The enigmatic Crimean green lizard (Lacerta viridis magnifica) is extinct but not valid: Mitogenomics of a 120-year-old museum specimen reveals historical introduction
Abstract
It has been proposed that Lacerta viridis magnifica Sobolevssky, 1930 represents an extinct species or subspecies of green lizard endemic to the southern Crimea. Using NGS protocols optimized for heavily degraded DNA, we sequenced the complete mitogenome of one of the originally formalin-preserved specimens collected in the late 19th century. A comparison with sequence data of other green lizards revealed that L. v. magnifica is a junior synonym of the northern subspecies of the western green lizard (L. b. bilineata Daudin, 1802), which occurs at least 1,500 km away, beyond the distribution ranges of other green lizards. In medieval times, a Genoese colony existed in the Crimean region where the extinct green lizards occurred. Until the early 20th century, close ties to Italy persisted, and locals of Genoese descent sent their children for education to Italy, where L. b. bilineata occurs. This suggests that the extinct Crimean green lizards have been introduced accidentally or intentionally from Italy. Our study exemplifies the value of historical formalin-preserved museum specimens for clarifying the status of questionable rare or extinct taxa.
1 INTRODUCTION
Green lizards of the Lacerta viridis complex have a wide distribution in the Western Palearctic, from northern Spain across France, continental Italy and Sicily, parts of western and eastern Germany, Austria, the Czech Republic, Slovakia, Hungary, and many southeastern European countries to central Ukraine and northern Asia Minor (Nettmann, 2001; Nettmann & Rykena, 1984). Since the influential studies by Rykena (1991) and Amann, Rykena, Joger, Nettmann, & Veith (1997), most authors assign western populations to the species Lacerta bilineata. Populations east of western Germany and Istria are thought to represent Lacerta viridis sensu stricto (see the review in Nettmann, 2001). However, the validity of these two morphologically cryptic species has been repeatedly doubted (Böhme et al., 2007; Brückner et al., 2001; Godinho, Crespo, Ferrand, & Harris, 2005; Mayer & Beyerlein, 2001). Using mtDNA, Marzahn et al. (2016) recently showed that green lizards correspond to ten terminal clades that cluster in four instead of two deeply divergent clades, challenging the current systematics. The distribution range of L. bilineata largely agrees with that of one of the four deeply divergent clades, while the range of L. viridis harbors three of the deeply divergent clades (Figure 1).
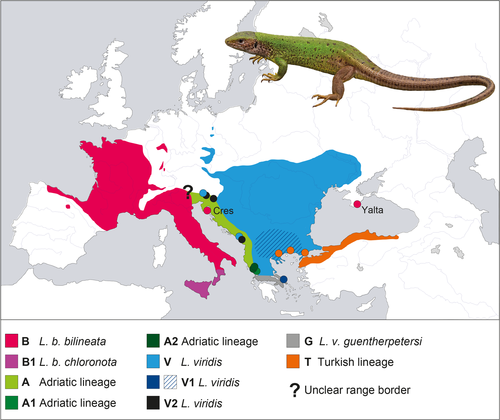
The easternmost European populations of green lizards are generally thought to occur in central mainland Ukraine (Nettmann, 2001; Nettmann & Rykena, 1984; Tarashchuk, 1959). Old records for the Crimean peninsula were meticulously reviewed by Nikolsky (1915), who concluded that they were in error or referred to misidentified sand lizards (L. agilis). However, during the time when Nikolsky’s (1915) book was compiled, Nikolsky worked in Kharkiv and had, according to his own words, no access to green lizards in the Zoological Museum of the Moscow University (ZMMU) that were collected in the southern Crimea in 1896 (Elpatjewsky & Sabanejew, 1907).
Based on four of these specimens, Sobolevssky [also sometimes transliterated Sobolevsky or Sobolewsky] (1930) described the subspecies Lacerta viridis magnifica. The type specimens and additional Crimean material originated from three different collectors. These green lizards were collected in 1896 and around 1900 near Yalta and Sevastopol. For two specimens, the exact collection date remains unknown. Later, no further specimens became known to science. Therefore, L. viridis was continued to be excluded from the Crimean fauna (Nettmann, 2001; Nettmann & Rykena, 1984; Shcherbak, 1966), and the taxonomic identity of L. v. magnifica remained unclear (Mertens & Wermuth, 1960).
The Crimean green lizards in the ZMMU were later re-examined by Kotenko (2010), who confirmed their identification and speculated about a historical extinction of an endemic species or subspecies. Until now, this vanished population remains enigmatic. The Crimea is a well-known glacial refuge (Bilton et al., 1998; Jablonski et al., 2019; Sommer & Beneke, 2005) with many relictual and some endemic taxa, among others an endemic sand lizard (Lacerta agilis tauridica; Kalyabina-Hauf et al., 2004), supporting the possibility that an endemic green lizard became extinct there. In the present paper, we use state-of-the-art NGS protocols optimized for ancient and historical samples to examine one originally formalin-preserved Crimean green lizard (Figure S1) to reveal its genetic identity.
2 MATERIALS AND METHODS
According to Sobolevssky (1930), the description of Lacerta viridis magnifica was based on four specimens from the southern Crimea. All of them are still present in the ZMMU collection, and Sobolevssky himself labeled these four lizards as types. In agreement with the specimen labels, Sobolevssky (1930) stated that the collector of two type specimens (ZMMU 2470) was Aleksandr Petrunkevich, then a student at Moscow University and later a renowned arachnologist and professor at Yale University (Hutchinson, 1991). In addition to the four types, there is a fifth specimen in the ZMMU bearing another collection number (ZMMU 1967; Figure S1). It was collected by Petrunkevich on the same date (May 1896) and at the same site as the specimens inventoried under ZMMU 2470 (Yalta, southern Crimea). This fifth specimen bears a field label with the number 34/9 immediately following the number 34/8 on a similar label of ZMMU 2470. According to another label in the same jar, ZMMU 1967 was examined later by the renowned Russian herpetologist Ilya Darevsky, who confirmed its determination as L. viridis. Even though Sobolevssky did not include ZMMU 1967 in the type series, all evidence indicates that this specimen belongs to the original material collected by Petrunkevich in the Crimea. We were allowed to sample this additional topotypic specimen of L. v. magnifica but not the types. ZMMU 1967 was originally formalin-fixed and is currently kept in ethanol.
Molecular work was conducted in the clean room facility of the Senckenberg Natural History Collections Dresden (MTD) according to established guidelines (Fulton, 2012). A negative control (water blank) was included during library preparation and screened for evidence of contamination. DNA was extracted from muscle tissue using the sbeadex forensic kit (LGC, Teddington), and 2.9 ng were converted into a single-indexed single-stranded Illumina sequencing library (Gansauge & Meyer, 2013; Korlević et al., 2015), including the removal of uracil residues. To enrich the library for mtDNA, we performed two rounds of in-solution hybridization capture with self-made baits from long-range PCR products of L. viridis (sample MTD 2350, Ukraine, Mygiya, 18 July 2004, leg. T. Kotenko; text of Supporting Information S1 and Table S1). Sequencing was conducted in-house on an Illumina MiSeq platform, generating c. 4 million 75-bp-paired-end raw reads. Adapter trimming, quality filtering, and duplicate removal reduced the readpool to c. 1.4 million reads. Assembly of the mt-genome was performed with 500,000 randomly selected reads using MITObim (Hahn, Bachmann, & Chevreux, 2013) with an allowed mismatch value of 2, and GenBank sequence AM176577 as the starting reference. The resulting high-quality contig was visualized and checked for assembly artefacts in Tablet 1.16.09.06 (Milne et al., 2013). The temporal authenticity of the mapped reads was tested with mapDamage 2.0 (Jónsson, Ginolhac, Schubert, Johnson, & Orlando, 2013), which accounts for nucleotide misincorporations due to DNA degradation. The contig was aligned to the complete mt-genomes of Lacerta agilis (KC990830), L. b. bilineata (KT722705, France), and L. v. viridis (AM176577, Lower Austria) and annotated accordingly. A second alignment corresponded to the cyt b gene only. It contained sequences of all 10 terminal clades clustering in four more inclusive clades, as identified by Marzahn et al. (2016). Each terminal clade was represented by two distinct sequences (Table S2). Phylogenetic reconstruction involved Maximum Likelihood and Bayesian inference approaches using RAxML 8.0.0 (Stamatakis, 2014) and MrBayes 3.2.6 (Ronquist et al., 2012), applied to an alignment of complete mitochondrial genomes of 17,086 bp length with sequences of four specimens, and a cyt b alignment of 1,143 bp with sequences of 22 specimens. Lacerta agilis served as the outgroup. Details are explained in the Supporting Information S1; alignments are available from TreeBase (http://purl.org/phylo/treebase/phylows/study/TB2:S24851).
3 RESULTS
The assembly process resulted in a single high-quality contig, consisting of 201,198 (40.2%) assembled reads, with a 694-fold average coverage and an average read length of 58 bp (35–143 bp). The complete mitogenome has 17,147 bp, without any ambiguous sites (the length exceeds that of the alignment for phylogenetic analyses because overlapping gene regions, etc., were excluded, see text of Supporting Information S1). The mitogenome includes 13 protein-coding genes (PCGs), two rRNA genes, the control region, and 22 tRNA genes and is deposited in the European Nucleotide Archive (LR694166). The scaled scaffold and length distribution of the assembled reads, as well as the fragment-misincorporation plot, are depicted in Figures S2–S4.
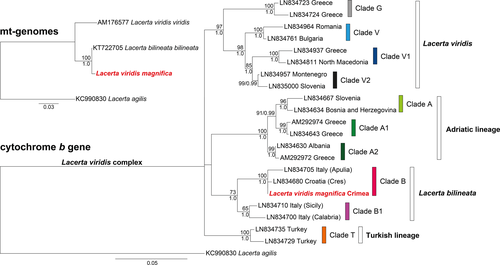
For each data set, the topologies of the two tree-building approaches were identical (Figure 2). In the analyses using complete mitogenomes, the sequence of Lacerta viridis magnifica did not cluster with that of a L. v. viridis but with maximum support with the mitogenome of a L. b. bilineata from France. The analyses using cyt b sequences retrieved the same ten well-supported terminal clades as already identified by Marzahn et al. (2016), representing four more inclusive deeply divergent clades. Again, the sequence of L. v. magnifica clustered with maximum support with L. bilineata (clade B in the terminology of Marzahn et al., 2016, corresponding to the nominotypical subspecies L. b. bilineata).
4 DISCUSSION
The near identity of the mitogenome of Lacerta viridis magnifica with that of L. b. bilineata is unexpected because the distribution range of L. bilineata is far away from the Crimea and beyond the distribution ranges of other green lizards (Figure 1). The populations occurring closest to the Crimea live in mainland Ukraine (Nettmann & Rykena, 1984), approximately 250 km away, and belong to L. viridis (clade V; Marzahn et al., 2016). The geographically closest record of L. bilineata is the isolated population of the nominotypical subspecies on Cres Island, Croatia (Marzahn et al., 2016), approximately 1,500 km away. The distribution range of L. bilineata embraces parts of northern Spain, most of France, parts of western Germany, the Apennine Peninsula, and Sicily (Nettmann, 2001). However, the majority of populations formerly assigned to this species from Slovenia and northwestern Croatia (Nettmann, 2001) belong to the so-called Adriatic lineage of the L. viridis complex (Marzahn et al., 2016), most likely a distinct but currently not recognized taxon. Marzahn et al. (2016) confirmed L. bilineata only for the Croatian island Cres, an isolated record embedded within the range of other genetic lineages. Lacerta bilineata shows phylogeographic structuring with two mitochondrial clades, one confined to Sicily and Calabria (clade B1) and the other occurring in the remaining distribution range (clade B). The latter is thought to represent the nominotypical subspecies, while clade B1 is currently identified with lizards belonging to the subspecies L. b. chloronota (Marzahn et al., 2016).
The sequence of L. v. magnifica clusters with maximum support with those of the northern subspecies L. b. bilineata. It is well known that lizards may be translocated incidentally or intentionally by humans. The most prominent case in Europe is the wall lizard Podarcis muralis that established more than 150 self-sustaining non-native populations (Schulte, 2008). Also introduced populations of L. bilineata are known, occurring far away of their native distribution range in Kansas, USA (Gubanyi, 2000), and the United Kingdom (Deichsel, Gleed-Owen, & Mayer, 2007).
Along the south coast of the Crimea, a Genoese colony (Gazara or Gazaria) with several settlements and trading posts existed from 1261 to 1464. Its area included both Yalta (then Caulita) and Balaklava (then Cembalo) near Sevastopol (Gavrilenko, Sivalnov, & Tsibulkin, 2017), that is, the region where the green lizards were collected. Even after the collapse of the Genoese colony, the connection to Italy was maintained for a long time. Until the early 20th century, locals of Genoese descent sent their children for education to Italy (Puzanov, 1960). This suggests that the green lizards were somewhen accidentally or intentionally introduced from Italy and established temporarily a population on the Crimea that vanished after the voucher specimens in the ZMMU were collected.
Our results clarify that L. v. magnifica Sobolevssky, 1930 is neither an extinct species nor an extinct subspecies of L. viridis (Laurenti, 1768), as suggested before (Kotenko, 2010), but a junior synonym of L. bilineata Daudin, 1802. Our study exemplifies the potential of NGS protocols for heavily degraded DNA for resolving the status of questionable rare or extinct taxa that are only available for study as formalin-preserved museum specimens.
ACKNOWLEDGEMENTS
The late Tatiana I. Kotenko inspired us to conduct the present study. Thanks go to Valentina Orlova (ZMMU) for access to specimens and allowing to sample ZMMU 1967 for the present study. Henrik Bringsøe donated the photograph shown in Figure 1. Laboratory work was conducted at Senckenberg Dresden (SGN-SNSD-Mol-Lab). Anke Müller (MTD) assisted during laboratory work.




