Diversification of Prochilodus in the eastern Brazilian Shield: Evidence from complete mitochondrial genomes (Teleostei, Prochilodontidae)
Rosiane P. Santos and Bruno F. Melo contributed equally to this article.
Abstract
The Neotropical fish genus Prochilodus includes five species occurring in the main drainages of the eastern Brazilian Shield: P. argenteus, P. costatus, P. lineatus, P. harttii, and P. vimboides. Multi and single locus phylogenies have questioned the monophyly of P. costatus and P. lineatus, and the biogeographic history of these species in the Brazilian Shield has never been explored. We characterized new mitogenomes for these species to reconstruct their evolutionary history, estimated the timing of Prochilodus cladogenesis, and discussed the implications of past geologic events on species diversification. The phylogeny supports the monophyly of both P. costatus and P. lineatus, and indicates a Miocene divergence of P. vimboides, much earlier than subsequent species diversification. The Early Pleistocene split of P. argenteus and P. harttii (~2.1 million years ago—MYA) is hypothesized to be related to recent Quaternary reactivations of the Rio Araçuaí Fault that promoted river captures between the eastern tributaries of the São Francisco basin and the upper Rio Jequitinhonha in the Serra do Espinhaço mountain range. The time-calibrated phylogeny also indicated a subsequent split of P. costatus and P. lineatus (~1.1 MYA) likely due to Quaternary activities of the Upper Rio São Francisco crustal discontinuity and Estrela Fault that catalyzed river captures between the upper Rio São Francisco and the Rio Grande of the Paraná basin in the Serra da Canastra mountain range. These results provide a temporal context for the diversification of Prochilodus and bring new insights to further study the historical biogeography of other riverine fish groups along the upland basins of the eastern Brazilian Shield.
1 INTRODUCTION
The freshwater fish order Characiformes includes approximately 2260 species, distributed in 24 families with about 90% being endemic to the Neotropical region with the others found in Africa (Betancur-R et al., 2018; Fricke et al., 2020; Oliveira et al., 2011). One of the Neotropical endemic clades is the family Prochilodontidae, popularly known as curimbatás, bocachicos, or flannel-mouthed characiforms, that is comprised of three genera: Ichthyoelephas Posada 1909 (two species), Prochilodus Agassiz 1929 (13 species), and Semaprochilodus Fowler 1941 (six species) (Castro & Vari, 2004). Prochilodus encompasses bottom-feeding, detritivorous species of medium-size and is morphologically characterized by highly specialized lips, teeth, and jaws (Castro & Vari, 2004; Melo & Sidlauskas, 2017). They occur in all major South American basins and are commercially important for industrial and regional fisheries due to their natural abundance (Batista & Petrere Jr, 2003; Castro & Vari, 2004).
The taxonomic revision of the family delimited 13 species of Prochilodus, with five being from southern and southeastern South America: (i) P. argenteus Spix & Agassiz 1829, (ii) P. costatus Valenciennes 1850, (iii) P. harttii Steindachner 1875, (iv) P. lineatus (Valenciennes 1837), and (v) P. vimboides Kner 1859, but their interspecific relationships were not resolved entirely (Castro & Vari, 2004). A recent molecular phylogeny revealed that P. vimboides from coastal Brazilian rivers is sister to all remaining Prochilodus, and the other four species were placed in two well-supported clades: one composed by P. argenteus (Rio São Francisco) sister to P. harttii (Rio Pardo and Rio Jequitinhonha), and another composed by P. lineatus (Paraná-Paraguay system and Rio Paraíba do Sul) nested within the paraphyletic P. costatus (Rio São Francisco) (Melo et al., 2016). Further analyses using a mtDNA dataset and a larger taxon sampling supported the non-monophyly of P. costatus and P. lineatus (Melo et al., 2018), but a recent study using the mitochondrial control region obtained the monophyly of these two species (Chagas et al., 2020). However, all these studies used a single or few loci resulting in low node support in many instances along the topologies (Chagas et al., 2020; Melo et al.,2016, 2018), and thus some clades in the phylogeny of Prochilodus remain to be explored by a large-scale molecular analysis.
Both complete and nearly complete mitochondrial genomes are available for characiform fishes (e.g., Landínez-García et al., 2016; Nakatani et al., 2011; Saitoh et al., 2003), but a combined phylogenetic analysis has not been conducted yet. For Prochilodus, complete mitogenomes have been generated for P. argenteus, P. costatus, and P. lineatus, all presenting similar gene annotations and phylogenetic analyses supporting the monophyly of the genus (Chagas et al., 2016; Carmo et al., 2016). However, these studies did not include more than one specimen to test the monophyly of the species nor did they use other prochilodontids. Thus, gathering mitogenome data from multiple specimens from each species is crucial to test the monophyletic status of P. costatus and P. lineatus and to assess the timing for diversification and biogeography of Prochilodus.
Although recent advances have been achieved in understanding the evolutionary relationships of Prochilodus (Chagas et al., 2020; Melo et al., 2016, 2018), the biogeographic history of the genus remains uncertain, especially in the eastern Brazilian Shield. A large-scale phylogeographic analysis among Prochilodus species from three hydrographic basins using mtDNA data showed that the phylogenetic structure is not associated with geography (Sivasundar et al., 2001). However, it is unclear which biogeographic events are related to the diversification of Prochilodus from the Rio São Francisco and those from adjacent basins (i.e., Paraná-Paraguay system and Pardo/Jequitinhonha rivers) or the geologic factors potentially driving speciation processes. Although some studies have tested the influence of geomorphological structures (e.g., faults and lineaments) in the current distribution of several freshwater fish taxa (Abreu et al., 2019; Buckup, 2011; Ribeiro, 2006), few empirical studies have addressed biogeographic hypotheses along the eastern Brazilian Shield (Camelier & Zanata, 2014; Pereira et al., 2013; Wendt et al., 2019), and none of these studies have directly combined time-calibrated phylogenies and geological evidence to explain species diversification in the eastern Brazilian Shield.
In this study, we characterized and analyzed mitochondrial genomes of five Prochilodus species from the eastern Brazilian Shield: P. argenteus, P. costatus, P. lineatus, P. harttii, and P. vimboides, with the mitogenomes of the latter two being described for the first time. Our aims were to (i) test the monophyly of P. costatus and P. lineatus, (ii) estimate the divergence time for Prochilodus diversification, and (iii) associate the timing of interspecific diversification with the influence of recent geologic reactivations of faults and lineaments on the distribution and speciation processes throughout the eastern Brazilian Shield.
2 MATERIAL AND METHODS
2.1 Sample collection, DNA extraction, and sequencing
Taxon sampling included six specimens encompassing five species of Prochilodus: two P. lineatus from the Rio Grande of the Paraná basin, one P. argenteus and one P. costatus from the Rio São Francisco basin, one P. harttii from the Rio Jequitinhonha basin, and one P. vimboides from the Rio Doce basin (Table 1). Muscle tissues were stored at −80°C, and DNA was extracted from the specimens using a modified salting-out method (Aljanabi & Martinez, 1997). Vouchers are deposited in the Laboratório Genética da Conservação at Pontifícia Universidade Católica, Belo Horizonte, Brazil (LGC-PUC), available through accession codes: LGC550 for P. argenteus; LGC1183 for P. costatus; LGC1764 for P. harttii; LGC58 for P. lineatus; LGC4515 for P. vimboides. Genomic libraries for each sample were prepared using a Nextera kit (Illumina) and underwent next-generation sequencing (NGS) using a HiSeq 2000 (Illumina) at the Instituto Nacional do Câncer (INCA), Rio de Janeiro, Brazil. Another P. lineatus specimen is deposited at the Universidade Federal de São João del-Rei (LARGE-UFSJ voucher 140000) and was subject to a parallel HiSeq 2000 pipeline outlined in Yazbeck et al. (2016). The other samples are stored at the UFMG-BDT collection of the Universidade Federal de Minas Gerais (deposit codes: UFMG-BDT-PP000001 for P. argenteus and UFMG-BDT-PP000002 for P. costatus).
| Species | Accession numbers and references | Locality | Geographic coordinates | Pair-end reads | mtDNA reads | Mitochondrial genome coverage | Mitogenome size (bp) | AT (%) |
|---|---|---|---|---|---|---|---|---|
| P. argenteus |
KY358753 This study |
Rio Urucuia, São Francisco basin, Urucuia MG, Brazil | 16°10'38.1"S 45°41'03.6"W | 14,270,680 | 6897 | 107.4 | 16,696 | 44 |
| P. argenteus |
NC_027689 Chagas et al. 2016 |
Rio Paraopeba, São Francisco basin, Felixlândia MG, Brazil | 18°52'23.9"S 44°46'52.8"W | 378 | 16,697 | 44 | ||
| P. costatus 1 |
KY358754 This study |
Rio São Francisco, Três Marias MG, Brazil | 18°08'9.7"S 45°14'41.7"W | 20,899,298 | 9639 | 105.3 | 16,699 | 45 |
| P. costatus 2 |
NC_027690.1 Chagas et al. 2016 |
Rio Paraopeba, São Francisco basin, Felixlândia MG, Brazil | 18°52'23.9"S 44°46'52.8"W | 345 | 16,699 | 44 | ||
| P. harttii |
NC037715 This study |
Rio Itacambiruçu, Jequitinhonha basin, Cristália MG, Brazil |
16°36'24"S 42°49'46"W |
21,326,856 | 1715 | 23.8 | 16,697 | 44 |
| P. lineatus 1 |
KY358755 This study |
Rio Grande, upper Paraná basin, Planura MG, Brazil | 20°07'33"S 48°34'20"W | 13,440,844 | 8567 | 69.9 | 16,698 | 45 |
|
P. lineatus 2 |
KY825189 This study |
Rio Grande, upper Paraná basin, Itutinga MG, Brazil | 21°17'40''S 44°37'23'’ W | 33,859,998 | 29,868 | 76.3 | 16,699 | 45 |
| P. lineatus 3 |
NC_024939 Carmo et al. 2016 |
Rio Grande, upper Rio Paraná basin, Miguelópolis SP, Brazil | 20°01'53.8"S 48°13'27.5"W | 296 | 16,699 | 44 | ||
| P. vimboides |
NC037712 This study |
Rio Açucena, Doce basin, Timóteo MG, Brazil | 19°35'39.9''S 42°36'09.9''W | 7,685,235 | 2020 | 29.2 | 16,696 | 45 |
2.2 Mitochondrial genome assembly
The MIRA genome assembler (Chevreux, 2007) was used through the MITObim workflow (Hahn et al., 2013) to perform the mitogenome assemblies by reference, whereas the SPADES program (Nurk et al., 2013) was used to perform de novo assemblies. The number of reads used in the assembly and the achieved coverage were obtained from the TABLET program (Milne et al., 2013). The annotation of the mitochondrial genomes was performed by the Mitos webserver (Bernt et al., 2013) and Mitoannotator (Iwasaki et al., 2013) using default parameters. All mitogenomes went through careful manual curation with the aid of the ARTEMIS program (Carver et al., 2008).
Whole mitochondrial genome comparisons were produced using a customized R script (R Core Team, 2020). All nine Prochilodus mitochondrial genomes were aligned with MAFFT v7.470 (Katoh & Standley, 2013), and the alignments were analyzed with tools from the R packages “ape” 5.4 (Paradis & Schliep, 2019), “spider” 1.5.0 (Brown et al., 2012), and “genebankr” 1.16.0 (Becker & Lawrence, 2020). A sliding window of 60 bp was used to assess the Kimura-2-parameters (K2P) distance and p-distance models for each region of the mitogenomes in comparison with a base mitogenome of P. lineatus. We used a 60 bp window to better visualize the polymorphic sites.
2.3 Phylogenetic and time - calibrated analyses
In order to perform a phylogenetic analysis and time calibrate the phylogeny with the available fossil record, 46 assembled mitogenomes belonging to 17 families of Characiformes and one sample of Siluriformes (Pseudoplatystoma corruscans Spix & Agassiz 1829) to root the trees were selected and downloaded from NCBI/GenBank (Table S1). These mitogenomes were submitted to the PHYLOMITO pipeline (https://github.com/igorrcosta/phylomito) for the construction of a supermatrix including 13 protein-coding mitochondrial genes (Carvalho et al., 2016; Dumans et al., 2017). The pipeline is based on three distinct steps: (i) alignment of nucleotides; (ii) retrotranslation; and (iii) concatenation of the contigs (i.e., set of aligned reads). DAMBE 5.2.31 was used to estimate the substitution saturation index (Iss) and to evaluate the occurrence of saturation by substitution (Xia & Xie, 2001; Xia et al., 2003). The matrix was partitioned by codons of each gene; that is, each of the codon for the 13 mitochondrial genes was considered an independent partition, and the best nucleotide substitution model for the best partition schemes was estimated with PartitionFinder 2.1.1 (Lanfear et al., 2017).
The divergence time estimation and the reconstruction of the phylogeny were performed in BEAST 2.6.3 (Bouckaert et al., 2019). We used a partitioned matrix as input (Supporting information 1), an uncorrelated relaxed clock, and a birth–death model for the speciation likelihood (Supporting information 2). We estimated the topology using four calibration priors. The first was †Leporinus scalabrinii, an incomplete, articulated skull from the Ituzaingó Formation of the Late Miocene in Argentina (Bogan et al., 2012), hypothesized to be closer to Abramites Fowler 1906 (Bogan et al., 2012) and to have morphological affinities with Megaleporinus Ramirez, Birindelli & Galetti 2017 (Ramirez, Birindelli, Carvalho, et al., 2017), which in turn has three living species with available mitogenomes (Bedore et al., 2017; Yazbeck et al., 2016). This fossil prior included a lognormal distribution (offset = 6.0; mean = 3.0; SD = 1.0). The second calibration was †Brycon avus, a full skeleton from Tremembé Formation of the Oligocene-Miocene boundary in Brazil (Woodward, 1898), which was included in the node of all analyzed species of Brycon Müller & Troschel 1844. This prior included a lognormal distribution (offset = 23.8; mean = 5.0; SD = 1.0). The third fossil was †Paleotetra entrecorregos from Entre-Córregos Formation of the Eocene-Oligocene boundary of Brazil (Weiss et al., 2012), either hypothesized as a stem Stevardiinae (Weiss et al., 2012) or a stem Characidae (Mirande, 2019). The prior was placed in the node with all analyzed characids and included a lognormal distribution (offset = 33.9; mean = 5.0; SD = 1.0). The fourth calibration was a constraint on the root as a normally distributed prior (offset = 130; sigma = 10.0) representing the split of all characiforms with the siluriform Pseudoplatystoma Bleeker, 1862. This constraint matches our current understanding of the time period split between Characiformes and the other otophysan orders at approximately 130 ± 20 Ma (Arroyave et al., 2013; Betancur-R et al., 2017; Near et al., 2012). MCMC runs were performed with five independent runs with 100 million generations each, and trees were saved every 10,000 generations. We used Tracer v1.7.1 (Rambaut et al., 2018) to check for stationarity and sufficient mixing of parameters (ESS > 200) and trace distributions. The Treeannotator v2.6.2 from BEAST2 package was used to process the 50,005 trees where the last 45,005 were used after a 10% burn-in, and to select the maximum clade credibility (MCC) tree. The final tree was visualized and edited with Figtree v1.4.4.
3 RESULTS
3.1 Data assembly
The number of reads clustered into mitogenomes per specimen ranged from 4027 to 15,984. The assembled mitochondrial genome coverage ranged from 29.2X to 107.4X, while the size of the complete mitogenomes showed very little variation, ranging from 16,696 bp to 16,699 bp. Data results are shown in Table 1. The assemblies evidenced the presence of all 37 known genes (13 protein-coding genes, 22 tRNAs, and two rRNAs) and the non-coding control region (D-loop), a pattern commonly observed in vertebrate mitogenomes (Boore, 1999). The complete annotations of the resulting mitogenomes are presented in Tables S4–S9.
3.2 Comparative mitogenome analyses
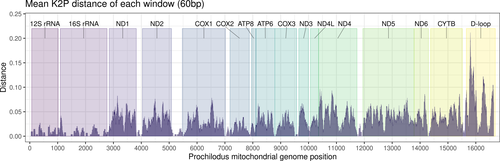
We analyzed two different and complete mitochondrial genomes for Prochilodus argenteus and P. costatus, and three for P. lineatus, comparing the newly generated mitogenomes with those previously available at GenBank. The number of differences (substitutions and indels) in the nucleotide sequences among different mitogenomes from the same species varied between 26 and 102 (Alignment S1). The mitogenomes of the sister species P. harttii and P. argenteus differed by 330 nucleotides. Complete mitogenome intraspecific variation ranged from 31 variable sites for P. costatus to 102 for P. lineatus. The pairwise K2P distances for all analyzed mitogenomes varied between 0.2% and 6.4% (Table S2) and p-distances varied between 0.16% and 6.9% (Table S3). The overall K2P divergence of mitogenomes from the same species ranged from 0.002 to 0.05, while the divergence among species varied from 0.009 to 0.064. As expected, pairwise distances for each region compared to the base genome (P. lineatus) revealed that the control region D-loop presented the highest number of polymorphisms followed by the ND4 gene (Figure 1). Results indicated a similar amount of variation in most genes (e.g., CYTB, ND1, and COX1) but very low variation in the 12S, 16S, COX2, and the rRNA ribosomal genes (Figure 1).
3.3 Phylogenetic relationships and timing of diversification
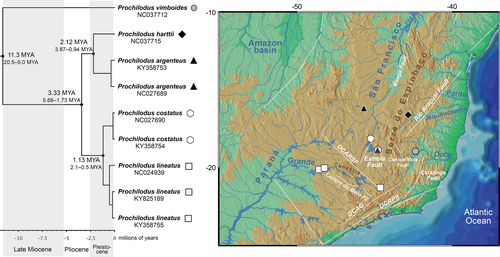
The aligned matrix contained 11,641 bp with 6909 variable sites (59%). DAMBE did not detect any saturation in transitions and transversions, indicating that the matrix was useful for phylogenetic analyses. The characiform tree based on mitogenomes shows high posterior probabilities for most nodes (Figure 2; Figure S1). The phylogeny reveals Prochilodontidae as sister to Curimatidae and this two-family clade being sister to Anostomidae and Chilodontidae, forming the traditional and well-established superfamily Anostomoidea (Vari, 1983) with Parodontidae as immediate sister group. It also shows the divergence time for the analyzed species of Characiformes (Figure 2; Figure S1). All represented families appear as monophyletic, and the time-calibrated analysis shows the split of Prochilodontidae and Curimatidae during the Eocene at around 42 MYA (61–25 MYA, highest posterior density, HPD), the earliest divergence in Prochilodontidae, that is, the split of Ichthyoelephas and Prochilodus, occurring at around 33 MYA (52–14 MYA, 95% HPD) (Figure 2). The split of P. vimboides is estimated to have occurred in the Miocene at around 11 MYA (20–5 MYA, 95% HPD), and the divergence between the clades from the eastern Brazilian Shield (P. argenteus+P. harttii and P. costatus+P. lineatus) occurring more recently in the Pliocene at approximately 3.3 MYA (5.6–1.7 MYA, 95% HPD) (Figures 2, 3). Within the two clades, we estimated that the interspecific divergences occurred contemporaneously during the Quaternary, more specifically in the Early Pleistocene: P. argenteus (Rio São Francisco) split from P. harttii (Rio Jequitinhonha) at around 2.1 MYA (3.8–0.9 MYA, 95% HPD), and P. costatus (Rio São Francisco) split from P. lineatus (Rio Grande/Paraná) at around 1.1 MYA (2.1–0.5 MYA, 95% HPD). Figure 2 shows the relationships among related characiform families, and Figure 3 depicts only Prochilodus relationships.

4 DISCUSSION
This study presents six new complete mitochondrial genomes for five Prochilodus species, with two being novel to science: P. harttii and P. vimboides. The combined mitogenomes allow the comparison of intraspecific features for the first time, as only one mitochondrial genome for each species was previously available. For example, the mitogenomes of P. lineatus presented more nucleotide variation than those of P. costatus, suggesting that the broad occurrence of the former in the Paraná-Paraguay system might help to increase intraspecific genetic variability. Among species, the highest interspecific divergence observed here (0.064) is still lower than that of the COX1 approach exclusively (0.103) (Melo et al., 2018), suggesting that some mtDNA regions other than COX1 might promote a reduction in the phylogenetic signal. In fact, our analyses indicate that all mitochondrial regions, except D-loop, present a genetic distance of no more than 0.10 among the five species (Figure 1). These results strengthen a previous hypothesis suggesting a very low genetic variation among Prochilodus species (Melo et al., 2018), likely due to their exceptional ability to perform long-distance migrations (Carolsfeld et al., 2003; Godinho & Kynard, 2006).
The phylogeny generated by new data of complete mitochondrial genomes exactly matches the relationships presented by the multilocus phylogeny of the family (Melo et al., 2016) by revealing Prochilodus vimboides as the sister clade to all other Prochilodus and the presence of two species pairs: P. argenteus sister to P. harttii, and P. costatus sister to P. lineatus (Figures 2, 3). The position of P. vimboides in the phylogeny of Prochilodus is biogeographically intriguing since the species has a restricted distribution in southeastern Brazil, while the other clade has species broadly distributed across South America from northern Colombia to the La Plata estuary (Castro & Vari, 2004; Melo et al., 2016). The time-calibrated phylogeny indicates that P. vimboides split from the remaining species during the Middle-Late Miocene boundary at around 11 MYA and suggests that the putative events related to this speciation process were certainly not the same as those for the other two species pairs from the eastern Brazilian Shield, which occurred later in the Early Pleistocene (Figure 3). However, the lack of mitogenomes for species of the remaining clades of Prochilodus from other Neotropical regions prevents any further exploration of biogeographic factors that may have promoted the diversification of P. vimboides relative to other species, leaving this topic an open question in the evolutionary history of the genus.
The cladogenetic event during the Pliocene at around 3.3 MYA splits the most recent common ancestor (MRCA) of all four species into two major clades: Prochilodus argenteus sister to P. harttii and P. costatus sister to P. lineatus. At first glance, this intuitively suggests that the MRCA lived in the Rio São Francisco region and had two subsequent dispersal paths: one toward the east to Rio Pardo and Rio Jequitinhonha (P. harttii) and another one southerly to Rio Grande/Paraná basin (P. lineatus). However, the phylogeny including all species of Prochilodontidae (Melo et al., 2016) indicates that these two clades appear in a successive arrangement with the clade P. costatus and P. lineatus being actually sister to the P. nigricans group, which is comprised of species mostly from Greater Amazonia (Amazon, Orinoco, and Guianas) and north-eastern Brazil (Melo et al., 2016). Furthermore, recent evidence from other Neotropical freshwater fish groups such as pimelodids (Tagliacollo et al., 2015), loricariids (Silva et al., 2016), bryconids (Machado et al., 2018), and curimatids (Melo et al. in review) indicates that most dispersal events occurred from the proto-Orinoco-Amazonas region to the craton drainages of the Brazilian and Guiana shields. In addition, the dispersal events to the Brazilian Shield were likely catalyzed by mega river captures between the wetlands of the proto-Orinoco-Amazonas and the proto-Paraguay basin (Albert et al., 2011, 2018; Tagliacollo et al., 2015). Therefore, the biogeographic scenario involving P. lineatus in the Paraná-Paraguay system and Rio Paraíba do Sul is probably more complex than we expected, and further studies including species of the P. nigricans group (Melo et al., 2016) from northern South America will reveal additional and complementary evidence to address this question.
We provide evidence that P. argenteus of the Rio São Francisco split from P. harttii of the Rio Jequitinhonha during the Early Pleistocene at around 2.1 MYA (Figure 3). The Serra do Espinhaço mountain chain divides the São Francisco basin of the several Atlantic-draining systems such as the Pardo, Jequitinhonha, Mucuri, and Doce rivers. The most recent geologic activities in the region are related to the Rio Araçuaí Fault during the Quaternary <1.6 MYA (Saadi et al., 2002), thus coinciding approximately with the split of P. argenteus and P. harttii in the time-calibrated phylogeny (Figure 3). Geologic evidence also suggests that post-Pliocene events in the Planalto Setentrional da Serra do Espinhaço reactivated thrust faults and, as a consequence, the upper reaches of the eastern tributaries of the São Francisco basin were captured by the upper Rio Jequitinhonha (Saadi, 1995). This is also supported, for example, by the relationship between Megaleporinus reinhardti (Lütken, 1875) from the São Francisco basin and M. garmani (Borodin, 1929) from Rio Jequitinhonha (Ramirez et al., 2017), and by the shared presence of native species in both basins such as Leporinus taeniatus Lütken, 1875 and Steindachnerina elegans (Steindachner, 1875) (Pugedo et al., 2016). In this context, we hypothesize that river captures promoted by Early Pleistocene reactivations of the Rio Araçuaí Fault in the Serra do Espinhaço may have promoted lineage split with subsequent speciation in this clade. Recent introductions have later established populations of P. argenteus into the Rio Jequitinhonha (Castro & Vari, 2004; Pugedo et al., 2016) that now hybridizes with its sister species P. harttii (Sales et al., 2018).
The mitogenome phylogeny supports the monophyly of both Prochilodus costatus from the Rio São Francisco and P. lineatus from the Rio Grande of the Paraná-Paraguay system (Figure 3). These results reject recent hypotheses using multilocus and barcoding studies that questioned their monophyly (Melo et al., 2016, 2018). Another recent study using the control region also found their monophyly (Chagas et al., 2020), although a marker such as this has extremely high levels of polymorphism (see Figure 1) meaning it is rather useful at a population level (e.g., Sivasundar et al., 2001; Machado et al., 2017). With this result, we corroborate the taxonomic revision and morphological phylogeny supporting their validity and monophyly (Castro & Vari, 2004) and reveal that complete mitochondrial genomes can provide distinct results compared to single or multilocus approaches. It is also possible that mitochondrial introgression and/or hybridization in Prochilodus (e.g., Sales et al., 2018) might affect the phylogenetic reconstruction. Finally, the lack of samples from the Rio Paraíba do Sul prevents a test for the monophyly of P. lineatus sampled in distinct drainages, although this has been addressed by the multilocus phylogeny (Melo et al., 2016).
Prochilodus costatus and P. lineatus split from each other during the Early Pleistocene at approximately 1.1 MYA (Figure 3). Prochilodus lineatus occurs broadly across immense regions of the Paraná-Paraguay and Paraíba do Sul basins, whereas P. costatus is endemic to the Rio São Francisco (Castro & Vari, 2004). These two basins are separated by the Serra da Canastra mountain range that divides the Rio Grande in the south, the upper Rio São Francisco and Rio Abaeté in the north/northeast and the Rio Araguari of the Paranaíba basin in the northwest (Figure 3). This region has the geologic influence of the Upper Rio São Francisco crustal discontinuity (DCARSF) dated to the Paleoproterozoic in the southern portion of the São Francisco craton (Saadi et al., 2002). According to Saadi et al. (2002), the most recent movements are dated to the Quaternary (<1.6 MYA) with several earthquakes occurring in the southeast margin of DCARSF, an area with the largest concentrations of seismic events in southeastern Brazil. In addition, the geomorphology of the Upper Rio São Francisco is also controlled by the Estrela Fault dated to the Neoproterozoic (Saadi et al., 2002). The intense geologic reactivations of these two faults may have promoted biotic exchange between the Rio Grande and the upper São Francisco systems with subsequent reduction of gene flow leading to speciation.
This river capture hypothesis is supported by (i) patterns of species richness with estimates of 63 species shared between the Upper Rio Paraná and Rio São Francisco (Buckup, 2011), by the (ii) presence of several genetic lineages in common (e.g., Pereira et al., 2011; Rossini et al., 2016; Serrano et al., 2019), and by (iii) instances of sister relationships. For example, Ramirez et al. (2020) found an Early Pleistocene divergence (~2 MYA) of the anostomids Schizodon nasutus Kner, 1858 from the Rio Paraná and S. knerii (Steindachner, 1875) from the Rio São Francisco. Machado et al. (2018) detected an intraspecific divergence among populations of the bryconid Salminus hilarii Valenciennes, 1850 from both basins at approximately 1.9 MYA. Therefore, the combined evidence reinforces our hypothesis of Early Pleistocene river capture in the Serra da Canastra region as the most plausible explanation for the cladogenesis of P. costatus and P. lineatus.
Overall, this study provides robust evidence for the monophyly of both Prochilodus costatus and P. lineatus, and reveals that interspecific diversification in the genus occurred during the Early Pleistocene when tectonic reactivations, associated especially with the Rio Araçuaí Fault in the Serra do Espinhaço and the Upper Rio São Francisco crustal discontinuity in the Serra da Canastra, may have promoted river captures between the Rio São Francisco and the adjacent drainages of the Jequitinhonha and Upper Paraná basins. We acknowledge, however, that earlier tectonic events surely occurred in the eastern Brazilian Shield and may have been important for other diversification processes (e.g., P. vimboides), but the available mitogenome data suggest that recent Quaternary reactivations had a more important role in the interspecific diversification of Prochilodus. Future studies will have the opportunity to (i) integrate mitogenomes of other prochilodontid species, (ii) investigate biogeographic and geomorphologic events associated with the diversification of P. vimboides along coastal Atlantic rivers such as those in the Mucuri, Doce and Paraíba do Sul basins, and (iii) test geodispersal events including other Prochilodus species, especially members of the P. nigricans group as well as other prochilodontids (Ichthyoelephas and Semaprochilodus), and the putative association of species from lowlands of the Amazon basin with the species of the Brazilian and Guiana shields.
ACKNOWLEDGEMENTS
We thank many colleagues for discussions on the evolution of prochilodontids: Brian L. Sidlauskas, Claudio Oliveira, Heraldo A. Britski, James S. Albert, and Ricardo M. C. Castro. We are grateful to Mateus Vidal and José M. Ribeiro, for helping with mitogenome assembly and annotation; and to Carolina Furtado (INCA) for library preparation and Illumina sequencing. Thanks to Tatiana M. Barroca for suggestions as a member of the master's evaluation committee (RPS). Part of the analysis was conducted on the Brycon & Zungaro servers at LBP/UNESP funded by FAPESP 14/26508-3. This study was funded by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (RPS), FAPESP grant 16/11313-8 (BFM), CNPq Universal 404991/2018-1 (BFM), CNPq DT-2 scholarship 310991/2017-0 (GMY), CNPq productivity fellowship 306155/ 2018-4 (DCC), and FAPERJ CNE grant 202.780/2018 (FP).




