidae gen. sp. | MW289086 | MW298485 | This study |
| JP_20_Selen_02 | Shikoku | Selenoribatidae gen. sp. | MW289087 | This study | |
| JP_20_Selen_03 | Shikoku | Selenoribatidae gen. sp. | MW289088 | This study | |
| JP_20_Selen_04 | Shikoku | Selenoribatidae gen. sp. | MW289089 | MW298486 | This study |
| JP_33_Aro_01 | Omishima | Arotrobates granulatus | MW289153 | MW298487 | This study |
| JP_33_Aro_02 | Omishima | Arotrobates granulatus | MW289154 | This study | |
| JP_33_Aro_03 | Omishima | Arotrobates granulatus | MW289155 | This study | |
| JP_33_Aro_04 | Omishima | Arotrobates granulatus | MW289156 | MW298488 | This study |
| JP_33_Aro_05 | Omishima | Arotrobates granulatus | MW289157 | This study | |
| JP_33_Aro_06 | Omishima | Arotrobates granulatus | MW289158 | This study | |
| JP_33_Aro_07 | Omishima | Arotrobates granulatus | MW289159 | This study | |
| JP_33_Aro_08 | Omishima | Arotrobates granulatus | MW289160 | MW298489 | This study |
| JP_33_Aro_09 | Omishima | Arotrobates granulatus | MW289161 | This study | |
| JP_33_Aro_10 | Omishima | Arotrobates granulatus | MW289162 | This study | |
| JP_33_Aro_11 | Omishima | Arotrobates granulatus | MW289163 | This study | |
| JP_33_Aro_12 | Omishima | Arotrobates granulatus | MW289164 | This study | |
| JP_33_Aro_13 | Omishima | Arotrobates granulatus | MW289165 | This study | |
| JP_33_Aro_14 | Omishima | Arotrobates granulatus | MW289166 | MW298490 | This study |
| JP_33_Aro_15 | Omishima | Arotrobates granulatus | MW289167 | This study | |
| JP_38_Indo_01 | Iriomote | Indopacifica taiyo sp. nov. | MW289090 | This study | |
| JP_38_Indo_02 | Iriomote | Indopacifica taiyo sp. nov. | MW289091 | This study | |
| JP_45_Aro_01 | Iriomote | Arotrobates granulatus | MW289168 | This study | |
| JP_45_Aro_02 | Iriomote | Arotrobates granulatus | MW289169 | This study | |
| JP_45_Aro_03 | Iriomote | Arotrobates granulatus | MW289170 | MW298491 | This study |
| JP_45_Aro_04 | Iriomote | Arotrobates granulatus | MW289171 | This study | |
| JP_45_Aro_05 | Iriomote | Arotrobates granulatus | MW289172 | This study | |
| JP_46_Indo_01 | Iriomote | Indopacifica taiyo sp. nov. | MW289092 | This study | |
| JP_46_Indo_02 | Iriomote | Indopacifica taiyo sp. nov. | MW289093 | MW298492 | This study |
| JP_46_Indo_03 | Iriomote | Indopacifica taiyo sp. nov. | MW289094 | This study | |
| JP_47_Indo_01 | Iriomote | Indopacifica taiyo sp. nov. | MW289095 | MW298493 | This study |
| JP_47_Indo_02 | Iriomote | Indopacifica taiyo sp. nov. | MW289096 | This study | |
| JP_47_Indo_03 | Iriomote | Indopacifica taiyo sp. nov. | MW289097 | This study | |
| JP_47_Indo_04 | Iriomote | Indopacifica taiyo sp. nov. | MW289098 | This study | |
| JP_47_Indo_05 | Iriomote | Indopacifica taiyo sp. nov. | MW289099 | MW298494 | This study |
| JP_47_Indo_06 | Iriomote | Indopacifica taiyo sp. nov. | MW289100 | This study | |
| JP_47_Indo_07 | Iriomote | Indopacifica taiyo sp. nov. | MW289101 | MW298495 | This study |
| JP_47_Indo_09 | Iriomote | Indopacifica taiyo sp. nov. | MW289102 | MW298496 | This study |
| JP_47_Indo_10 | Iriomote | Indopacifica taiyo sp. nov. | MW289103 | This study | |
| JP_50_Indo_01 | Iriomote | Indopacifica taiyo sp. nov. | MW289104 | This study | |
| JP_50_Indo_02 | Iriomote | Indopacifica taiyo sp. nov. | MW289105 | This study | |
| JP_50_Indo_03 | Iriomote | Indopacifica taiyo sp. nov. | MW289106 | MW298497 | This study |
| JP_50_Indo_04 | Iriomote | Indopacifica taiyo sp. nov. | MW289107 | This study | |
| JP_50_Indo_05 | Iriomote | Indopacifica taiyo sp. nov. | MW289108 | This study | |
| JP_56_Indo_01 | Yonaguni | Indopacifica taiyo sp. nov. | MW289109 | MW298498 | This study |
| JP_56_Indo_02 | Yonaguni | Indopacifica taiyo sp. nov. | MW289110 | This study | |
| JP_56_Indo_03 | Yonaguni | Indopacifica taiyo sp. nov. | MW289112 | This study | |
| JP_56_Indo_04 | Yonaguni | Indopacifica taiyo sp. nov. | MW289113 | MW298499 | This study |
| JP_56_Indo_05 | Yonaguni | Indopacifica taiyo sp. nov. | MW289114 | This study | |
| JP_56_Indo_06 | Yonaguni | Indopacifica taiyo sp. nov. | MW289115 | MW298500 | This study |
| JP_56_Indo_07 | Yonaguni | Indopacifica taiyo sp. nov. | MW289116 | This study | |
| JP_56_Indo_08 | Yonaguni | Indopacifica taiyo sp. nov. | MW289117 | This study | |
| JP_56_Indo_09 | Yonaguni | Indopacifica taiyo sp. nov. | MW289118 | MW298501 | This study |
| JP_56_Indo_10 | Yonaguni | Indopacifica taiyo sp. nov. | MW289119 | This study | |
| JP_57_Indo_01 | Yonaguni | Indopacifica taiyo sp. nov. | MW289120 | This study | |
| JP_57_Indo_02 | Yonaguni | Indopacifica taiyo sp. nov. | MW289121 | This study | |
| JP_57_Indo_03 | Yonaguni | Indopacifica taiyo sp. nov. | MW289122 | This study | |
| JP_59_Indo_01 | Yonaguni | Indopacifica tyida sp. nov. | MW289123 | MW298502 | This study |
| JP_59_Indo_02 | Yonaguni | Indopacifica tyida sp. nov. | MW289111 | This study | |
| JP_59_Indo_03 | Yonaguni | Indopacifica tyida sp. nov. | MW289124 | This study | |
| JP_59_Indo_04 | Yonaguni | Indopacifica tyida sp. nov. | MW289125 | MW298503 | This study |
| JP_59_Indo_05 | Yonaguni | Indopacifica tyida sp. nov. | MW289126 | This study | |
| JP_59_Indo_06 | Yonaguni | Indopacifica tyida sp. nov. | MW289127 | This study | |
| JP_59_Indo_07 | Yonaguni | Indopacifica tyida sp. nov. | MW289128 | MW298504 | This study |
| JP_59_Indo_08 | Yonaguni | Indopacifica tyida sp. nov. | MW289129 | This study | |
| JP_59_Indo_09 | Yonaguni | Indopacifica tyida sp. nov. | MW289130 | MW298505 | This study |
| JP_59_Indo_10 | Yonaguni | Indopacifica tyida sp. nov. | MW289131 | This study | |
| JP_62_Indo_01 | Ishigaki | Indopacifica taiyo sp. nov. | MW289132 | This study | |
| JP_62_Indo_02 | Ishigaki | Indopacifica taiyo sp. nov. | MW289134 | This study | |
| JP_62_Indo_03 | Ishigaki | Indopacifica taiyo sp. nov. | MW289133 | This study | |
| JP_62_Indo_04 | Ishigaki | Indopacifica taiyo sp. nov. | MW289135 | This study | |
| JP_62_Indo_05 | Ishigaki | Indopacifica taiyo sp. nov. | MW289136 | This study | |
| JP_62_Indo_06 | Ishigaki | Indopacifica taiyo sp. nov. | MW289137 | This study | |
| JP_62_Indo_07 | Ishigaki | Indopacifica taiyo sp. nov. | MW289138 | This study | |
| JP_62_Indo_08 | Ishigaki | Indopacifica taiyo sp. nov. | MW289139 | This study | |
| JP_68_Aro_01 | Okinawa | Arotrobates granulatus | MW289173 | MW298506 | This study |
| JP_68_Aro_02 | Okinawa | Arotrobates granulatus | MW289174 | This study | |
| JP_68_Aro_03 | Okinawa | Arotrobates granulatus | MW289175 | This study | |
| JP_71_Aro_01 | Okinawa | Arotrobates granulatus | MW289176 | This study | |
| JP_71_Aro_02 | Okinawa | Arotrobates granulatus | MW289177 | This study | |
| JP_71_Aro_03 | Okinawa | Arotrobates granulatus | MW289178 | MW298507 | This study |
| JP_71_Aro_04 | Okinawa | Arotrobates granulatus | MW289179 | This study | |
| JP_71_Aro_05 | Okinawa | Arotrobates granulatus | MW289180 | MW298508 | This study |
| JP_72_Indo_01 | Okinawa | Indopacifica taiyo sp. nov. | MW289140 | This study | |
| JP_72_Indo_02 | Okinawa | Indopacifica taiyo sp. nov. | MW289141 | This study | |
| JP_72_Indo_03 | Okinawa | Indopacifica taiyo sp. nov. | MW289142 | This study | |
| JP_72_Indo_04 | Okinawa | Indopacifica taiyo sp. nov. | MW289143 | This study | |
| JP_72_Indo_05 | Okinawa | Indopacifica taiyo sp. nov. | MW289144 | This study | |
| JP_72_Indo_06 | Okinawa | Indopacifica taiyo sp. nov. | MW289145 | This study | |
| JP_72_Indo_07 | Okinawa | Indopacifica taiyo sp. nov. | MW289146 | This study | |
| JP_72_Indo_08 | Okinawa | Indopacifica taiyo sp. nov. | MW289147 | This study | |
| JP_72_Indo_09 | Okinawa | Indopacifica taiyo sp. nov. | MW289148 | This study | |
| JP_72_Indo_10 | Okinawa | Indopacifica taiyo sp. nov. | MW289149 | This study | |
| JP_73_Aro_01 | Okinawa | Arotrobates granulatus | MW289150 | This study | |
| JP_73_Aro_02 | Okinawa | Indopacifica taiyo sp. nov. | MW289151 | This study | |
| JP_73_Aro_03 | Okinawa | Arotrobates granulatus | MW289152 | This study | |
| JP_76_Aro_01 | Okinawa | Arotrobates granulatus | MW289181 | This study | |
| JP_76_Aro_02 | Okinawa | Arotrobates granulatus | MW289182 | This study | |
| JP_76_Aro_03 | Okinawa | Arotrobates granulatus | MW289183 | This study | |
| JP_82_Aro_01 | Amami | Arotrobates granulatus | MW289184 | MW298509 | This study |
| JP_82_Aro_02 | Amami | Arotrobates granulatus | MW289185 | This study | |
| JP_82_Aro_03 | Amami | Arotrobates granulatus | MW289186 | This study | |
| JP_82_Aro_04 | Amami | Arotrobates granulatus | MW289187 | MW298510 | This study |
| JP_82_Aro_05 | Amami | Arotrobates granulatus | MW289188 | This study | |
| JP_82_Aro_06 | Amami | Arotrobates granulatus | MW289189 | MW298511 | This study |
| JP_82_Aro_07 | Amami | Arotrobates granulatus | MW289190 | This study | |
| JP_82_Aro_08 | Amami | Arotrobates granulatus | MW289191 | This study | |
| JP_82_Aro_09 | Amami | Arotrobates granulatus | MW289192 | MW298512 | This study |
| JP_82_Aro_10 | Amami | Arotrobates granulatus | MW289193 | This study | |
| JP_84_Aro_01 | Amami | Arotrobates granulatus | MW289194 | This study | |
| JP_85_Aro_01 | Amami | Arotrobates granulatus | MW289195 | This study | |
| JP_85_Aro_02 | Amami | Arotrobates granulatus | MW289196 | This study | |
| JP_86_Aro_01 | Kagoshima | Arotrobates granulatus | MW289197 | This study | |
| JP_86_Aro_02 | Kagoshima | Arotrobates granulatus | MW289198 | This study | |
| JP_86_Aro_03 | Kagoshima | Arotrobates granulatus | MW289199 | MW298513 | This study |
| JP_86_Aro_04 | Kagoshima | Arotrobates granulatus | MW289200 | This study | |
| JP_90_Aro_03 | Ishigaki | Arotrobates granulatus | MW289201 | This study | |
| JP_90_Aro_07 | Ishigaki | Arotrobates granulatus | MW289202 | This study | |
| JP_90_Aro_11 | Ishigaki | Arotrobates granulatus | MW289203 | This study | |
| T_BA_30_2 | Barbados | Thalassozetes barbara | MW289085 | MW298484 | This study |
| S_MY_17_6 | Penang | Indopacifica pantai | MH285652 | MH285691 | Pfingstl, Lienhard, et al. (2019) |
| S_MY_17_8 | Penang | Indopacifica pantai | MH285654 | MH285692 | Pfingstl, Lienhard, et al. (2019) |
| S2_TH_09_1 | Khao Lak | Indopacifica parva | MH285671 | MH285690 | Pfingstl, Lienhard, et al. (2019) |
Open Research
Data availability statement
Gene sequence data are available at GenBank-NCBI; https://www.ncbi.nlm.nih.gov/genbank/; accession numbers MW289085 – MW289203 (COI) and MW298484 – MW298513 (18S).




