Borrowing the Pentatomomorpha tome from the DNA barcode library: Scanning the overall performance of cox1 as a tool
Abstract
Pentatomomorpha (Hemiptera: Heteroptera) occurs worldwide with almost 19,000 species within six superfamilies. Their wide morphological, behavioral, and ecological diversity is remarkable, making them subject of basic and applied studies. The mitochondrial cytochrome c oxidase subunit I (cox1) proposed as a DNA barcode for Metazoa is used in species discovery and identification, relying on threshold values to split intra- and interspecific sequences and a comprehensive library to accurate identification. Here, we scanned all the pentatomomorphan cox1 sequences on Barcode of Life Data System library aiming to provide an overview of available data; verify barcoding gaps at generic level; infer individual empirical threshold values to distinguish congeneric species; and test efficiency of cox1 based on the Probability of Correct Identification (PCI) analysis. Our final dataset comprised 12,189 sequences, covering five superfamilies, 32 families, 460 genera, and 1068 species. The dataset abundance and composition were biased to families with economic importance, that is, Pentatomidae, Lygaeidae, Scutelleridae, Coreidae, and Rhyparochromidae. Barcode gaps were detected for most of the analyzed genera, reaffirming the efficiency of cox1 for Pentatomomorpha. We inferred threshold values for 131 genera and found a global PCI of 74.33%, suggesting that one out of four analyzed species suffer from operational biases or hide cryptic species. We brought examples to illustrate how cox1 can be used to flag inconsistencies, refine, and shed light onto future studies on Pentatomomorpha. We emphasize the efficiency of cox1 as DNA barcode for these true bugs, advocating for its combined use with, for example, morphology in integrative approaches.
1 INTRODUCTION
Pentatomomorpha (Heteroptera, the true bugs) are part of the most successful radiation of non-holometabolous insects, the Hemiptera (Weirauch & Schuh, 2011). They occur in all zoogeographical regions, except Antarctica, and thrive in tropical zones (Panizzi & Grazia, 2015). This infraorder encompasses six superfamilies (sensu Schuh & Weirauch, 2020): Aradoidea, Coreoidea, Idiostoloidea, Lygaeoidea, Pentatomoidea, and Pyrrhocoroidea, including almost 19,000 species (Henry, 2017). The wide morphological, behavioral, and ecological diversity disclosed in Pentatomomorpha is remarkable (Figure 1).

These terrestrial true bugs comprise mycophagous, hematophagous, predators, and phytophagous species (Weirauch et al., 2019). Several predatory species are used as biocontrol agents, and phytophagous species—the majority of pentatomomorphans—feed on roots to flowers and seeds, with many species being serious crop pests around the globe. Invasive pentatomomorphans intensify economic damages, mainly related to crops (McPherson, 2018). Since the Pentatomomorpha are subject of basic and applied studies, there is an urge to understand their lineage splits, diversification, and accurate identification.
Morphology is the foundation of pentatomomorphan description and identification. But the morphological nuances that distinguish closely related species might be so complex that even expert taxonomists struggle in their diagnosis (Figure 1). Plastic phenotypes, sex-dependent characters, and the little knowledge for the identification of eggs and immature stages may further hinder this process. Alternative data sources have been expanding taxonomic studies, providing novelties to accurate identification, supporting synonyms, and consistently unveiling cryptic diversity (e.g., Hickmann et al., 2019; Zhang et al., 2017). In this scenario, molecular tools integrated to morphology may aid in organism identification and can be used to assess biodiversity quickly. DNA barcoding—the standardized use of short nucleotide sequences for specimen identification (Hebert et al., 2003)—was already proved useful in pentatomomorphan taxonomy (Park et al., 2011; Tembe et al., 2014).
Since the proposition of the 5′ end of the mitochondrial cytochrome c oxidase subunit I (cox1) as a standard DNA barcode for Metazoa (Hebert et al., 2003), some studies built up sequence libraries for Pentatomomorpha, providing reference sequences to hundreds of species (e.g., Gwiazdowski et al., 2015; Raupach et al., 2014). Moreover, vast amounts of pentatomomorphan barcoding cox1 sequences are generated yearly (e.g., Gowande et al., 2018; Grebennikov & Heiss, 2014). These data supply Barcode of Life Data System (BOLD), a database dedicated to assembling barcode records that meet a series of criteria (Ratnasingham & Hebert, 2007). DNA barcoding as a tool impacts directly on pest management, forensics, conservation, environmental ecology, phylogeography, and many other biological disciplines. Since species are the fundamental unit for many biological subjects, DNA barcoding strongly improves integrative taxonomy (Goulding & Dayrat, 2016). The increase of studies using cox1 as a barcode mirrors the versatility of the marker and the method (DeSalle & Goldstein, 2019).
DNA barcoding manages two fundamental tasks: to diagnose species (i.e., specimen identification) and to discover new species (i.e., species delimitation) (DeSalle et al., 2005). Specimen identification is accomplished through comparisons with a reference database. In contrast, species discovery and delimitation use threshold values based on the interval between intra- and interspecific distances (i.e., the barcoding gap; Meyer & Paulay, 2005). Moreover, the barcoding gap may disclose outliers within a pool of congeneric samples, serving as a taxonomic red flag (e.g., Gonçalves et al., 2021). Since the availability of representative species and specimens (i.e., sequences) directly impact on DNA barcoding performance (Meyer & Paulay, 2005), threshold and gap values should be reviewed regularly whenever new samples are deposited in the databases.
Here, we mined all pentatomomorphan cox1 sequences deposited on BOLD, aiming to (1) provide an overview of information available in BOLD; (2) verify the existence of a barcoding gap at generic level in Pentatomomorpha; (3) set up a threshold value that allows distinguishing intra- and interspecific taxa; and (4) test efficiency of cox1 based on the Probability of Correct Identification (PCI) analysis. We also brought empirical data to illustrate how cox1 can be used to flag inconsistencies and clue future studies on Pentatomomorpha.
2 MATERIALS AND METHODS
2.1 Data acquisition and filtering
Sequences were retrieved from BOLD on August 17, 2020, generating separate datasets for each pentatomomorphan family. We followed several filtering steps to ensure robust analyses (summarized in Figure 2). First, we removed sequences belonging to other markers (or regions) than COI-5P (e.g., COI-3P, COXII) or without species-level identification (e.g., “Euschistus,” “Pentatomidae sp.”). Then, we made preliminary alignments of each dataset using MAFFT 7.0 (Katoh et al., 2019), keeping the parameters in default, and looking for non-sense mutations, insertions, and deletions. These sequences were removed from the dataset since we assumed they resulted from low-quality sequencing, erroneous amplification, or laboratory contamination. Additionally, misaligned sequences were used as a query for comparison with the NCBI database (http://www.ncbi.nlm.nih.gov/) using the Basic Local Alignment Search Tool—Nucleotide (BLASTn) to verify their identity. Non-pentatomomorphan sequences were purged from the dataset.
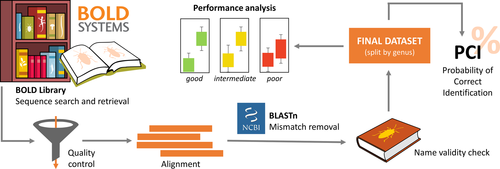
After these steps, we realigned the sequences (same parameters as above). The software AliView (Larsson, 2014) was used to visualize the alignments, verify the reading frame, and trim the sequences to restrict them to the canonical barcode region of cox1 (Ratnasingham & Hebert, 2007). Sequences shorter than 400 base pairs were removed. As a final filtering step, we double-checked scientific names seeking for misspellings and non-valid names (i.e., synonyms). Then, we ran a final alignment for each dataset.
2.2 Data analyses
For the barcoding gap analysis, we generated separate FASTA files for each genus represented in the datasets. We only analyzed genera featuring at least two species, and with at least one of the species being represented by more than one sequence, to ensure intra- and interspecific comparisons. For each genus, the R package SPIDER (Brown et al., 2012) was used to estimate uncorrected p-distances. Output values were visualized in a boxplot and we followed Badotti et al., (2017) to rank cox1 efficiency based on three categories: good, intermediate, and poor. Efficiency was considered good when boxplots disclosed a gap between intra- and interspecific distances, intermediate whenever the whiskers of intra- and interspecific distances overlapped, and poor when the boxes overlapped. We additionally used the function “localMinima” implemented in SPIDER to determine a threshold value for genus with a good cox1 efficiency. This function optimizes putative threshold values from a density plot of the genetic distances (Brown et al., 2012).
Since the success of DNA barcoding for specimen identification is not necessarily related to a barcoding gap (see Discussion), we calculated the PCI (Hollingsworth et al., 2009) to measure the identification success when using cox1 sequences. For each species, we considered the maximum intraspecific distance and the minimum interspecific distance (i.e., nearest-neighbor distance). If the maximum intraspecific distance of a species was less than its minimum interspecific distance, identification for that species was considered a success (Hollingsworth et al., 2009). PCI was calculated as the percentage of species correctly identified. Species represented by only one sequence were not analyzed, since it was not possible to obtain intraspecific distances in such cases. PCI rates were then graphically represented in a scatter plot, as suggested by Collins and Cruickshank (2012).
Since some family groups and their rankings are not consensual with current classification, we opted to present data for Thaumastellinae and Thyreocorinae, treated as subfamilies of Cydnidae for some authors (e.g., Schuh & Weirauch, 2020) but as independent families in other classifications (e.g., Grazia et al., 2008; Lis et al., 2017; Rider et al., 2018). Here, we adopted the classification of Pentatomomorpha and numbers of species and genera per family presented by Schuh and Weirauch (2020).
3 RESULTS
Our raw dataset consisted of 18,250 sequences. Removing sequences of non-barcode markers or without species-level identification reduced this number to 12,892. During the preliminary alignment step, we found a small number of sequences with insertions and deletions; we also detected nine sequences matching with non-pentatomomorphan taxa (e.g., bacteria, lepidopterans, and dipterans) in BLASTn. The removal of these entries led to 12,761 sequences. After purging sequences shorter than 400 bp, our final dataset comprised 12,189 pentatomomorphan sequences representing five superfamilies, 32 families, 460 genera, and 1068 species (Table S1). The global alignment of the analyzed sequences is available as Alignment S1.
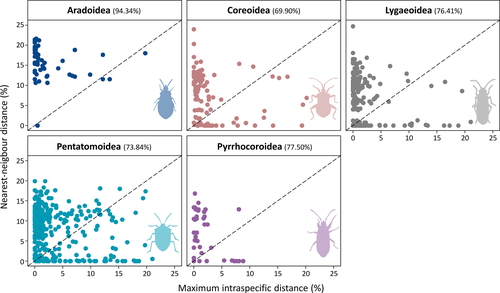
The state of knowledge of sequences filtered from BOLD library cover 6% of the valid species within Pentatomomorpha. Sequence coverage by species range from 1 to 1116, with 82.30% of sampled species being represented by less than 10 sequences. The available sequences for families and the number of species sampled increase with their respective species richness (Table 1). The families with higher coverage of species were Stenocephalidae (20.00%), Rhopalidae (14.00%), Cymidae (13.00%), and Berytidae (11.00%). On the other hand, 10 of 40 families lack any sampled species. Regarding dataset composition, the recovered sequences display a bias toward Pentatomoidea, representing almost half of analyzed sequences (47.05%), followed by Lygaeoidea (34.19%), Coreoidea (14.55%), Pyrrhocoroidea (2.55%), and Aradoidea (1.67%). The only sequence retrieved for Idiostoloidea (Trisecus sp.; GBA29199-16) was filtered out due to the lack of specific identification. The families with the greatest contribution of sequences for our final dataset were Pentatomidae (n = 4160), Lygaeidae (n = 2450), Rhyparochromidae (n = 1192), Scutelleridae (n = 892), and Coreidae (n = 866); for the total richness of 1070 species were Pentatomidae (S = 361), Rhyparochromidae (S = 106), Coreidae (S = 95), Lygaeidae (S = 77), and Aradidae (S = 77). See Table S1 for a list of all sampled species and Table S2 for a list of all sampled sequences.
| Taxon | Data | Genera | Species | |||
|---|---|---|---|---|---|---|
| Raw | Final | Valid | Sampled | Valid | Sampled | |
| Aradoidea | ||||||
| Aradidae | 544 | 203 | 233 | 29 | 1931 | 77 |
| Termitaphididae | 1 | 0 | 2 | 0 | 9 | 0 |
| Coreoidea | ||||||
| Alydidae | 675 | 480 | 45 | 11 | 359 | 21 |
| Coreidae | 1150 | 866 | 267 | 52 | 3178 | 95 |
| Hyocephalidae | 0 | 0 | 2 | 0 | 3 | 0 |
| Rhopalidae | 556 | 422 | 21 | 12 | 295 | 36 |
| Stenocephalidae | 53 | 5 | 1 | 1 | 16 | 4 |
| Idiostoloidea | ||||||
| Henicocoridae | 0 | 0 | 1 | 0 | 1 | 0 |
| Idiostolidae | 1 | 0 | 3 | 0 | 4 | 0 |
| Lygaeoidea | ||||||
| Artheneidae | 33 | 33 | 8 | 1 | 20 | 1 |
| Berytidae | 197 | 140 | 36 | 10 | 174 | 21 |
| Blissidae | 78 | 32 | 51 | 3 | 436 | 8 |
| Colobathristidae | 12 | 2 | 23 | 1 | 84 | 1 |
| Cryptorhamphidae | 0 | 0 | 2 | 0 | 4 | 0 |
| Cymidae | 113 | 79 | 9 | 2 | 54 | 9 |
| Geocoridae | 164 | 78 | 25 | 2 | 280 | 15 |
| Heterogastridae | 10 | 8 | 24 | 2 | 100 | 2 |
| Lygaeidae | 3194 | 2450 | 102 | 28 | 968 | 77 |
| Malcidae | 25 | 4 | 3 | 2 | 29 | 2 |
| Meschiidae | 0 | 0 | 2 | 0 | 5 | 0 |
| Ninidae | 1 | 1 | 5 | 1 | 13 | 1 |
| Oxycarenidae | 79 | 26 | 26 | 4 | 150 | 11 |
| Pachygronthidae | 107 | 105 | 13 | 2 | 78 | 4 |
| Piesmatidae | 24 | 17 | 6 | 2 | 44 | 3 |
| Rhyparochromidae | 3170 | 1192 | 372 | 59 | 1850 | 106 |
| Pentatomoidea | ||||||
| Acanthosomatidae | 407 | 363 | 57 | 13 | 317 | 52 |
| Canopidae | 1 | 0 | 1 | 0 | 16 | 0 |
| Cydnidae | 196 | 82 | 120 | 13 | 765 | 18 |
| Thaumastellinae | 2 | 2 | 1 | 1 | 3 | 2 |
| Thyreocorinae | 68 | 39 | 30 | 2 | 224 | 8 |
| Dinidoridae | 28 | 17 | 20 | 3 | 110 | 7 |
| Lestoniidae | 1 | 1 | 1 | 1 | 2 | 1 |
| Megarididae | 1 | 0 | 2 | 0 | 18 | 0 |
| Pentatomidae | 5368 | 4158 | 945 | 140 | 4855 | 361 |
| Phloeidae | 1 | 0 | 3 | 0 | 4 | 0 |
| Plataspidae | 475 | 135 | 66 | 4 | 604 | 7 |
| Saileriolidae | 0 | 0 | 3 | 0 | 4 | 0 |
| Scutelleridae | 961 | 892 | 102 | 34 | 531 | 54 |
| Tessaratomidae | 43 | 20 | 62 | 5 | 250 | 6 |
| Urostylididae | 39 | 26 | 9 | 4 | 177 | 8 |
| Pyrrhocoroidea | ||||||
| Largidae | 100 | 41 | 13 | 6 | 106 | 13 |
| Pyrrhocoridae | 372 | 270 | 33 | 10 | 340 | 37 |
| Total | 18,250 | 12,189 | 2750 | 460 | 18,411 | 1068 |
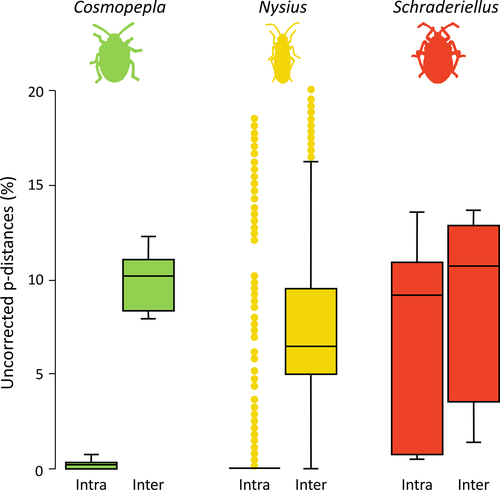
For most of the evaluated genera, cox1 efficiency was considered good (75.14%), although in some instances performance was intermediate (12.15%) or poor (12.71%). Figure 3 shows examples of such performances; boxplots for each genus are available as Data S1. Intra- and interspecific distances varied considerably among taxa, which directly affected the threshold values detected with “localMinima.” Threshold values ranged from 0.08% to 13.24%, with an average of 5.49%. For a detailed classification of each genus efficiency and the determined threshold values, see Appendix 1.
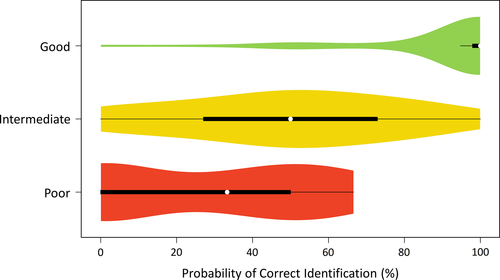
The PCI was estimated using a trimmed dataset, consisting of 11,853 sequences belonging to 339 genera and 737 species. The number of sequences decreased since we only considered species represented by more than one sequence, to guarantee intraspecific comparisons. The global PCI was 74.33%, but this value varied greatly among superfamilies and families (Figure 4, Appendix 1). Overall, genera that cox1 efficiency was good disclosed higher PCI, whereas genera with poor performance showed lower values (Figure 5).
4 DISCUSSION
In this paper, we scrutinized the BOLD library to detach the whole data related to the cox1 barcode region of Pentatomomorpha. This broad approach detected barcode gaps for most of the analyzed genera, reaffirming the transversal efficiency of this marker as DNA barcode for Pentatomomorpha. These findings also allowed us to infer individual empirical thresholds values for 129 genera (Appendix 1), with a broad range of minimum interspecific distance and local minimum. For most genera, this is the first time that a threshold value is evaluated based on empirical data. The PCI analyses exposed a high number of sequences associated with potentially misidentified taxa—one out of four species on BOLD presents specimens wrongly labeled.
4.1 How is the Pentatomomorpha tome sorted in the BOLD library?
The available data on BOLD are biased to families with economic importance. Economic losses leveraged molecular studies through cox1 to refine the knowledge on crop pest species (e.g., Barman et al., 2017; Gariepy et al., 2019; Soares et al., 2018). For instance, established biosafety and quarantine protocols use DNA barcoding to assess pest diversity on a global scale (Boykin et al., 2012; Lee et al., 2019). For pentatomomorphans, of which egg and immature identification is a challenging task (Lis et al., 2013; Matesco et al., 2014), the diagnosis using this molecular tool may promote a fast and accurate species identification, playing a pivotal role in crop pest management.
Besides being the most speciose family within Pentatomomorpha (Henry, 2017), Pentatomidae encompasses numerous crop pests (Schaefer & Panizzi, 2000). The pentatomids brown marmorated stink bug [Halyomorpha halys (Stål)], green stink bug [Chinavia hilaris (Say)], dusky stink bug [Euschistus tristigmus (Say)], and Neotropical brown stink bug [Euschistus heros (Fabricius)] sum alone almost 1400 sequences in our final datasets. They are aggressive crops in different regions. Chinavia hilaris and E. tristigmus occur in North America, while E. heros occurs mostly in South America (Schaefer & Panizzi, 2000; Smaniotto & Panizzi, 2015). The H. halys is native from Asia and has invaded other continents [i.e., America (Hoebeke & Carter, 2003) and Europe (Wermelinger et al., 2008)] The brown marmorated stink bug shows rapid spreading throughout invaded territories (Zhu et al., 2012) and seems to be the most prominent pentatomid pest in the past two decades.
The second most representative family in our sample was Lygaeidae, with Nysius Dallas comprising more than half of the sequences for the family, and in fact, being the genus with most analyzed sequences (1695 sequences) among our datasets. Although Nysius groups around a hundred valid species (Malipatil, 2010), our dataset presents only 17 species labels. Nysius binotatus (Germar) (as commonly found on BOLD) is currently a subspecies of Nysius senecionis (Schilling) (treated here only by the binomen), and this is the species with the greatest contribution of sequences for our dataset (1116 sequences). These data are from exploratory efforts on Saharo-Arabian region with year-long deployment of Malaise traps and show the high abundance of N. senecionis in the region. Nysius species occur worldwide and feed on the seeds of a broad range of economically important host plants (Ge & Li, 2019; Schaefer & Panizzi, 2000). The birch catkin bug [Kleidocerys resedae (Panzer)] is another lygaeid species prevalent in our datasets (396 sequences). This species has a Holarctic distribution and is a pest of birch and alder trees, feeding and breeding on the seed catkin (Dioli et al., 2019; Schaefer & Panizzi, 2000).
Only some small families lack cox1 data in BOLD (Table 1). Most of them are endemic to one zoogeographical region, are rare, and have little knowledge of their natural histories (e.g., Canopidae, Cryptorhamphidae, Hyocephalidae). Although Termitaphididae is widespread, occurring in Australian, Afrotropical, Neotropical, and Oriental regions (Schuh & Weirauch, 2020), it is also a small family, with nine species without barcoding data. The lack of barcoding data for Idiostoloidea mirrors the scientific knowledge for its six species: Little is known concerning their natural history, systematics, and evolution (Schuh & Weirauch, 2020). Within the Pentatomomorpha tome, many taxa are blank pages waiting to be filled with barcode sequences.
4.2 Using thresholds to discover species by the book
Some DNA barcoding analyses (e.g., gap discovery, threshold values) perform better if based on curated libraries (Meyer & Paulay, 2005) due to their sample dependency. The singular coalescent depths of each lineage (e.g., genus, species, population) produce a practical challenge to cluster/split sequences in operational taxonomic units (Fujita et al., 2012; Zinger & Philippe, 2016). Hence, assuming a fixed threshold value for higher taxonomic levels is unrealistic and broadly accepted that a comprehensive haplotype sampling and species coverage leads to more reliable values (e.g., DeSalle & Goldstein, 2019; Meyer & Paulay, 2005). The use of empirical data is the best first step to look for barcoding gaps and threshold values (e.g., Gonçalves et al., 2021). The amassing of data for a lineage will progressively reduce inaccurate identification, false positives, false negatives (Meyer & Paulay, 2005) and shed light on cryptic species and specimen identification accuracy. It is convenient to review these values whenever new samples are available in databases (see Qing et al., 2020).
Arbitrary fixed threshold values have been used and tested in Pentatomomorpha (e.g., Park et al., 2011; Zhao et al., 2018). Previous research with Cletus Stål species showed that the use of popular fixed threshold values [e.g., 1% BOLD (Ratnasingham & Hebert, 2007) and 2% (Hebert et al., 2003)] would overestimate the genus diversity (Zhang et al., 2017). The authors suggest a threshold value above 7% based on the minimum interspecific distance, which would help to solve the overestimation of diversity. Our result for the minimum interspecific distance was similar to that (6.86%), and the local minimum was 3.30%. Here we provide both minimum interspecific distance and local minima for three out of four tested genera (Appendix 1). Future studies may benefit from these empirical values, since they are more reliable than an arbitrary fixed threshold. As the taxonomic knowledge for the taxa under study is refined and new sequences are generated, reassessing these values will increase their reliability and improve their practical applications,
The local minimum represents the barcoding gap, where a consistent dip in the density of genetic distances split intra- and interspecific distances is observed (Brown et al., 2012). Although the use of minimum interspecific distance is widespread in the literature as a threshold value (e.g., Meier et al., 2008; Song, Lin, Wang, & Wang, 2018), this may result in misidentifications when sequences from different species have distance values falling below this threshold, increasing the chance of false negatives. Since we understand DNA barcoding as a source of evidence and not the only way to make taxonomic decisions, the local minimum provides more rigor to identify molecular entities (see Casiraghi et al., 2010). It would flag more samples to be explored with other evidence, allowing more robust taxonomic decisions (e.g., identification, species description, synonyms).
The PCI and barcoding gap analysis are methods based on genetic distances that work in distinct scopes. While the PCI identifies discrepancies on the cohesion of the data and points out putative misidentification, the barcoding gap analysis shows the distribution of intra- and interspecific distances disclosing the existence (or not) of a gap. Higher PCI values tend to lead to good barcoding performances (Figure 5), but the relationship between both analyses is not obvious. For instance, Leptocorisa Latreille (Alydidae), Catorhintha Stål (Coreidae), Neortholomus Hamilton (Lygaeidae), and Padaeus Stål (Pentatomidae) presented 0% of PCI although a good barcoding performance. It means the pool of samples for each taxa presented a clear gap, although these specimens seem to be wrongly labeled. The opposite scenario is also possible, where DNA barcoding can be used for specimen identification without a consistent barcoding gap (see Collins & Cruickshank, 2012). Thus, these methods are complementary to taxonomic approaches using barcoding.
Most superfamilies analyzed here presented PCI values around 70%, but Aradoidea showed 94.34%. Grebennikov and Heiss (2014) and Grebennikov and Heiss (2018) are responsible for most of Aradidae sequences deposited in BOLD. The authors built a reference barcoding library for flat bugs (Aradidae) species and identified many cryptic new species exploring the Tanzanian fauna. High PCI values reflect rigorous taxonomic treatment. Although DNA barcoding may speed up the taxonomy, this is only possible (or at least should be) when molecular data meet taxonomic experts—but the training of such taxonomists is a long-term commitment that usually takes many years (Coleman & Radulovici, 2020). The slow increase in taxonomists is incompatible with the increase in articles dependent on taxonomic services (Wang et al., 2020). Specimen identification by non-specialists can be risky, and it is a source for many misidentification in databases.
Misidentification is an issue inherent to any public databases, where there is a claim for accurate identifications and rigorous curation (van den Burg et al., 2020). Ideally, all sequences in BOLD should be derived from vouchered specimens identified by taxonomic experts (Lis et al., 2016; Meiklejohn et al., 2019) and obtained through rigorous laboratory protocols. DNA barcoding success may also be affected by non-biological processes, referred by Mutanen et al., (2016) as sources of operational bias. These include inaccurate reference taxonomy, misidentifications, amplification of non-target sequences, alignment errors, erroneous sequencing due to contamination, and other methodological issues. Operational biases impact the reliability of the libraries and complicate the taxonomy and systematics of species (Mutanen et al., 2016).
We emphasize that the threshold values suggested here are not a “magic bullet” for species discrimination, but a source of evidence to this. Species delimitation based on DNA barcodes is hampered, for instance, in scenarios of hybridization or introgression, incomplete lineage sorting, and bacterial endosymbionts changing pathways of mtDNA inheritance (Magoga et al., 2018; Mutanen et al., 2016). Thus, this kind of approach as the single evidence for taxonomic decisions should be avoided. We advocate for the use of integrative approaches, which produce stronger species hypotheses (Padial et al., 2010).
4.3 Empirical examples: How may these data clue future studies?
4.3.1 Green flags: Fixing synonyms improves performance
We tracked the improvement of the results during filtering steps, and we found that misspellings and synonyms impacted the finding of barcoding gaps for some genera. On BOLD, Cosmopepla Stål is represented by four species labels. Before our last filter (checking synonyms and looking for misspellings), the barcoding gap analysis for this genus disclosed a poor efficiency (data not shown). However, the two-spotted stink bug [Cosmopepla bimaculata (Thomas)] labels 38 sequences. This name is a junior synonym of Cosmopepla lintneriana (Kirkaldy), which in our final dataset presented 44 sequences due to the lumping of these species labels. Another taxonomic issue in Cosmopepla was the misspelling of Cosmopepla intergressus, which probably refers to Cosmopepla intergressa (Uhler), a valid name. After solving these taxonomic issues, the final analysis for Cosmopepla resulted in good efficiency (Figure 3).
Misspellings and synonyms will inevitably worsen the performance of PCI and barcoding gap analyses. In both cases, disparate names for the same taxonomic unit will often lead to interspecific distance being masked as intraspecific distances. Thus, PCI would fail to distinguish the two putative names (i.e., species), suggesting a problematic identification or delimitation in the pool of sampled specimens; at the same time, it would distort the boxplots, decreasing the efficiency to detect a true barcoding gap.
4.3.2 Yellow flags: Are speciose genera a bad omen?
We found that the efficiency of the barcoding gap was intermediate for some genera (e.g., Euschistus Dallas, Geocoris Fallen, Leptoglossus Leach). Some Pentatomomorpha need extensive efforts and expertize to accurate identification (e.g., Cioato et al., 2015; Packauskas & Schaefer, 2001). For instance, Nysius (Figure 3) was described to contain 11 species (Dallas, 1852). Since then, some species were transferred to other genera, and many others have been added to, resulting in Nysius as a speciose taxon (Barber, 1947; Malipatil, 2010). The interspecific morphological similarities and intraspecific coloration mislead taxonomists and have given taxa of this genus the reputation of being “hard to identify” (Nakatani, 2015). Reports of taxonomic misidentification are spread in the literature for decades (e.g., Barber, 1947; Beardsley, 1977; Nakatani, 2015; Schaefer & Panizzi, 2000; Swezey, 1942). The PCI results corroborate this chaotic scenario, suggesting that only three out of the 17 analyzed species have a pool of correctly identified specimens. The intermediate efficiency of barcoding may be related to a relatively high number of misidentification, noising a putative barcoding gap among species.
4.3.3 Red flags: Poor performance—Don't panic!
While the intermediate efficiency of barcoding gap mirrors relatively scarce misidentification, the poor efficiency suggests gross taxonomic inconsistencies in the dataset (Figure 5). Most of the genera included in this rank of efficiency were, again, those with historically problematic taxonomy, such as Blissus Burmeister (Blissidae), Mozena Amyot & Serville (Coreidae), and Physopelta Amyot & Serville (Largidae) (Brailovsky & Barrera, 2001; Henry et al., 2015; Stehlík, 2013). Besides these emblematic scenarios, we also found the case of Schraderiellus Rider.
Schraderiellus is represented in our sample by specimens from Costa Rica of Schraderiellus cinctus (Ruckes) and Schraderiellus hughesae (Ruckes). These sequences were deposited in BOLD in 2012. At the time, only these two species were valid in the genus. The barcoding gap analysis resulted in poor efficiency (Figure 3), and the PCI also failed to distinguish species (Appendix 1). These analyses raise a taxonomic red flag to this genus, indicating, for example, cryptic species or operational biases. Roell and Campos (2018) reviewed Schraderiellus and described five new species. Currently, this genus occurs from Nicaragua to Ecuador, being most species recorded to Costa Rica (Roell & Campos, 2018).
Checking the voucher pictures available in BOLD, we found misidentifications. The records ASIHE211-12, ASIHE214-12, ASIHE215-12, and ASIHE260-12 are identified as S. hughesae; however, only ASIHE211-12 seems to be correctly identified; the remain specimens indeed belong to other species (S. cinctus or Schcraderiellus rufilineatus Roell & Campos). The records ASIHE196-12, ASIHE200-12, and ASIHE203-12 are identified as S. cinctus. Again, only the first seems to be correctly identified, while the last two likely belong to other species (i.e., S. rufilineatus) (T. Roell pers. comm.). This case exemplifies how barcoding analyses may clue taxonomic problems (e.g., misidentification, cryptic species) for in-depth studies, as already pointed out by many authors (e.g., Lis et al., 2016; Zhang et al., 2017).
4.4 Closing the book, for now
Molecular data changed drastically the way science produces and evaluates biological knowledge. Since the description and understanding of biodiversity are remarkably incomplete (Costello et al., 2013; Sheth & Thaker, 2017), the approaches to optimize robust species delimitation and specimen identification are desired. Here, we provide an overview of the available cox1 information for Pentatomomorpha deposited in BOLD. After inspecting more than 12,000 sequences, we verified for the first time the existence of barcoding gaps and provided individual threshold values for 132 genera, also casting doubt on the label on many specimens. Our results may be used to mitigate the Linnean shortfall—the knowledge gap between formally described species and the number of extant species. The Linnean shortfall underpins the biological knowledge, since the underlying recognition of the biological units precedes any study (see Hortal et al., 2015).
The use of cox1 as a taxonomic tool is copious, and our empirical examples reaffirm the efficiency of DNA barcoding to identify taxonomic inconsistencies. A careful handle of the data (i.e., samplings, specimens identification, laboratory protocols, or deposit of sequences in libraries) and interpretation of the results are mandatory. The simple act to double-check for invalid names and misspellings may denoise analyses and improve results. Here, we flag many genera for further investigations based on the state of knowledge of this infraorder. Future studies, including new sequences and species, may refine values and sharpen the accuracy of barcode tools.
ACKNOWLEDGEMENTS
We are thankful to Dr Paul Masonick and Dr Emília Wendt for the critical reading of this manuscript; and Dr Pablo Dellapé and MSc Wanessa Costa for helping us to compose Figure 1. We also would like to thank Dr Talita Roell for the comments on Schraderiellus and the two anonymous reviewers for their valuable comments. FMB is supported by a postdoctoral fellowship from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (PNPD/CAPES)—Finance code 001; LTG is supported by a doctoral fellowship from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq).
APPENDIX 1
Performance category, maximum intraspecific distance (limit of upper intraspecific whisker), minimum interspecific distance (limit of lower interspecific whisker), and local minima for all analyzed genera. In parentheses, the Probability of Correct Identification (PCI). Values are in percent
| Taxa (PCI) | Performance | Maximum intraspecific distance | Minimum interspecific distance | Local minima |
|---|---|---|---|---|
| Aradoidea (94.34) | ||||
| Aradidae (94.34) | ||||
| Afropictinus (50) | Intermediate | 13.37 | 11.55 | |
| Aneurus (88.88) | Intermediate | 19.73 | 18.02 | |
| Aradus (100) | Good | 2.24 | 10.64 | 4.01 |
| Brachyrhynchus (100) | Good | 9.88 | 12.69 | 8.57 |
| Ctenoneurus (100) | — | — | — | |
| Dysodius (100) | Good | 6.69 | 11.40 | 6.79 |
| Hesus (100) | Good | 0.25 | 17.48 | 7.69 |
| Isodermus (100) | — | — | — | |
| Jacobsaptera (100) | — | — | — | |
| Libiocoris (100) | — | — | — | |
| Malgasyaptera (100) | — | — | — | |
| Mauricicoris (100) | — | — | — | |
| Mezira (100) | Good | 0.00 | 17.17 | 6.67 |
| Neochelonoderus (100) | Good | 0.25 | 21.62 | 11.14 |
| Neuroctenus (100) | Good | 7.14 | 12.18 | 9.59 |
| Overlatiella (100) | — | — | — | |
| Pericartaptera (100) | — | — | — | |
| Pseudomezira (100) | — | — | — | |
| Stysaptera (100) | Good | 0.30 | 12.77 | 5.61 |
| Usumbaraia (100) | — | — | — | |
| Wuiessa (100) | — | — | — | |
| Coreoidea (69.90) | ||||
| Alydidae (40) | ||||
| Alydus (40) | Intermediate | 4.12 | 0.00 | |
| Heegeria (0) | — | — | — | |
| Leptocorisa (0) | Good | 2.58 | 7.20 | 4.18 |
| Megalotomus (100) | — | — | — | |
| Noliphus (100) | — | — | — | |
| Protenor (100) | — | — | — | |
| Riptortus (33.33) | Good | 0.61 | 5.21 | 0.08 |
| Tollius (0) | — | — | — | |
| Coreidae (74.55) | ||||
| Acanthocephala (100) | Good | 1.16 | 12.52 | |
| Amblypelta (100) | — | — | — | |
| Anasa (50) | Good | 0.81 | 11.84 | |
| Anoplocnemis (100) | Good | 0 | 23.97 | 8.75 |
| Arenocoris (100) | — | — | — | |
| Bathysolen (100) | — | — | — | |
| Catorhintha (0) | Good | 10.92 | 11.09 | 5.81 |
| Ceraleptus (100) | Good | 0.76 | 8.97 | 6.42 |
| Chelinidea (100) | Good | 10.60 | 14.67 | 4.58 |
| Cletus (44.44) | Good | 2.78 | 6.86 | 3.10 |
| Coreus (100) | — | — | — | |
| Coriomeris (66.66) | Poor | 7.13 | 3.19 | |
| Enoplops (100) | — | — | — | |
| Euthochtha (100) | — | — | — | |
| Gonocerus (100) | Good | 0.30 | 8.36 | 4.32 |
| Homoeocerus (100) | Good | 1.22 | 16.37 | 4.78 |
| Hydaropsis (100) | — | — | — | |
| Hypselonotus (66.66) | Poor | 1.22 | 10.76 | |
| Leptoglossus (83.33) | Intermediate | 2.97 | 1.96 | |
| Madura (100) | — | — | — | |
| Mictis (100) | — | — | — | |
| Mozena (33.33) | Poor | 0.46 | 0.30 | |
| Notopteryx (100) | — | — | — | |
| Petillia (100) | — | — | — | |
| Piezogaster (100) | Good | 1.37 | 3.80 | 13.24 |
| Plinachtus (100) | Good | 0.65 | 13.40 | 7.48 |
| Prismatocerus (100) | Good | 0.00 | 12.77 | 6.29 |
| Spathocera (100) | — | — | — | |
| Syromastus (100) | — | — | — | |
| Thasus (100) | — | — | — | |
| Villasitocoris (0) | — | — | — | |
| Rhopalidae (75) | ||||
| Arhyssus (20) | Good | 2.23 | 9.86 | 5.01 |
| Aufeius (100) | — | — | — | |
| Boisea (0) | Poor | 2.78 | 0 | |
| Brachycarenus (100) | — | — | — | |
| Corizus (100) | — | — | — | |
| Harmostes (100) | Good | 2.21 | 11.25 | 3.76 |
| Leptocoris (100) | Good | 1.99 | 10.06 | 4.45 |
| Liorhyssus (100) | Good | 2.65 | 0 | 4.87 |
| Myrmus (100) | — | — | — | |
| Rhopalus (100) | Good | 0.33 | 10.97 | 5.69 |
| Stictopleurus (75) | Intermediate | 2.67 | 0.46 | |
| Stenocephalidae (100) | ||||
| Dicranocephalus (100) | Good | 0 | 10.26 | 2.28 |
| Lygaeoidea (76.41) | ||||
| Artheneidae (100) | ||||
| Chilacis (100) | — | — | — | |
| Berytidae (78.95) | ||||
| Berytinus (100) | Good | 0.50 | 9.27 | 4.98 |
| Gampsocoris (100) | Good | 0.46 | 14.24 | 6.66 |
| Hoplinus (100) | — | — | — | |
| Jalysus (60) | Good | 0.74 | 7.37 | 3.67 |
| Metatropis (100) | Good | 0.17 | 5.88 | 3.02 |
| Neides (100) | — | — | — | |
| Neoneides (100) | — | — | — | |
| Yemma (0) | — | — | — | |
| Yemmalysus (0) | — | — | — | |
| Blissidae (50) | ||||
| Blissus (0) | Poor | 1.38 | 0.15 | |
| Ischnodemus (100) | Good | 2.46 | 16.89 | 8.32 |
| Phaenacantha (100) | — | — | — | |
| Cymidae (87.50) | ||||
| Cymus (87.50) | Good | 0.37 | 7.13 | 3.53 |
| Geocoridae (60) | ||||
| Geocoris (60) | Intermediate | 1.99 | 0 | |
| Heterogastridae (100) | ||||
| Heterogaster (100) | — | — | — | |
| Platyplax (100) | — | — | — | |
| Lygaeidae (53.85) | ||||
| Arocatus (25) | Good | 1.22 | 8.55 | 5.87 |
| Aspilocoryphus (100) | — | — | — | |
| Belonochilus (100) | — | — | — | |
| Graptostethus (100) | Good | 1.85 | 5.62 | Na |
| Kleidocerys (50) | Intermediate | 6.09 | 5.10 | |
| Lygaeospilus (100) | Good | 1.91 | 6.98 | 4.33 |
| Lygaeus (50) | Good | 1.22 | 10.91 | 8.06 |
| Melacoryphus (75) | Good | 2.28 | 6.99 | 4.20 |
| Melanocoryphus (100) | — | — | — | |
| Melanopleurus (100) | Good | 3.00 | 13.32 | 5.92 |
| Neortholomus (0) | Good | 1.28 | 8.75 | 2.16 |
| Nithecus (100) | — | — | — | |
| Nysius (20) | Intermediate | 0 | 0 | |
| Ochrimnus (100) | Good | 0.46 | 9.97 | 5.47 |
| Oncopeltus (100) | Good | 0.30 | 11.83 | 6.75 |
| Orsillus (100) | Good | 0 | 5.50 | 11.00 |
| Ortholomus (50) | Poor | 13.02 | 0.16 | |
| Spilostethus (33.33) | Poor | 21.04 | 7.25 | |
| Tropidothorax (100) | — | — | — | |
| Xyonysius (100) | — | — | — | |
| Malcidae (100) | ||||
| Chauliops (100) | — | — | — | |
| Malcus (100) | — | — | — | |
| Oxycarenidae (83.33) | ||||
| Oxycarenus (80) | Intermediate | 0.77 | 0 | |
| Pachygronthidae (100) | ||||
| Oedancala (100) | Good | 0.32 | 10.94 | 5.77 |
| Phlegyas (100) | Good | 1.23 | 10.57 | 6.27 |
| Piesmatidae (100) | ||||
| Parapiesma (100) | Good | 0.74 | 13.59 | 7.76 |
| Piesma (100) | — | — | — | |
| Rhyparochromidae (89.16) | ||||
| Acompus (100) | — | — | — | |
| Antillocoris (100) | Good | 0.15 | 8.04 | 4.45 |
| Atrazonotus (100) | — | — | — | |
| Beosus (100) | — | — | — | |
| Cnemodus (100) | — | — | — | |
| Cryphula (100) | — | — | — | |
| Drymus (100) | Good | 0.40 | 10.36 | 5.59 |
| Elasmolomus (100) | — | — | — | |
| Emblethis (100) | Good | 9.83 | 11.09 | 11.06 |
| Eremocoris (50) | Intermediate | 0 | 0 | |
| Gastrodes (100) | Good | 0.74 | 10.62 | 5.85 |
| Graptopeltus (100) | — | — | — | |
| Heraeus (100) | — | — | — | |
| Ischnocoris (100) | Good | 0.35 | 4.61 | 2.36 |
| Kolenetrus (100) | — | — | — | |
| Lanchnophorus (100) | — | — | — | |
| Lamprodema (100) | — | — | — | |
| Laryngodus (100) | — | — | — | |
| Ligyrocoris (33.33) | Intermediate | 0.48 | 0.17 | |
| Macrodema (100) | — | — | — | |
| Malezonotus (100) | Good | 1.40 | 9.27 | 4.72 |
| Megalonotus (75) | Good | 0.38 | 0.75 | 5.02 |
| Metochus (100) | — | — | — | |
| Myodocha (100) | — | — | — | |
| Neolethaeus (100) | — | — | — | |
| Neopamera (100) | — | — | — | |
| Neosuris (100) | — | — | — | |
| Ozophora (100) | — | — | — | |
| Pachybrachius (100) | — | — | — | |
| Paromius (100) | Good | 0 | 7.45 | 3.87 |
| Perigenes (100) | — | — | — | |
| Peritrechus (100) | Good | 3.69 | 9.27 | 6.84 |
| Plinthisus (100) | Good | 0 | 9.72 | 5.16 |
| Pseudocnemodus (0) | — | — | — | |
| Pseudopachybrachius (100) | Good | 0.49 | 5.52 | 3.08 |
| Pterotmetus (100) | — | — | — | |
| Raglius (100) | Good | 0.61 | 15.42 | 8.39 |
| Remaudiereana (100) | — | — | — | |
| Rhyparochromus (100) | Good | 0.61 | 9.22 | 5.20 |
| Scolopostethus (100) | Good | 3.71 | 4.10 | 5.61 |
| Sisamnes (100) | — | — | — | |
| Slaterobius (0) | — | — | — | |
| Sphragisticus (0) | — | — | — | |
| Stygnocoris (100) | Good | 1.32 | 12.09 | 6.66 |
| Trapezonotus (0) | Poor | 4.74 | 0.15 | |
| Uhleriola (100) | — | — | — | |
| Valtissius (100) | — | — | — | |
| Xanthochilus (100) | — | — | — | |
| Zeridoneus (100) | — | — | — | |
| Pentatomoidea (73.84) | ||||
| Acanthosomatidae (85) | ||||
| Acanthosoma (100) | Good | 1.32 | 8.25 | 2.97 |
| Cyphostethus (100) | Good | 0.26 | 9.70 | 4.34 |
| Elasmostethus (100) | Good | 1.33 | 1.65 | 1.48 |
| Elasmucha (50) | Good | 0.59 | 3.03 | 2.69 |
| Lindbergicoris (100) | Good | 0.82 | 4.53 | 8.57 |
| Sastragala (100) | Good | 5.91 | 7.22 | Na |
| Cydnidae (92.86) | ||||
| Adrisa (100) | — | — | — | |
| Aethus (100) | — | — | — | |
| Amnestus (100) | Good | 0.74 | 15.43 | 3.56 |
| Geotomus (100) | — | — | — | |
| Macroscytus (66.66) | Poor | 3.35 | 1.42 | |
| Melanaethus (100) | — | — | — | |
| Microporus (100) | Good | 3.65 | 17.63 | 7.84 |
| Pangaeus (100) | — | — | — | |
| Sehirus (100) | — | — | — | |
| Tritomegas (100) | Good | 3.19 | 13.98 | 7.19 |
| Thyreocorinae (66.67) | ||||
| Corimelaena (80) | Poor | 1.41 | 10.65 | |
| Galgupha (0) | Poor | 13.51 | 0.30 | |
| Dinidoridae (60) | ||||
| Coridius (50) | Poor | 13.95 | 3.27 | |
| Cyclopelta (66.66) | Poor | 3.08 | 1.57 | |
| Pentatomidae (72.13) | ||||
| Ablaptus (100) | — | — | — | |
| Acesines (100) | — | — | — | |
| Acrosternum (0) | Poor | 13.84 | 5.80 | |
| Adevoplitus (100) | — | — | — | |
| Aelia (100) | Good | 1.83 | 11.21 | 7.41 |
| Aeliomorpha (100) | — | — | — | |
| Agonoscelis (100) | Good | 0.71 | 13.27 | 7.34 |
| Agroecus (100) | — | — | — | |
| Amaurochrous (100) | — | — | — | |
| Amyotea (100) | — | — | — | |
| Ancyrosoma (100) | — | — | — | |
| Andrallus (100) | — | — | — | |
| Antestiopsis (100) | — | — | — | |
| Antheminia (100) | — | — | — | |
| Apateticus (100) | Good | 3.71 | 10.96 | 6.32 |
| Arma (50) | Poor | 3.30 | 0.00 | |
| Arocera (100) | Good | 2.48 | 14.14 | 6.59 |
| Arvelius (100) | Good | 9.18 | 14.14 | 5.16 |
| Bagrada (100) | Good | 1.39 | 12.72 | 1.81 |
| Banasa (42.85) | Good | 2.23 | 10.44 | 3.90 |
| Berecynthus (100) | — | — | — | |
| Boea (100) | — | — | — | |
| Brachystethus (50) | Good | 1.98 | 14.92 | 8.57 |
| Braunus (100) | — | — | — | |
| Brepholoxa (100) | — | — | — | |
| Brochymena (80) | Good | 1.24 | 3.67 | 2.51 |
| Carbula (50) | Good | 1.22 | 11.70 | 6.16 |
| Carpocoris (80) | Good | 1.53 | 7.34 | 2.31 |
| Cermatulus (100) | — | — | — | |
| Chinavia (50) | Good | 1.06 | 6.77 | 1.35 |
| Chlorochroa (28.57) | Good | 0.92 | 3.50 | 2.78 |
| Chlorocoris (50) | Intermediate | 1.70 | 7.84 | |
| Codophila (100) | Good | 0.33 | 15.98 | 8.53 |
| Coenus (100) | — | — | — | |
| Cosmopepla (100) | Good | 0.75 | 7.95 | 1.36 |
| Cuspicona (100) | — | — | — | |
| Cyptocephala (100) | — | — | — | |
| Dalpada (100) | Good | 1.98 | 12.28 | 5.41 |
| Degonetus (100) | — | — | — | |
| Dendrocoris (100) | Intermediate | 11.55 | 10.00 | |
| Dictyotus (100) | — | — | — | |
| Dinocoris (0) | Intermediate | 10.92 | 0 | |
| Dinorhynchus (100) | — | — | — | |
| Dolycoris (50) | Poor | 4.04 | 0.54 | |
| Dryptocephala (100) | — | — | — | |
| Edessa (91.66) | Good | 1.83 | 11.57 | 3.32 |
| Eludocoris (100) | — | — | — | |
| Eocanthecona (100) | — | — | — | |
| Eritrachys (100) | — | — | — | |
| Erthesina (100) | Good | 0.76 | 6.39 | 3.52 |
| Eurydema (16.66) | Good | 1.17 | 2.13 | 2.97 |
| Euschistus (75) | Intermediate | 17.69 | 4.84 | |
| Eysarcoris (80) | Good | 6.03 | 8.16 | 6.80 |
| Glaucias (100) | Good | 0.18 | 9.17 | 4.80 |
| Gonopsis (100) | Good | 0.00 | 7.46 | 2.72 |
| Graphosoma (50) | Intermediate | 3.19 | 0.00 | |
| Gynenica (100) | — | — | — | |
| Halyomorpha (50) | Good | 1.46 | 12.32 | 1.65 |
| Halys (0) | Intermediate | 8.81 | 5.93 | |
| Holcogaster (100) | — | — | — | |
| Holcostethus (60) | Good | 0.85 | 9.88 | 6.55 |
| Hymenarcys (100) | Good | 2.04 | 16.92 | 9.23 |
| Kermana (0) | — | — | — | |
| Loxa (0) | Poor | 23.57 | 2.98 | |
| Macropygium (100) | — | — | — | |
| Mecidea (100) | — | — | — | |
| Menecles (100) | — | — | — | |
| Menida (100) | Good | 0.31 | 13.39 | 7.54 |
| Mormidea (60) | Good | 2.73 | 9.44 | 4.90 |
| Murgantia (50) | Good | 3.20 | 15.67 | 9.32 |
| Neotibilis (0) | — | — | — | |
| Neottiglossa (100) | Good | 0.76 | 8.69 | 3.98 |
| Nezara (0) | Intermediate | 7.84 | 4.69 | |
| Odmalea (100) | — | — | — | |
| Oebalus (100) | Good | 2.58 | 10.42 | 6.76 |
| Oechalia (0) | — | — | — | |
| Padaeus (0) | Good | 0.33 | 4.87 | 2.89 |
| Pallantia (100) | — | — | — | |
| Palomena (100) | Good | 0.74 | 6.81 | 2.33 |
| Pantochlora (100) | — | — | — | |
| Parabrochymena (0) | Poor | 0.46 | 0.00 | |
| Parastalius (3) | — | — | — | |
| Pellaea (100) | Good | 11.66 | 18.11 | 6.33 |
| Pentatoma (100) | Good | 0.30 | 10.06 | 3.58 |
| Perillus (100) | Good | 1.83 | 2.69 | 1.84 |
| Peromatus (50) | Poor | 14.46 | 9.74 | |
| Pharypia (100) | — | — | — | |
| Picromerus (100) | Good | 0.00 | 10.86 | 4.70 |
| Piezodorus (75) | Good | 1.74 | 4.40 | 3.27 |
| Placosternum (100) | — | — | — | |
| Plautia (66.66) | Good | 1.03 | 8.78 | 3.92 |
| Podisus (25) | Intermediate | 2.34 | 1.80 | |
| Prionosoma (100) | — | — | — | |
| Proxys (100) | Good | 1.24 | 9.81 | 5.34 |
| Ramosiana (100) | — | — | — | |
| Rhaphigaster (100) | — | — | — | |
| Rhyncholepta (100) | — | — | — | |
| Rhyssocephala (100) | Good | 0.20 | 12.90 | 6.14 |
| Rio (100) | Good | 0.99 | 10.42 | 5.12 |
| Roferta (100) | — | — | — | |
| Rubiconia (100) | — | — | — | |
| Sarju (100) | — | — | — | |
| Schraderiellus (0) | Poor | 13.61 | 1.38 | |
| Sciocoris (100) | Good | 1.85 | 15.26 | 8.94 |
| Scotinophara (100) | Good | 0.22 | 7.22 | 3.23 |
| Sibaria (100) | — | — | — | |
| Spermatodes (100) | — | — | — | |
| Stagonomus (100) | Good | 0 | 8.30 | 4.29 |
| Stiretrus (100) | — | — | — | |
| Taurocerus (100) | Good | 2.13 | 11.11 | 5.82 |
| Thyanta (16.66) | Intermediate | 1.38 | 0.00 | |
| Tolumnia (100) | Good | 0.66 | 7.91 | 4.25 |
| Trichopepla (50) | Intermediate | 7.44 | 5.63 | |
| Troilus (100) | — | — | — | |
| Vulsirea (100) | — | — | — | |
| Zicrona (100) | — | — | — | |
| Plataspidae (60) | ||||
| Brachyplatys (100) | — | — | — | |
| Coptosoma (100) | Good | 0.75 | 7.95 | 9.64 |
| Megacopta (0) | Poor | 2.13 | 0.15 | |
| Scutelleridae (69.57) | ||||
| Augocoris (0) | — | — | — | |
| Choerocoris (100) | — | — | — | |
| Chrysocoris (0) | Poor | 4.48 | 0.17 | |
| Coleotichus (0) | — | — | — | |
| Eucorysses (100) | — | — | — | |
| Eurygaster (50) | Intermediate | 0.87 | 2.11 | |
| Homaemus (75) | Good | 3.66 | 7.89 | 5.16 |
| Hotea (100) | — | — | — | |
| Odontotarsus (100) | Good | 6.23 | 10.94 | 8.28 |
| Pachycoris (100) | Good | 2.88 | 12.78 | 6.28 |
| Phimodera (100) | — | — | — | |
| Solenostethium (100) | Good | 1.58 | 18.32 | 9.68 |
| Stethaulax (100) | — | — | — | |
| Vanduzeeina (100) | Good | 0.46 | 13.01 | 6.18 |
| Tessaratomidae (100) | ||||
| Eusthenes (100) | — | — | — | |
| Piezosternum (100) | — | — | — | |
| Tessaratoma (100) | Good | 0.00 | 11.29 | 5.91 |
| Urostylididae (83.33) | ||||
| Urochela (50) | Poor | 19.79 | 15.66 | |
| Urostylis (100) | Good | 1.72 | 14.29 | 7.08 |
| Pyrrhocoroidea (77.50) | ||||
| Largidae (80) | ||||
| Arhaphe (100) | Good | 2.43 | 9.12 | 5.26 |
| Delacampius (100) | — | — | — | |
| Iphita (100) | Good | 0.48 | 10.53 | 5.14 |
| Largus (100) | Good | 1.37 | 1.98 | 4.96 |
| Macrocheraia (100) | — | — | — | |
| Physopelta (100) | Poor | 2.29 | 0.00 | |
| Pyrrhocoridae (74.19) | ||||
| Antilochus (100) | Good | 1.90 | 11.08 | 6.16 |
| Dindymus (100) | Good | 0.16 | 28.12 | 13.01 |
| Dysdercus (53.33) | Good | 1.86 | 3.16 | 2.41 |
| Euscopus (100) | Good | 1.82 | 12.27 | 6.81 |
| Melamphaus (100) | Good | 8.04 | 12.90 | 4.88 |
| Probergrothius (100) | Good | 0.32 | 11.92 | 6.64 |
| Pyrrhocoris (100) | — | — | — | |
| Pyrrhopeplus (100) | Good | 3.09 | 3.34 | 0.82 |
| Scantius (100) | — | — | — | |




