Telomere structure in insects: A review
Abstract
Telomeres are terminal regions of chromosomes, which protect them from fusion with other chromosomes and stabilize their structure. Telomeres usually contain specific DNA repeats (motifs), which are maintained by telomerase, a kind of reverse transcriptase. In this review, we survey the current state of knowledge of telomere motifs in insects. Among Hexapoda, data on telomere composition are available for more than 350 species from 108 families and 25 orders. The telomere motif (TTAGG)n is considered ancestral for the class Insecta. However, certain insects have different and often unknown telomeric sequences. This apparently happens because telomerase-dependent mechanisms usually coexist with various means of alternative lengthening of telomeres as backup mechanisms of telomere maintenance. This coexistence can explain losses and reappearances of the TTAGG repeat and telomerase-dependent telomere replication in insect evolution. For example, a few higher taxa, such as Heteroptera (Hemiptera) and Hymenoptera, show presence of the (TTAGG)n motif in their basal clades as well as a subsequent loss and, at least in the Hymenoptera, independent reappearance of this repeat in some advanced groups. Analogously, most members of Coleoptera also retain the TTAGG repeat, although it is changed to TCAGG in certain families. Furthermore, the (TTAGG)n motif seems to have been irreversibly lost in the order Diptera. In this group, telomeric sequences are represented either by long terminal repeats or by retrotransposons. Retrotransposons are also interspersed with other telomeric sequences in many groups of insects. The accumulating data demonstrate that the class Insecta is substantially diverse in terms of its telomere structure.
1 INTRODUCTION
Telomeres are nucleoprotein complexes located at the ends of eukaryotic chromosomes that ensure integrity and replication of chromosome ends and are therefore essential for genome stability (Blackburn, 1991; Zakian, 1995, 2012). Telomeres usually contain short tandem DNA repeats and associated proteins. In most eukaryotes, telomere arrays are composed of 5–10 bp repeats, and synthesis and maintenance of telomeric DNA involve telomerase, a telomere-specific reverse transcriptase that elongates terminal sections of DNA using a small region of its RNA subunit as a template (Greider & Blackburn, 1989).
A phylogenetic analysis of short telomeric repeats (motifs), across eukaryotes shows that they are largely conserved among most organisms, and usually mark higher taxa (Frydrychová, Grossmann, Trubač, Vítková, & Marec, 2004; Krupp, Klapper, & Parwaresch, 2000; Lukhtanov & Kuznetsova, 2010; Mason, Randall, & Capkova Frydrychova, 2016). For example, TTAGGG, TTTAGGG, TTAGGC, and TTAGG repeats are characteristic of vertebrates, flowering plants, nematodes, and arthropods respectively (Vítková, Král, Traut, Zrzavý, & Marec, 2005). Within Arthropoda, all major subphyla, i.e., Chelicerata (except for spiders), Myriapoda, Crustacea, and most Hexapoda, have the (TTAGG)n telomere motif (Mandrioli, Monti, & Manicardi, 2012; Traut et al., 2007). Using various techniques, with fluorescence in situ hybridization (FISH) being the most important, distribution of the so-called “insect” telomere motif (TTAGG)n was extensively explored across different orders. This repetitive pentanucleotide telomeric sequence appeared to be characteristic of the majority of insects studied to date. It is thus considered an ancestral DNA motif of insect telomeres which apparently evolved from the “vertebrate” one, (TTAGGG)n, that is also found in many lower eukaryotes (Anokhin & Kuznetsova, 2018; Frydrychová et al., 2004; Frydrychová & Marec, 2002; Mandrioli et al., 2012; Sahara, Marec, & Traut, 1999; Traut et al., 2007; Vítková et al., 2005).
Nevertheless, structure of telomeric repeats can vary within a few insect groups. For example, different families and even genera of the order Coleoptera have either (TTAGG)n or (TCAGG)n motif (Frydrychová & Marec, 2002; Mravinac, Meštrović, Čavrak, & Plohl, 2011). Moreover, some other taxa, primarily those belonging to one of the most successful lineages of insects, the order Diptera, are characterized by telomerase-independent mechanisms of telomere maintenance, known as alternative lengthening of telomeres (ALT) (Biessmann & Mason, 1997). Specifically, these mechanisms include transposition of non-long terminal repeat (non-LTR) retrotransposons in the fruit fly genus Drosophila Fallén 1823, at least in D. melanogaster (Meigen, 1830) and closely related species (reviewed by Mason, Capkova Frydrychova, & Biessmann, 2008), or presence of long terminal repeats (LTRs) in the non-biting midge Chironomus dilutus Shobanov, Kiknadze & Butler, 1999 (Rosén & Edström, 2000) and the mosquito Anopheles gambiae Giles, 1902 (Walter, Bozorgnia, Maheshwari, & Biessmann, 2001). The mechanism of transposition of non-LTR retrotransposons is very similar to that of telomere elongation by telomerase. Moreover, telomerase and retrotransposons are presumed to have evolved from a common ancestral retroelement (Arkhipova, Pyatkov, Meselson, & Evgen'ev, 2003; Servant & Deininger, 2016). If this is true, these two mechanisms of telomere elongation are highly likely to coexist from the very beginning. For example, this lends further support from the fact that both these mechanisms were detected in such distant organisms like yeasts and humans (Roth, Kobeski, Walter, & Biessmann, 1997). In most insects, however, telomerase-mediated maintenance of chromosome ends remains predominant, whereas ALT operates as a backup mechanism with a similar role (Servant & Deininger, 2016). The latter mechanism therefore usually activates when normal functioning of the telomerase becomes problematic (Osanai, Kojima, Futahashi, Yaguchi, & Fujiwara, 2006). Nevertheless, both telomeric repeats and telomerase (as well as the telomerase-coding gene) lack in all studied members of the order Diptera (Garavís, González, & Villasante, 2013; Rosén, Kamnert, & Edström, 2002) suggesting that this mechanism is fully replaced by ALT.
Recent research on insect telomeres has revealed several examples of heterogeneity for presence/absence of the (TTAGG)n motif within, e.g., Hemiptera: Heteroptera (Angus, Jeangirard, Stoianova, Grozeva, & Kuznetsova, 2017; Chirino, Dalíková, Marec, & Bressa, 2017; Golub, Golub, & Kuznetsova, 2015, 2017, 2018; Grozeva, Kuznetsova, & Anokhin, 2010; Kuznetsova, Grozeva, & Anokhin, 2012; Pita et al., 2016) and Hymenoptera (Gokhman, Anokhin, & Kuznetsova, 2014; Gokhman & Kuznetsova, 2018; Menezes, Bardella, Cabral-de-Mello, Lucena, & Almeida, 2017; Menezes et al., 2013), and even between closely related species of the order Odonata (Kuznetsova, Maryańska-Nadachowska, Shapoval, Anokhin, & Shapoval, 2017). Insects that lack canonical telomeric repeats probably constitute about 10%–15% of all animal species (Mason et al., 2016). Nevertheless, structure of telomeres in many TTAGG-negative insects remains unknown. In general, these insects are considerably diverse in terms of telomere structure, but modern literature lacks a thorough review of different types of telomere composition and insect taxa associated with them. The last comprehensive review on this subject covering all main branches of the insect phylogenetic tree was published in 2004 when only about 60 species were studied using a single primer polymerase reaction, Southern blot hybridization (SBH) of genomic DNA, and/or FISH (Frydrychová et al., 2004; see also Mandrioli et al., 2012).
Since this compilation, the number of species and higher taxa targeted by telomere studies experienced a manifold increase. New data became available for many previously poorly studied or unexplored orders such as Odonata (Kuznetsova et al., 2017), Orthoptera (Buleu, Jetybayev, Chobanov, & Bugrov, 2019; Kociński, Grzywacz, Chobanov, & Warchałowska-Śliwa, 2018), Phasmatodea (Liehr, Buleu, Karamysheva, Bugrov, & Rubtsov, 2017; Scali et al., 2016), Mantophasmatodea (Lachowska-Cierlik, Maryańska-Nadachowska, Kuznetsova, & Picker, 2015), Hemiptera (Anjos, Rocha, Paladini, Mariguela, & Cabral-de-Mello, 2016; Chirino et al., 2017; Grozeva, Kuznetsova, & Anokhin, 2011; Kuznetsova, Grozeva, Hartung, & Anokhin, 2015; Luan, Sun, Fei, & Douglas, 2018; Mandrioli, Zanasi, & Manicardi, 2014; Maryańska-Nadachowska, Kuznetsova, Golub, & Anokhin, 2018; Mohan, Rani, Kulashreshta, & Kadandale, 2011), Neuroptera (Kuznetsova, Khabiev, & Anokhin, 2016), Coleoptera (Mravinac et al., 2011), Hymenoptera (Gokhman et al., 2014; Gokhman & Kuznetsova, 2018; Menezes et al., 2017) and others (see Figure 1, Appendix 1). All this information was mainly obtained by FISH and is therefore more reliable if compared with SBH, because, when (TTAGG)n sequences are detected by the latter technique, they are not necessarily telomeric, as the specific probe could also hybridize with repetitive sequences located at non-telomeric sites (Meyne et al., 1990; Sahara et al., 1999).
Canonical telomeric repeats are located at the terminal regions of chromosomes, but interstitial telomeric sequences (ITSs) have also been observed in many eukaryotes, from yeasts to humans. These repeats are often considered relicts of the tandem fusion of ancestral chromosomes (reviewed by Lin & Yan, 2008). Futhermore, ITSs were suggested to represent “hot spots” for chromosome repatterning relevant to karyotype evolution and speciation (Ruiz-Herrera, Nergadze, Santagostino, & Giulotto, 2008). In insects, ITSs containing the (TTAGG)n motif have so far been identified in a few species generally showing derived karyotypes when compared to their ancestral complements (Chirino et al., 2017; Jetybayev, Bugrov, Karamysheva, Camacho, & Rubtsov, 2012; Rego & Marec, 2003).
TTAGG-specific telomerase activity was studied in a number of insect orders (Korandová, Krůček, Vrbová, & Frydrychová, 2014; Sasaki & Fujiwara, 2000). Distribution of this nucleotide sequence in insects is predominantly, but not universally, consistent with that of telomerase activity, and therefore suggests that the TTAGG-telomerase system is functional in this group. Telomerase activity has been found in several phylogenetically distant orders including Isoptera, Blattodea, Lepidoptera, Hymenoptera, Trichoptera, Coleoptera, and Hemiptera (Aphidoidea). Nevertheless, similar studies did not detect telomerase activity in Orthoptera, Zygentoma, and Phasmatodea, although these insects do possess the (TTAGG)n motif (Frydrychová et al., 2004).
These contradictory results could be explained by a number of factors, e.g., the low amount of telomerase (Korandová et al., 2014). Also, the presence/absence of the TTAGG telomeric repeat coincides with the presence/absence of a candidate telomerase gene in several model insect species (reviewed in Robertson & Gordon, 2006). Taken together, all this information indicates that the above-mentioned problem deserves further study.
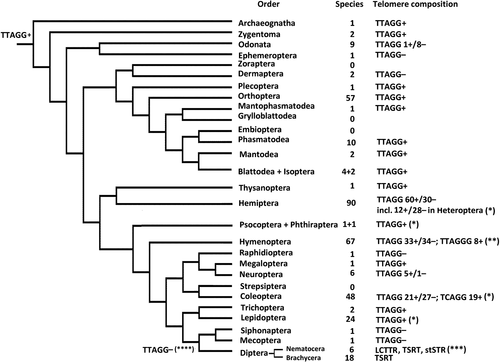
In this paper, we will review the current state of knowledge of the telomere structure and function across all orders of the class Insecta (=Ectognatha; Appendix 1, Figure 1) given the fact that the structure of telomeres in other Hexapoda, i.e., in the class Entognatha, which comprises the orders Protura, Collembola, and Diplura, is completely unknown. Our review will summarize the data previously scattered among many papers and often having a narrow taxonomic focus. For the purposes of classification of various groups of Insecta, we generally use the system developed by Misof et al. (2014) which is based on phylogenomic data. For some orders, we also follow Beutel et al. (2011), Peters et al. (2014), Mason et al. (2016), Blackmon, Ross, and Bachtrog (2017) and a few other papers.
2 TELOMERE STRUCTURE IN THE CLASS INSECTA
2.1 “Thysanura s.l.” (primary wingless insects)
The class Insecta contains about two dozen orders of winged insects (Pterygota) as well as two orders of primary wingless insects (“Thysanura”), i.e., Archaeognatha and Zygentoma. The latter is the putative sister group of pterygote insects (Giribet, Edgecombe, Carpenter, D'Haese, & Wheeler, 2004).
2.1.1 Archaeognatha (=Microcoryphia)
Archaeognatha (bristletails) are the most primitive among all living insects. They constitute the sister group of Zygentoma + Pterygota. The order harbors about 500 species and is composed of two extant families, Meinertellidae and Machilidae. Many authors consider the former family as more advanced than Machilidae (see Sturm & Machida, 2001 and references therein). In turn, Machilidae are subdivided into three subfamilies, Petrobiellinae, Petrobiinae, and Machilinae, and the last one shows apparently less advanced morphological characters (Sturm & Machida, 2001). In Lepismachilis sp. (Machilinae), the only species studied until now, both FISH and SBH revealed the TTAGG telomeric repeat (Frydrychová et al., 2004).
2.1.2 Zygentoma (=Thysanura s.str.)
This is a small order with about 400 species that belong to 90 genera and five families. Both species studied up to now, i.e., a silverfish, Lepisma saccharina Linnaeus, 1758, and a firebrat, Thermobia domestica Packard, 1873, are from the same large family Lepismatidae. FISH (as well as SBH in the case of Th. domestica) revealed the TTAGG telomeric repeat in both those insects (Frydrychová et al., 2004). Nevertheless, real-time quantitative telomeric repeat amplification protocol (RTQ-TRAP) did not detect telomerase activity in L. saccharina (Korandová et al., 2014); however, the authors concluded that this failure could be explained either by a low amount of telomerase or by the presence of inhibitors of TRAP assays.
2.1.3 Concluding comment
Since Archaeognatha and Zygentoma branch off the very base of the insect phylogenetic tree, the (TTAGG)n telomeric sequence found in these insects most likely represents an ancestral telomere motif in the class Insecta in general (Frydrychová et al., 2004; Vítková et al., 2005).
2.2 Non-holometabolous insects
2.2.1 Palaeoptera (basal winged insects)
This monophyletic group comprises the orders Odonata and Ephemeroptera. Taken together, they probably represent the earliest taxa of winged insects which are derived from a common ancestor (Ogden et al., 2009; Ogden & Whiting, 2005; Wheeler, Whiting, Wheeler, & Carpenter, 2001). According to molecular data, main palaeopteran lineages originated about 360 Mya (Misof et al., 2014).
Odonata (damselflies and dragonflies)
This is one of the oldest orders of winged insects, with fossils dating back to the Permian (Grimaldi & Engel, 2005). Extant Odonata include about 6,000 species worldwide in two suborders, Zygoptera (damselflies) and Anisoptera (dragonflies), comprising almost 3,000 species each. The third relict suborder, Anisozygoptera (damseldragons, or ancient dragonflies), contains just a few extant species and is often considered as a family of Anisoptera (Rehn, 2003; Schorr & Paulson, 2017). Within Zygoptera and Anisoptera, up to 21 and 11 families, respectively, are currently recognized (Dijkstra et al., 2013; Rehn, 2003; Schorr & Paulson, 2017). So far, data on the telomere structure have been obtained only for the suborder Anisoptera in which nine species belonging to eight genera of the families Coenagrionidae, Corduliidae and Libellulidae were studied (Frydrychová et al., 2004; Kuznetsova et al., 2017). Interestingly, most species were shown to lack the (TTAGG)n telomeric motif, while in Sympetrum vulgatum (Linnaeus, 1758) clear hybridization signals were seen at the ends of almost all chromosomes as well as in spermatids (Kuznetsova et al., 2017). These data suggest that the TTAGG repeat represents the major component of the telomeres in this species; an important point is that another member of the same genus, Sympetrum danae (Sulzer, 1776), showed no signals. This finding raises the question as to whether the (TTAGG)n telomeric motif is actually lost in Odonata, as hypothesized by Frydrychová et al. (2004), or it presents in just a few copies and therefore cannot be detected by FISH (Kuznetsova et al., 2017). On the other hand, Odonata could initially lack the TTAGG telomeric sequence. If this is true, the (TTAGG)n motif independently reappears in S. vulgatum (Kuznetsova et al., 2017); an analogous situation occurs, e.g., in aculeate Hymenoptera (see Menezes et al., 2017). An alternative explanation is that this repeat represents a persisting but non-functional relict of the ancestral telomeric motif/subtelomeric region, as suggested by Korandová et al. (2014) for similar cases.
Ephemeroptera (mayflies)
This order represents an ancient lineage of pterygote insects encompassing over 3,000 species and more than 400 genera, constituting at least 42 described families (Barber-James, Gattolliat, Sartori, & Hubbard, 2008). The only species studied so far, Cloeon dipterum (Linnaeus, 1761), belongs to the most species-rich family Baetidae which is considered the sister group to all other mayflies (Barber-James et al., 2008; Ogden et al., 2009; Ogden & Whiting, 2005). C. dipterum appeared to be TTAGG-negative in different analyses, including a single-primer PCR of genomic DNA, SBH with whole-genome DNA, and FISH (Frydrychová et al., 2004).
Concluding comment
The absence of the (TTAGG)n telomere motif in both Ephemeroptera and almost all Odonata may represent their synapomorphy, which additionally supports the monophyly of Palaeoptera. We can therefore speculate that the first loss of the ancestral “insect” telomere motif in evolution of Pterygota has occurred in the common ancestor of Odonata + Ephemeroptera, with the TTAGG repeat being replaced by another telomeric sequence. Nevertheless, this hypothesis contradicts the fact that the mayflies and dragonflies have strongly different types of chromosomes, monocentric and holocentric (=holokinetic), respectively (Kiauta & Mol, 1977; Nokkala, Laukkanen, & Nokkala, 2002). However, there is also a possibility that Odonata could independently acquire holokinetic chromosomes, i.e., chromosomes without localized centromeres. It would be of interest to examine both types of chromosomes and telomere motifs in the recently discovered family Siphluriscidae (Zhou & Peters, 2003), which is currently considered a phylogenetic intermediate between Odonata and Ephemeroptera (Ogden et al., 2009).
2.2.2 POLYNEOPTERA (ORTHOPTEROID INSECTS)
This is a monophyletic group that comprises 11 orders, i.e., Zoraptera, Dermaptera, Plecoptera, Orthoptera, Embioptera, Phasmatodea, Mantophasmatodea, Grylloblattodea, Blattodea, Mantodea, and Isoptera. According to molecular data, the main polyneopteran lineages originated about 300 Mya (Misof et al., 2014).
Zoraptera (angel insects)
Angel insects are an enigmatic order representing one of the most challenging and uncertain concerns in ordinal-level phylogenies of the insects. This order consists of a single family, Zorotypidae, with just two recognized genera (Engel, 2003). The only species, Zorotypus hubbardi Caudell, 1918, has so far been karyotyped (Kuznetsova, Nokkala, & Shcherbakov, 2002); its chromosomes appeared to be holokinetic but their telomere composition remains unknown.
Dermaptera (earwigs)
A comparatively small order comprising about 2,000 species distributed throughout the world (except the polar regions), with the greatest diversity in the tropics. Virtually every polyneopteran order was proposed as a sister group to Dermaptera. The currently best-supported clades suggested by transcriptomic analysis are Dermaptera + Zoraptera (Misof et al., 2014) or Dermaptera + Plecoptera (Letsch & Simon, 2013). Moreover, the former clade is well supported by the fact that earwigs and zorapterans are the only two polyneopteran groups possessing holokinetic chromosomes (Kuznetsova et al., 2002; White, 1971), which could evolve during the divergence of Dermaptera and Zoraptera from a “monocentric” ancestor. The two studied species of the genus Forficula Linnaeus, 1758 (Forficulidae), F. auricularia Linnaeus, 1758 and F. scudderi de Bormans, 1888, appeared to be negative for the (TTAGG)n motif (Frydrychová et al., 2004; Okazaki, Tsuchida, Maekawa, Ishikawa, & Fujiwara, 1993).
Plecoptera (stoneflies)
A moderately diverse order with more than 3,500 described species in 16 families (Fochetti & de Figueroa, 2007). In the only examined stonefly species, Perla burmeisteriana Claassen, 1936, very strong hybridization signals at the ends of metaphase chromosomes unambiguously prove the presence of the TTAGG telomeric sequence (Frydrychová et al., 2004).
Orthoptera (grasshoppers, locusts and crickets)
This order harbors more than 25,700 species worldwide, and it is thus the most diverse among the polyneopteran insect lineages (Grimaldi & Engel, 2005). Orthoptera are subdivided into two suborders, i.e., Caelifera, with approximately 11,000 species, including grasshoppers and locusts (Acrididae), and Ensifera (about 9,000 species), which includes crickets and katydids (Song et al., 2015). Up to now, telomere structure of 59 species (including three subspecies of a particular species) in 34 genera belonging to both Caelifera (the families Acrididae, Pamphagidae, and Pyrgomorphidae) and Ensifera (Raphidiophoridae, Gryllidae, and Tettigoniidae) was studied (Bueno, Palacios-Gimenez, & Cabral-de-Mello, 2013; Buleu et al., 2019; Frydrychová et al., 2004; Jetybayev, Bugrov, Dzuybenko, & Rubtsov, 2018; Jetybayev et al., 2012; Kociński et al., 2018; Kojima, Kubo, & Fujiwara, 2002; López-Fernández, Arroyo, Fernández, & Gosálvez, 2006; López-Fernández et al., 2004; Okazaki et al., 1993; Sahara et al., 1999; Vitturi, Lannino, Mansueto, Mansueto, & Colomba, 2008; Warchałowska-Śliwa, Grzywacz, Maryańska-Nadachowska, Hemp, & Hemp, 2015; Warchałowska-Śliwa et al., 2009; Yoshimura, Nakata, Mito, & Noji, 2006). The (TTAGG)n telomere motif apparently presents in all members of both major orthopteran lineages, and is therefore presumed the ancestral telomere motif in the order in general.
Embioptera (web spinners)
A comparatively small monophyletic group with about 2,000 species inhabiting tropical and subtropical regions of the world (Ross, 2000). No data on the telomere structure in web spinners are currently available.
Phasmatodea (leaf and stick insects)
This order consists of more than 3,000 phytophagous species in about 450 genera and 13 families that range mostly throughout the tropics and subtropics, and are less common in the temperate zone (Brock & Marshall, 2011). Leaf and stick insects are an ancient group of Hemimetabola, with many lineages evolved in the last 200 Mya (Misof et al., 2014). So far, telomeres of 11 species from seven genera of the only family Phasmatidae were studied. All of them were shown to be positive for the (TTAGG)n telomere motif (Frydrychová et al., 2004; Liehr et al., 2017; Scali et al., 2016). In some species, e.g., the Annam walking stick, Medauroidea extradentata Brunner von Wattenwyl, 1907, as well as Leptynia montana Scali, 1996 and L. attenuata Pantel, 1890, ITSs were present on certain chromosomes, in addition to telomeres (Liehr et al., 2017; Scali et al., 2016). On the contrary, no pericentromeric ITSs were detected on the neo-X chromosome of L. attenuata, suggesting that they were either inactivated or eliminated from the fused chromosome (Scali et al., 2016).
Mantophasmatodea (gladiators or heelwalkers)
This recently described order of apterous carnivorous insects is endemic to arid portions of southern Africa (Klass, Zompro, Kristensen, & Adis, 2002). Its phylogenetic relationships remain ambiguous. Data from multiple sources have supported the sister-group relationship between Mantophasmatodea and Grylloblattodea (thus forming a single order Notoptera) and confirmed that Mantophasmatodea represent one of the major lineages of the “lower Neoptera” (Klass, Picker, Damgaard, van Noort, & Tojo, 2003). The order currently comprises 19 described species, classified within the two families, namely, Mantophasmatidae and comparatively more derived Austrophasmatidae (Buder & Klass, 2013; Klass et al., 2003, 2002; Terry & Whiting, 2005; Wipfler et al., 2014). Recently, first cytogenetic data were published for Mantophasmatodea, namely for Karoophasma biedouwense Klass et al., 2003 (Austrophasmatidae). This species was reported to have 2n = 12 + X (in the male), monocentric chromosomes and the (TTAGG)n telomeric sequence (Lachowska-Cierlik et al., 2015).
Grylloblattodea (ice crawlers)
This is a small, cryophilic order with 25 extant species in five genera confined to northwestern North America and northeastern Asia. The order is considered a sister taxon of Mantophasmatodea (Klass et al., 2003). No data on telomere structure have been reported for the ice crawlers.
Mantodea (praying mantids)
A highly diverse and charismatic predatory order comprising approximately 2,300 described species in more than 430 genera and 15 families (Ehrmann, 2002). Two species from two families, the Indian flower mantis, Creobroter pictipennis (Wood-Mason, 1878) (Hymenopodidae), and Hierodula sp. (Mantidae), were studied using SBH and FISH, respectively; the (TTAGG)n telomere motif was detected in both species (Frydrychová et al., 2004).
Blattodea (cockroaches)
Cockroaches form one of the smaller insect orders with about 4,600 living species, the majority of which are found in tropical forests, distributed into six families (Beccaloni, 2014). Four species from the families Blattidae, Ectobiidae, Blattellidae, and Blaberidae studied using SBH and/or FISH were identified as positive for the (TTAGG)n telomere motif (Frydrychová et al., 2004; Marziliano, 1999; Okazaki et al., 1993). In a particular species, Periplaneta americana (Linnaeus, 1758), telomerase activity was detected using both the conventional TRAP (Sasaki & Fujiwara, 2000) and RTQ-TRAP (Korandová et al., 2014).
Isoptera (termites)
The world fauna of termites contains approximately 2,300 species that belong to seven families. Recent research (Inward, Beccaloni, & Eggleton, 2007) has shown that termites bear sufficient similarities with cockroaches (e.g., they occupy similar habitats, share the same type of food resources, the same intestinal symbionts, etc.) and must therefore be considered as a family of Blattodea, i.e., Termitidae s.l. Two species from the families Rhinotermitidae and Termopsidae respectively were studied using SBH and/or FISH; both appeared to be positive for the (TTAGG)n motif (Frydrychová et al., 2004; Okazaki et al., 1993). In a lab stock of another species of the family Rhinotermitidae, Prorhinotermes simplex (Hagen, 1858), telomerase activity was detected using RTQ-TRAP (Korandová et al., 2014).
Concluding comment
Except for Dermaptera, the TTAGG telomeric repeat was found in all studied Polyneoptera. Since Plecoptera and Dermaptera differ in terms of presence/absence of the (TTAGG)n motif, this does not support their sister-group relationship which was proposed by Letsch and Simon (2013). The absence of the (TTAGG)n in earwigs can have at least two possible explanations. Specifically, this repeat can be ancestral for all Polyneoptera, and in this case, its lack in Dermaptera represents an autapomorphy of this order (Frydrychová et al., 2004). Alternatively, since Dermaptera are currently considered a sister group to all other Polyneoptera (Misof et al., 2014), the absence of the TTAGG repeat can reflect its initial loss in the latter group with a subsequent reappearance in more advanced Polyneoptera. Moreover, taken together with the apparent lack of the (TTAGG)n motif in almost all studied Palaeoptera, this might even mean its loss during early evolution of Pterygota, followed by independent reappearance of this repeat in Polyneoptera and the remaining Neoptera (see below).
2.2.3 PARANEOPTERA (HEMIPTEROID ASSEMBLAGE)
Paraneopteran insects (=Acercaria) include more than 120,000 described species. They are subdivided into four orders: Hemiptera, Thysanoptera, Psocoptera, and Phthiraptera. Many paraneopteran insects are agricultural pests, animal parasites, and disease vectors. The monophyly of Paraneoptera is widely accepted and supported by numerous studies (Grimaldi & Engel, 2005).
Hemiptera (aphids, cicadas, planthoppers, true bugs, etc.)
It is an ancient lineage with fossils known since the Early Permian. With approximately 82,000 described species, this is the fifth largest insect order after Coleoptera, Diptera, Hymenoptera, and Lepidoptera (Grimaldi & Engel, 2005). The higher level relationships within Hemiptera have been debated for over centuries. Traditionally, Hemiptera have been subdivided into Homoptera and Heteroptera, sometimes with ordinal status. At present, they are considered as a monophyletic group comprising four major suborders, Sternorrhyncha (Psylloidea, Aleyrodoidea, Aphidoidea, and Coccoidea), Auchenorrhyncha (Cicadomorpha and Fulgoromorpha), Coleorrhyncha, and Heteroptera (Li et al., 2017).
Auchenorrhyncha (cicadas, leafhoppers, treehoppers, planthoppers, and froghoppers). The suborder Auchenorrhyncha is subdivided into two major lineages: the infraorder Cicadomorpha with the superfamilies Cicadoidea (cicadas), Cercopoidea (spittlebugs), Membracoidea (leafhoppers and treehoppers), and Myerslopioidea (ground-dwelling leafhoppers), as well as the infraorder Fulgoromorpha with the single superfamily Fulgoroidea (planthoppers) (Aguin-Pombo & Bourgoin, 2012; Szwedo, Bourgoin, & Lefebvre, 2004). More than 42,000 valid species of Auchenorrhyncha have been reported worldwide (Deitz, 2008). Depending on the used classification, these species can be grouped into 30–40 families. Data on telomeric DNA sequences are currently available for 33 species and 14 genera from the families Cicadidae, Myerslopiidae, Cicadellidae, Aphrophoridae, Cercopidae, Delphacidae, and Issidae, which therefore represent both infraorders and all superfamilies (Anjos et al., 2016; Frydrychová et al., 2004; Golub, Kuznetsova, & Rakitov, 2014; Kuznetsova, Maryańska-Nadachowska, Anokhin, & Aguin-Pombo, 2015; Maryańska-Nadachowska, Anokhin, Gnezdilov, & Kuznetsova, 2016; Maryańska-Nadachowska, Kuznetsova, & Karamysheva, 2013; Okazaki et al., 1993). All the species studied were shown to share the (TTAGG)n motif, thus suggesting its ancestral nature in the suborder Auchenorrhyncha in general.
Sternorrhyncha (Aphidoidea, Coccoidea, Psylloidea, and Aleyrodoidea)
- Aphidoidea (aphids)
- At present, about 5,000 species of aphids have been recorded in the world fauna (Favret & Eades, 2009). The most comprehensive taxonomic catalog (Remaudière & Remaudière, 1997) places all recent “true aphids” in the single family Aphididae (in contrast to 6–13 families accepted by some other authors; see Gavrilov-Zimin, Stekolshchikov, & Gautam, 2015), while the so-called “non-true aphids” are classified into two families, Adelgidae and Phylloxeridae. At least in four species of Aphididae, the TTAGG telomeric repeat was detected using both SBH and FISH (Bizzaro, Mandrioli, Zanotti, Giusti, & Manicardi, 2000; Mandrioli et al., 2014; Monti, Manicardi, & Mandrioli, 2011; Spence, Blackman, Testa, & Ready, 1998). On the contrary, no hybridization signals were observed in the Russian wheat aphid, Diuraphis noxia (Kurdjumov, 1913), in the experiments using FISH and SBH with the (TTAGG)n telomeric probes (Novotna, Havelka, Starý, Koutecký, & Vítkova, 2011). However, the same probes successfully hybridized to telomeres of the Mediterranean flour moth, Ephestia kuehniella (Zeller, 1879) (Lepidoptera, Pyralidae) which was used simultaneously as a positive control (Novotna et al., 2011). In a cosmopolitan aphid species, Acyrthosiphon pisum (Harris, 1776), a telomerase-coding gene was detected (Monti, Giusti, Bizzaro, Manicardi, & Mandrioli, 2011). Futhermore, TTAGG repeats in this species as well as in Myzus persicae (Sulzer, 1776) are interspersed with insertions of non-LTR retrotransposons (Monti, Serafini, Manicardi, & Mandrioli, 2013; The International Aphid Genomics Consortium, 2010). In addition, it was demonstrated in TTAGG-positive M. persicae that its telomerase could produce telomeres de novo to stabilize chromosome fragments (Mandrioli et al., 2014; Monti, Giusti, et al., 2011; Monti, Mandrioli, Rivi, & Manicardi, 2012). Yet, the genome of this species, as well as that of some other arthropods, lacks the genes coding for shelterin, a sequence-specific protein complex which stabilizes telomeres that are maintained by telomerase (Capkova Frydrychova & Mason, 2013; Mandrioli et al., 2014).
- Coccoidea (scale insects)
- Information on more than 8,200 valid species is currently recorded in the most comprehensive catalog of Coccoidea (García Morales et al., 2016). In a particular mealybug species, Planococcus lilacinus (Cockerell, 1905), the (TTAGG)n telomere motif was discovered using single-primer PCR and SBH (Mohan et al., 2011). Moreover, the same repeat was detected at interstitial sites of chromosomes, although with smaller copy numbers. In this species, telomerase activity was also observed at the sites of chromosome breaks initiated by radiation. This activity leads to de novo telomere formation at the ends of the fragmented chromosomes (Mohan et al., 2011).
- Psylloidea (jumping plant lice)
- A medium-sized group that currently includes about 3,600 species described from across all biogeographic regions of the world, with most of them recorded from the tropical and subtropical regions (Ouvrard, 2018; Percy et al., 2018). The most recent classification of jumping plant lice recognizes eight families (Burckhardt & Ouvrard, 2012). Approximately 175 species and 55 genera of all families except for the small family Phacopteronidae have been analyzed cytogenetically (Maryańska-Nadachowska, 2002). Data on telomere structure are currently known for only five species and four genera from the families Psyllidae and Aphalaridae studied by FISH; these species appeared to have the (TTAGG)n telomere motif (Maryańska-Nadachowska et al., 2018). In two species, Cacopsylla mali (Schmidberger, 1836) and C. sorbi (Linnaeus, 1767), neo-sex chromosomes showed no interstitial hybridization signals of the telomeric probe, although these chromosomes originate via several X-autosome fusions. Similarly, in another species, Baeopelma foersteri Flor, 1861, no ITSs are present on a pair of huge chromosomes originated from fusions between at least five pairs of ancestral autosomes. In all these species, the absence of ITSs is attributed either to the preceding loss of telomeric repeats or to their subsequent inactivation. Alternatively, these sequences could have too low copy number to be detected by FISH (Maryańska-Nadachowska et al., 2018).
- Aleyrodoidea (whiteflies)
- This is a smaller infraorder with a single family Aleyrodidae that includes about 1,600 described species in the world fauna (Ouvrard & Martin, 2018). The (TTAGG)n motif was detected using SBH in the only studied species, Trialeurodes vaporariorum (Westwood, 1856) (Frydrychová et al., 2004). Another evidence of telomere structure comes from the results of genomic analysis of Bemisia tabaci (Gennadius, 1889) (Luan et al., 2018). The latter experiments investigated bacteriocytes (insect cells harboring symbiotic bacteria) in this species, and were aimed to study bacteriocyte inheritance. The bacteriocyte genome of B. tabaci was revealed to contain the TTAGG repeat. Moreover, it was shown to express telomere maintenance genes. Thus, data on T. vaporariorum and B. tabaci suggest the presence of the canonical insect motif (TTAGG)n at the ends of whitefly chromosomes, although it still has to be strictly proven.
Coleorrhyncha (moss bugs). This group is an ancient lineage of moss-feeding insects, which contains more than 30 extant species and 17 genera belonging to a single recent family, the Gondwanan relict Peloridiidae (Burckhardt, 2009). The phylogenetic relationships of peloridiids within Hemiptera have been a matter of long lasting debate. Among several competing hypotheses, a sister-group relationship between Coleorrhyncha and Heteroptera is best supported by the morphological and phylogenomic data (Wang et al., 2019) as well as by an exclusive cytogenetic synapomorphy, namely an inverted sequence of sex chromosome divisions in male meiosis, the so-called sex chromosome post-reduction (Kuznetsova, Grozeva, et al., 2015). FISH has revealed the TTAGG telomeric repeat in the only hitherto studied species, Peloridium pomponorum Shcherbakov, 2014 (Kuznetsova, Grozeva, et al., 2015).
Heteroptera (true bugs). This suborder represents the largest and most diverse group of hemimetabolous insects, which includes more than 45,000 described species in 91 families distributed into seven infraorders (Henry, 2017). Heteroptera are relatively heterogeneous in respect to their telomere structure. Specifically, some species have the canonical TTAGG telomeric repeat, and some others apparently do not. Telomeric DNA sequences were studied in 40 species, 27 genera, and nine families belonging to the relatively basal infraorder Nepomorpha as well as to more derived sister infraorders Cimicomorpha and Pentatomomorpha (=the Geocorisae). The (TTAGG)n telomeric sequence is present in all hitherto studied families of Nepomorpha, namely Belostomatidae and Nepidae (Angus et al., 2017; Chirino et al., 2017; Kuznetsova et al., 2012). Taken together with the recent detection of the same telomeric sequence in Coleorrhyncha (Kuznetsova, Grozeva, et al., 2015), this suggests the ancestral nature of the TTAGG repeat in the Hemiptera in general. On the contrary, this motif was not found in all but one (the Reduviidae; Pita et al., 2016) families of the sister infraorders Cimicomorpha and Pentatomomorpha (Frydrychová et al., 2004; Golub, Golub, & Kuznetsova, 2015; Golub, Golub, & Kuznetsova, 2017; Golub, Golub, & Kuznetsova, 2018; Grozeva et al., 2011 etc.). Moreover, the dot blot experiments ruled out a number of alternative telomerase-based repeats in tested TTAGG-negative species, and therefore the telomeric sequence(s) in these groups remains unknown (Grozeva et al., 2011). Indeed, a recent survey of sequenced genomes of several pentatomomorphan and cimicomorphan species (Mason et al., 2016) demonstrated the lack of the TTAGG telomeric repeat. Furthermore, it was suggested that these taxa had a defective version of the telomerase gene. Nevertheless, Pita et al. (2016) showed that the cimicomorphan family Reduviidae, namely members of the subfamily Triatominae, possessed the telomeric motif (TTAGG)n. Phylogeny and relationships within and between families of Cimicomorpha are still uncertain (Li et al., 2017; Schuh, Weirauch, & Wheeler, 2009). Nevertheless, the DNA sequence data, alone and in concert with morphology, invariably treat the Reduviidae as part of the Cimicomorpha, with Reduvioidea (Reduviidae + Pachynomidae) apparently being basal within the group (Schuh et al., 2009). If it is true, a most plausible explanation is that Reduviidae inherited the ancestral (TTAGG)n telomere motif followed by its independent losses by other families of the Geocorisae.
Thysanoptera (thrips)
This is a smaller insect order, which harbors about 6,100 described species (Adler & Foottit, 2017). Telomere structure was studied in the only member of the group, the palm thrips, Parthenothrips dracaenae (Heeger, 1854), using SBH; presence of the (TTAGG)n telomere motif was detected (Frydrychová et al., 2004).
Psocoptera (booklice)
This is another smaller order of insects with more than 5,600 known species (Adler & Foottit, 2017). SBH has revealed the TTAGG telomeric repeat in the only studied member of the order, Stenopsocus lachlani (Kolbe, 1880) (Frydrychová et al., 2004).
Phthiraptera (true lice)
A smaller order of insects which are exclusive parasites of vertebrates (birds and mammals, including humans); it includes about 5,200 described species (Adler & Foottit, 2017). Based on genomic data, telomeric DNA of the human body louse, Pediculus humanus humanus Linnaeus, 1758 (Pediculidae), was suggested to consist of the TTAGG repeats with insertions of non-LTR retrotransposons (Kirkness et al., 2010).
Concluding comment
Despite an apparent loss in many advanced lineages of true bugs (Heteroptera), the (TTAGG)n motif is characteristic of all studied major clades of Paraneoptera, which strongly supports its ancestral nature in this monophyletic group.
2.3 Holometabolous insects
2.3.1 Oligoneoptera
This is an enormous group with more than 880,000 described species in the world fauna (Adler & Foottit, 2017). It harbors 11 orders, including the most numerous ones, i.e., members of the so-called “big four”—Coleoptera, Lepidoptera, Hymenoptera, and Diptera.
Neuropterida
This clade, which is sometimes considered as a superorder, harbors somewhat 6,400 species (Adler & Foottit, 2017). It contains members of three smaller orders, i.e., Raphidioptera, Megaloptera, and Neuroptera.
- Raphidioptera (snakeflies)
- A small order with about 250 described species in the world fauna (Adler & Foottit, 2017). SBH with the (TTAGG)n motif did not detect this repeat on chromosomes of the only studied species, Phaeostigma (Magnoraphidia) major (Burmeister, 1839) (Frydrychová et al., 2004).
- Megaloptera (dobsonflies)
- This is another small order, which harbors just about 370 described species (Adler & Foottit, 2017). The TTAGG telomeric repeat was found in the single examined species, the alder fly, Sialis lutaria (Linnaeus, 1758), using both FISH and SBH (Frydrychová et al., 2004).
- Neuroptera (antlions, owlflies, lacewings, etc.)
- The largest order of Neuropterida with more than 5,800 described species (Adler & Foottit, 2017). Telomere structure was previously studied using SBH in the common green lacewing, Chrysoperla carnea (Stephens, 1836) (Chrysopidae), as well as in an antlion, Protidricerus japonicus (McLachlan, 1891) (Myrmeleontidae) (Frydrychová et al., 2004; Okazaki et al., 1993). However, the (TTAGG)n telomeric motif was detected only in the latter species. Besides, recent data (Kuznetsova et al., 2016) suggest that the TTAGG telomeric repeat is characteristic of both Myrmeleontidae (antlions) and Ascalaphidae (owlflies) as well as of a common ancestor of those two.
Strepsiptera (twisted-wing parasites)
A small order of exclusively parasitic insects, which harbors slightly more than 600 described species (Adler & Foottit, 2017). No data on telomere structure in this order are known to date.
Coleoptera (beetles)
This is reputedly the largest order of insects with about 390,000 described species (Bouchard et al., 2017). Telomere structure in this group shows considerable diversity. Specifically, telomeres of many higher taxa of beetles contain the TTAGG canonical insect repeat (see Mora, Vela, Sanllorente, Palomeque, & Lorite, 2015), although this telomere motif is absent from chromosomes of certain coleopteran families, e.g., Scarabaeidae s.l. In general, telomerase-dependent telomere maintenance is supposed to have been lost five or six times during evolution of the beetles; the losses of (TTAGG)n sequence were reported in both the main coleopteran suborders, Adephaga and Polyphaga, and apparently happened irrespective of the phylogenetic relationships of the corresponding taxa (Frydrychová & Marec, 2002). Moreover, presence/absence of the (TTAGG)n motif can vary either between or even within several groups of Coleoptera (see Appendix 1). In addition, an alternative telomeric repeat, TCAGG, was found in all studied members of the large family Tenebrionidae as well as in some Mycetophagidae and Meloidae (Mravinac et al., 2011; see also Osanai et al., 2006 and Richards et al., 2008). Furthermore, telomeric repeat-specific non-LTR retrotransposons were found to be inserted into the proximal regions of the TCAGG arrays in the flour beetle, Tribolium castaneum (Herbst, 1797), which also belongs to the family Tenebrionidae (Osanai et al., 2006).
Hymenoptera (wasps, ants, bees etc.)
This is one of the largest orders of insects including more than 150,000 described species (Huber, 2017). Analogously to the hemipteran suborder Heteroptera (see above), our views on the distribution of the TTAGG telomeric repeat in the Hymenoptera dramatically changed during the last years. Specifically, the (TTAGG)n motif was initially considered characteristic of this order in general (Frydrychová et al., 2004). However, at that time this motif was detected only on chromosomes of certain aculeate Hymenoptera, i.e., the honeybee, Apis mellifera Linnaeus, 1758, from the family Apidae as well as about 20 ant species (Formicidae), using either FISH or SBH (Lorite, Carrillo, & Palomeque, 2002; Meyne & Imai, 1995; see also Korandová et al., 2014; Okazaki et al., 1993; Pereira, dos Reis, Cardoso, & Cristiano, 2018; Wurm et al., 2011). Interestingly, the telomeres of A. mellifera appeared to be a mosaic of short TTAGG repeats interspersed with TCAGGCTGGG, TCAGGCTGGGTTGGG, and TCAGGCTGGGTGAGGATGGG higher order repeat arrays (Garavís et al., 2013). Furthermore, the first fully sequenced parasitoid genome of Nasonia vitripennis (Walker, 1836) (Chalcidoidea, Pteromalidae), together with genomes of two other members of the same genus, was found to lack the TTAGG telomeric repeat, although the seemingly intact telomerase gene was detected in Nasonia vitripennis (The Nasonia Genome Working Group, 2010). A few years later, Gokhman et al. (2014) also showed absence of this repeat in several superfamilies of parasitoid Hymenoptera including Chalcidoidea. Futhermore, Menezes et al. (2013) and Menezes et al. (2017) have demonstrated absence of the (TTAGG)n motif in most aculeate Hymenoptera and independent reappearance of this repeat in the families Formicidae and Apidae. Nevertheless, in contrast to Apocrita (=Parasitica + Aculeata), presence of the TTAGG telomeric repeat in the only other existing suborder of this group, i.e., Symphyta, was not investigated until the last years. Recently, the existence of this motif in two members of the family Tenthredinidae, Tenthredo omissa (Förster, 1844) and Taxonus agrorum (Fallén, 1808), has been demonstrated by Gokhman and Kuznetsova (2018). Taken together with this detection of the TTAGG telomeric repeat in Eusymphyta, a sister group to all remaining Hymenoptera (i.e., to the clade Unicalcarida), all published results suggest the ancestral nature of this repeat in the order Hymenoptera and its subsequent loss somewhere within Unicalcarida. In addition, another telomeric repeat, TTAGGG, was detected in some ants, but its significance for defining telomere structure in these insects remains unclear (Menezes et al., 2017).
Amphiesmenoptera
This is a large superorder of insects with about 175,000 described species (Goldstein, 2017; Morse, 2017). It includes members of the two orders, Trichoptera and Lepidoptera.
- Trichoptera (caddisflies)
- This is a moderate-sized order with exclusively aquatic immature stages. It harbors more than 15,000 described species (Morse, 2017). Both FISH and SBH detected the (TTAGG)n telomere motif in two studied species, Limnephilus decipiens (Kolenati, 1848) and Stenopsyche marmorata Navás, 1920 (=S. japonica Martynov, 1926) (Frydrychová et al., 2004; Okazaki et al., 1993) that belong to the families Limnephilidae and Stenopsychidae, respectively.
- Lepidoptera (butterflies and moths)
- An enormous order of insects, which includes about 160,000 described species (Goldstein, 2017). The TTAGG telomeric repeat was detected using different techniques (FISH, SBH, etc.) in approximately two dozen studied species of butterflies and moths (Frydrychová et al., 2004; Mandrioli, 2002; Okazaki et al., 1993; Rego & Marec, 2003; Sahara et al., 1999; Sasaki & Fujiwara, 2000; Vershinina, Anokhin, & Lukhtanov, 2015; Yoshido, Marec, & Sahara, 2005, etc.). In the subgenus Polyommatus (Agrodiaetus) Hübner, 1822 (Lycaenidae) which members drastically differ in their chromosome numbers ranging from n = 10 to n = 108, de novo telomere formation is suggested in small chromosomes that evolved via fragmentations (Vershinina et al., 2015). Nevertheless, various members of the order can differ in respect to the levels of telomerase activity in their cells (Gong et al., 2015; Sasaki & Fujiwara, 2000). In the silk moth, Bombyx mori (Linnaeus, 1758) (Bombycidae), the telomerase activity was shown to be weak, and, as in the beetle T. castaneum, aphid M. persicae, and true louse P. humanus (see above), the canonical TTAGG telomeric repeats are regularly interspersed with insertions of non-LTR retrotransposons (Fujiwara, Osanai, Matsumoto, & Kojima, 2005; Okazaki, Ishikawa, & Fujiwara, 1995). The insertions of the retroelements into telomere regions might represent a putative evolutionary intermediate between canonical and retrotransposon-based telomeres (Mason et al., 2016).
Antliophora
A particular clade of holometabolous insects with more than 160,000 described species (Adler & Foottit, 2017; Courtney, Pape, Skevington, & Sinclair, 2017). It includes members of the three recent orders, i.e., Mecoptera, Siphonaptera, and Diptera.
- Mecoptera (scorpionflies)
- This is a small insect order with about 700 described species (Adler & Foottit, 2017). In the only studied species, Panorpa communis Linnaeus, 1758, both FISH and SBH did not detect the TTAGG telomeric repeat (Frydrychová et al., 2004).
- Siphonaptera (fleas)
- This moderate-sized order of insects, exclusively parasitizing warm blooded vertebrates, harbors about 2,200 described species (Adler & Foottit, 2017). As in the previous order, SBH did not reveal the TTAGG telomeric repeat on chromosomes of the only studied species, Ctenocephalides canis (Curtis, 1826) (Frydrychová et al., 2004).
- Diptera (true flies-midges, mosquitoes, flies etc.)
-
This is one of the most diverse and largest insect orders with about 160,000 described species (Courtney et al., 2017). According to the recent estimates, this order originated in the Permian, ca. 160 Mya (Misof et al., 2014). In all studied members of this group, the canonical insect TTAGG (or analogous) DNA repeat is not found up to now, and in most species, no telomerase activity was also detected (Biessmann & Mason, 1997; Casacuberta & Pardue, 2003; Frydrychová et al., 2004; Kamnert, López, Rosén, & Edström, 1997; Levis, Ganesan, Houtchens, Tolar, & Sheen, 1993; Mason et al., 2016; Okazaki et al., 1993; Sasaki & Fujiwara, 2000). Taken together with the absence of the (TTAGG)n motif in the examined species of Mecoptera and Siphonaptera, these data suggest an apparent loss of this repeat in a common ancestor of all Antliophora (Biessmann & Mason, 1997; Frydrychová et al., 2004; Pardue, Danilevskaya, Traverse, & Lowenhaupt, 1997). Alternatively, in all studied members of the suborder Nematocera (=lower Diptera) that belong to the genera Chironomus Meigen, 1803 (Chironomidae) and Anopheles Meigen, 1818 (Culicidae), long complex terminal tandem repeats (LCTTR) were found (Biessmann, Kobeski, Walter, Kasravi, & Roth, 1998; Carmona et al., 1985; Lopez, Nielsen, & Edström, 1996; Martínez Guitarte, de la Fuente, & Morcillo, 2014; Nielsen & Edström, 1993; Rosén & Edström, 2000, 2002; Rosén et al., 2002; Roth et al., 1997; Saiga & Edström, 1985; Zhang, Kamnert, López, Cohn, & Edström, 1994). For example, Chironomus tentans Fabricius, 1805 has 350-bp repeats at its telomeres (Rosén & Edström, 2000). At least in certain Chironomus species, some of these repeats are believed to evolve from short telomeric ones of other eukaryotic organisms, probably by insertion of transposons (Nielsen & Edström, 1993). These telomeres are reported to replicate by homologous recombination (Roth et al., 1997). In most examined members of the only other existing suborder, Brachycera (=higher Diptera), i.e., in some species of the genus Drosophila (Drosophilidae) further modification of telomere structure occured (Bachmann, Raab, & Sperlich, 1990; Biessmann, Zurovcova, Yao, Lozovskaya, & Walter, 2000). Specifically, telomeres of these insects contain multiple copies of certain retrotransposable elements in place of complex terminal arrays characteristic of other Diptera (Danilevskaya, Tan, Wong, Alibhai, & Pardue, 1998; Danilevskaya, Traverse, Hogan, DeBaryshe, & Pardue, 1999). At least in D. melanogaster, they are represented by long tandem head-to-tail arrays of retrotransposons identified as HeT-A, TART, and TAHRE, with HeT-A being the most abundant and comprising about 80%–90% of the total number of those elements (Capkova Frydrychova, Biessmann, & Mason, 2008; Mason et al., 2008). The telomeric retrotransposons are reverse transcribed directly onto the end of the chromosome, extending the end by successive transposition (Pardue & DeBaryshe, 2008). Moreover, the telomerase gene was not detected in D. melanogaster genome, which also can be explained by the deep modification of the process of telomere replication in this species (Osanai et al., 2006).
Interestingly, short telomeric repeats (14, 16, and 22 bp) and RaTART, which is apparently related to Drosophila's TART, were also detected in a particular member of Nematocera, Rhynchosciara americana (Wiedemann, 1821) that belongs to the family Sciaridae (dark-winged fungus gnats) (Fernandes, Madalena, & Gorab, 2012; Madalena, Amabis, & Gorab, 2010; Madalena, Fernandes, Villasante, & Gorab, 2010). In the same species, reverse transcriptase-related proteins were also found at non-centromeric chromosome ends (Gorab, 2003), which likely represents a unique character state within Nematocera. Although many details of telomere structure in Rh. americana remain uncertain, this situation probably models the origin of telomeric retrotransposons in advanced dipterans, e.g., Drosophila.
All accumulated data show that the order Diptera predominantly lacks short telomeric repeats characteristic of most insects and other eukaryotic organisms. In this order, alternative mechanisms of telomere replication, like homologous recombination or retrotransposition, apparently took over the conventional ones (Biessmann & Mason, 1997). Since these processes are not sequence-specific, it is not surprising that telomeric repeats demonstrate extensive variation between different species of the order Diptera, and even between different chromosomes of the same species (Rosén & Edström, 2000). Moreover, telomeres are protected with sequence-specific shelterin in most eukaryotic organisms, but at least in D. melanogaster chromosome ends are capped with terminin, another telomere protecting protein complex, which consequently lacks sequence specificity (Capkova Frydrychova et al., 2008; Capkova Frydrychova & Mason, 2013; Mason et al., 2008).
Concluding comment
The (TTAGG)n motif is characteristic of most major clades of Oligoneoptera except for Antliophora. The accumulated data therefore support the ancestral nature of the TTAGG telomeric repeat in the Oligoneoptera in general and its subsequent loss in Antliophora as well as in some other advanced lineages of Oligoneoptera. Specifically, in the order Diptera, the canonical insect telomeric repeat TTAGG was first substituted by characteristic long repeats and then (in the genus Drosophila) by certain retrotransposons.
3 EVOLUTION OF TELOMERE STRUCTURE IN THE CLASS INSECTA
Insects are the most diverse animal group with more than 1,000,000 described species (Adler & Foottit, 2017; Grimaldi & Engel, 2005). Data on telomere structure are presently available for more than 350 species and about 240 genera from 108 families and 25 insect orders (Appendix 1, Figure 1). In the subphylum Hexapoda, these data are still absent for the orders Zoraptera, Embioptera, Grylloblattodea, and Strepsiptera as well as for the three orders of Entognatha (Protura, Collembola, and Diplura). All accumulated information supports the hypothesis suggested by Frydrychová et al. (2004) that the TTAGG telomeric repeat is ancestral in the class Insecta. However, this motif was either changed to another sequences (e.g., TCAGG in some beetles) or lost many times along various branches of the insect phylogenetic tree, as shown by Frydrychová et al. (2004) and Mason et al. (2016). Furthermore, these authors listed the existing data on particular backup mechanisms of telomere maintenance in this group (ALT). Coexistence of different (either active or disabled) mechanisms of telomere replication in most insects can therefore explain losses and reappearances of the TTAGG repeat in insect evolution, including an apparent absence of this motif in Antliophora as well as the change of this repeat to TCAGG in certain clades of the order Coleoptera. In addition, a few higher taxa, e.g., Heteroptera (Hemiptera) and Hymenoptera, show presence of the (TTAGG)n motif in their basal clades as well as a subsequent loss and, at least in the Hymenoptera, independent reappearance of this motif in some advanced groups (Angus et al., 2017; Chirino et al., 2017; Gokhman & Kuznetsova, 2018; Menezes et al., 2017; Pita et al., 2016). This also shows the importance of phylogenetic inference for reconstructing evolutionary transformations of telomere structure. Moreover, although particular types of telomere composition are generally characteristic of higher taxa of insects, these patterns can substantially change between certain families, genera and even species (Kuznetsova et al., 2017 and Novotna et al., 2011). The accumulating data therefore demonstrate that the class Insecta is much more diverse in terms of its telomere structure than it was supposed just about 10 or 15 years ago (Frydrychová et al., 2004). It is also noteworthy that the sole presence of a particular telomere motif does not necessarily mean its main role in the process of telomere replication. For example, Bombyx mori, together with all other studied members of the order Lepidoptera, does have the (TTAGG)n telomere motif (Frydrychová et al., 2004). Yet, certain non-LTR retrotransposons, that are also present in the telomere regions of B. mori, are actively transcribed and retrotransposed into TTAGG repeats in a highly sequence-specific manner; however, TTAGG-specific telomerase retains its activity at a very low level, but it is apparently not involved in telomere replication in this species (Fujiwara et al., 2005). Although, these data contradict the earlier opinion on the apparent absence of telomerase activity in B. mori expressed by Sasaki and Fujiwara (2000) and later cited by Frydrychová et al. (2004). We believe that telomere structure in B. mori, as well as in a few similar cases, suggests a possible way of transition from a canonical insect telomere containing the TTAGG repeat to that characteristic of certain Diptera (see above).
We can also assume that certain telomeric motifs sometimes cannot be detected by FISH due to e.g., their low copy number. Nevertheless, this is not the case when the results of the full genome sequencing are available, e.g., in the parasitic wasp N. vitripennis (The Nasonia Genome Working Group, 2010) which definitely lacks the TTAGG telomeric repeat. In addition, if the copy number of a particular telomere motif is low enough, it will apparently be unable to ensure effective replication of the telomeres. Anyway, structures underlying the process of telomere replication in similar cases are yet to be discovered.
In conclusion, we would like to express our sincere belief that future research will integrate much more taxon sampling, and will therefore certainly provide greater insight into the evolution of telomere structure in Insecta.
ACKNOWLEDGEMENTS
We are very grateful to the editor and two anonymous reviewers for their thorough revision of the manuscript and helpful comments. We also thank Dr. Nazar A. Shapoval and Natalia S. Khabazova (Zoological Institute RAS, St. Petersburg) for critical comments on the discussion of terminology concerning the techniques applied in Appendix 1 and careful technical assistance respectively. The present study was supported by the research project no. AAAA-A19-119020790106-0 (VGK) and by the RFBR research grants no. 17-04-00828 (VGK) and 18-04-00611 (VEG).
APPENDIX 1 A:
| Species | Telomere composition | References | Techniques applied |
|---|---|---|---|
| Archaeognatha | |||
| Machilidae | |||
| Lepismachilis sp. | TTAGG + | Frydrychová et al. (2004) | FISH, SBH |
| Zygentoma | |||
| Lepismatidae | |||
| Lepisma saccharina Linnaeus, 1758 | TTAGG + | Frydrychová et al. (2004) | SBH |
| Thermobia domestica (Packard, 1873) | TTAGG + | Frydrychová et al. (2004) | FISH, SBH |
| Odonata | |||
| Coenagrionidae | |||
| Ischnura elegans (Vander Linden, 1820) | TTAGG – | Frydrychová et al. (2004) | FISH, SBH |
| Corduliidae | |||
| Cordulia aenea (Linnaeus, 1758) | TTAGG – | Kuznetsova et al. (2017) | FISH |
| Epitheca bimaculata (Charpentier, 1825) | TTAGG – | Kuznetsova et al. (2017) | FISH |
| Libellulidae | |||
| Leucorrhinia rubicunda (Linnaeus, 1758) | TTAGG – | Kuznetsova et al. (2017) | FISH |
| Libellula depressa Linnaeus, 1758 | TTAGG – | Kuznetsova et al. (2017) | FISH |
| L. quadrimaculata quadrimaculata Linnaeus, 1758 | TTAGG – | Kuznetsova et al. (2017) | FISH |
| Orthetrum cancellatum Linnaeus, 1758 | TTAGG – | Kuznetsova et al. (2017) | FISH |
| Sympetrum danae (Sulzer, 1776) | TTAGG – | Kuznetsova et al. (2017) | FISH |
| S. vulgatum (Linnaeus, 1758) | TTAGG + | Kuznetsova et al. (2017) | FISH |
| Ephemeroptera | |||
| Baetidae | |||
| Cloeon dipterum (Linnaeus, 1761) | TTAGG – | Frydrychová et al. (2004) | FISH, SBH |
| Dermaptera | |||
| Forficulidae | |||
| Forficula auricularia Linnaeus, 1758 | TTAGG – | Frydrychová et al. (2004) | FISH, SBH |
| F. scudderi de Bormans, 1880 | TTAGG – | Okazaki et al. (1993) | SBH |
| Plecoptera | |||
| Perlidae | |||
| Perla burmeisteriana Claassen, 1936 | TTAGG + | Frydrychová et al. (2004) | FISH, SBH |
| Orthoptera | |||
| Caelifera | |||
| Acrididae | |||
| Arcyptera fusca (Pallas, 1773) | TTAGG + | López-Fernández et al. (2004) | FISH |
| TTAGG + | Jetybayev et al. (2012) | FISH | |
| A. tornosi Bolívar, 1884 | TTAGG + | López-Fernández et al. (2004) | FISH |
| Bryodema gebleri (Fischer-Waldheim, 1836) | TTAGG + | Jetybayev et al. (2018) | FISH |
| Chorthippus albomarginatus (De Geer, 1773) | TTAGG + | Jetybayev et al. (2012) | FISH |
| Ch. apricarius (Linnaeus, 1758) | TTAGG + | Jetybayev et al. (2012) | FISH |
| Ch. biguttulus (Linnaeus, 1758) | TTAGG + | Jetybayev et al. (2012) | FISH |
| Ch. binotatus (Charpentier, 1825) | TTAGG + | López-Fernández et al. (2004) | FISH |
| Ch. hammarstroemi (Miram, 1907) | TTAGG + | Jetybayev et al. (2012) | FISH |
| Ch. intermedius (Bey-Bienko, 1926) | TTAGG + | Jetybayev et al. (2012) | FISH |
| Ch. jacobsoni (Ikonnikov, 1911) | TTAGG + | Jetybayev et al. (2012), Jetybayev et al. (2018) | FISH |
| Ch. jucundus (Fischer, 1853) | TTAGG + | López-Fernández et al. (2004) | FISH |
| Ch. karatavicus Bey-Bienko, 1936 | TTAGG + | Jetybayev et al. (2012) | FISH |
| Dociostaurus hispanicus Bolívar, 1898 | TTAGG + | López-Fernández et al. (2004) | FISH |
| Eyprepocnemis plorans (Charpentier, 1825) | TTAGG + | López-Fernández et al. (2004) | FISH |
| TTAGG + | Jetybayev et al. (2018) | FISH | |
| Gomphocerippus rufus (Linnaeus, 1758) | TTAGG + | Jetybayev et al. (2012) | FISH |
| Gomphocerus sibiricus (Linnaeus, 1767) | TTAGG + | Jetybayev et al. (2012) (as Aeropus) | FISH |
| Locusta migratoria (Linnaeus, 1758) | TTAGG + | Okazaki et al. (1993) | SBH |
| TTAGG + | Sahara et al. (1999) | FISH, SBH | |
| TTAGG+ | Sasaki and Fujiwara (2000) | FISH | |
| Mesasippus kozhevnikovi (Tarbinsky, 1925) | TTAGG + | Jetybayev et al. (2012) | FISH |
| Omocestus viridulus (Linnaeus, 1758) | TTAGG + | Jetybayev et al. (2012) | FISH |
| Podisma pedestris (Linnaeus, 1758) | TTAGG + | López-Fernández et al. (2004) | FISH |
| P. sapporensis Shiraki, 1910 | TTAGG + | Jetybayev et al. (2018) | FISH |
| Podismopsis altaica (Zubovski, 1900) | TTAGG + | Jetybayev et al. (2012) | FISH |
| Pseudochorthippus parallelus (Zetterstedt, 1821) | TTAGG + | López-Fernández et al. (2004) (as Chorthippus) | FISH |
| Ramburiella hispanica (Rambur, 1838) | TTAGG + | López-Fernández et al. (2004) | FISH |
| Stauroderus scalaris (Fischer-Waldheim, 1846) | TTAGG + | López-Fernández et al. (2004) | FISH |
| TTAGG + | Jetybayev et al. (2012) | FISH | |
| Stenobothrus eurasius Zubovski, 1898 | TTAGG + | Jetybayev et al. (2012) | FISH |
| Pamphagidae | |||
| Acinipe hesperica lepineyi Chopard, 1943 | TTAGG + | Buleu et al. (2019) | FISH |
| Asiotmethis heptapotamicus songoricus Shumakov, 1949 | TTAGG + | Jetybayev et al. (2018) | FISH |
| Eunapiodes granosus Stål, 1876 | TTAGG + | Buleu et al. (2019) | FISH |
| Euryparyphes rungsi Massa, 2013 | TTAGG + | Buleu et al. (2019) | FISH |
| Paracinipe alticola Werner, 1932 | TTAGG + | Buleu et al. (2019) | FISH |
| P. crassicornis Bolívar, 1907 | TTAGG + | Buleu et al. (2019) | FISH |
| P. dolichocera Bolívar, 1907 | TTAGG + | Buleu et al. (2019) | FISH |
| P. theryi Werner, 1931 | TTAGG + | Buleu et al. (2019) | FISH |
| Paraeumigus parvulus Bolívar, 1907 | TTAGG + | Buleu et al. (2019) | FISH |
| Pamphagus ortolaniae Cusimano & Massa, 1977 | TTAGG + | Vitturi et al. (2008) | FISH |
| Pseudoglauia tarudantica Bolívar, 1914 | TTAGG + | Buleu et al. (2019) | FISH |
| Tmethis cisti Fabricius, 1787 | TTAGG + | Buleu et al. (2019) | FISH |
| Pyrgomorphidae | |||
| Pyrgomorpha conica (Olivier, 1791) | TTAGG + | López-Fernández et al. (2006), López-Fernández et al. (2004) | FISH |
| Ensifera | |||
| Gryllidae | |||
| Brachyteleogryllus occipitalis Serville, 1838 | TTAGG + | Sasaki and Fujiwara (2000) (as Teleogryllus taiwanemma) | FISH, TRAP-Seq |
| TTAGG + | Kojima et al. (2002) | FISH, SBH | |
| Teleogryllus emma (Ohmachi & Matsuura, 1951) | TTAGG + | Sasaki and Fujiwara (2000) | FISH |
| Rhaphidophoridae | |||
| Diestrammena japanica Blatchey, 1920 | TTAGG + | Okazaki et al. (1993) (as D. japonica) | SBH |
| Tettigoniidae | |||
| Abracris flavolineata (De Geer, 1773) | TTAGG + | Bueno et al. (2013) | FISH |
| Gampsocleis abbreviata ebneri Uvarov, 1921 | TTAGG + | Kociński et al. (2018) | FISH |
| G. abbreviata renei Miksic, 1973 | TTAGG + | Kociński et al. (2018) | FISH |
| G. abbreviata subsp. indet. | TTAGG + | Kociński et al. (2018) | FISH |
| G. glabra Herbst, 1786 | TTAGG + | Kociński et al. (2018) | FISH |
| G. gratiosa Brunner von Wattenwyl, 1888 | TTAGG + | Kociński et al. (2018) | FISH |
| G. sedakovii sedakovii (Fischer-Waldheim, 1846) | TTAGG + | Kociński et al. (2018) | FISH |
| G. ussurensis Adelung, 1910 | TTAGG + | Kociński et al. (2018) | FISH |
| Saga campbelli campbelli Uvarov, 1921 | TTAGG + | Warchałowska-Śliwa et al. (2009) | FISH |
| S. hellenica Kaltenbach, 1967 | TTAGG + | Warchałowska-Śliwa et al. (2009) | FISH |
| S. natoliae Serville, 1838 | TTAGG + | Warchałowska-Śliwa et al. (2009) | FISH |
| S. rammei Kaltenbach, 1965 | TTAGG + | Warchałowska-Śliwa et al. (2009) | FISH |
| S. rhodiensis Salfi, 1929 | TTAGG + | Warchałowska-Śliwa et al. (2009) | FISH |
| Spalacomimus verruciferus (Karsch, 1887) | TTAGG + | Warchałowska-Śliwa et al. (2015) | FISH |
| S. talpa (Gerstaecker, 1869) | TTAGG + | Warchałowska-Śliwa et al. (2015) | FISH |
| Phasmatodea | |||
| Phasmatidae | |||
| Extatosoma tiaratum (Macleay, 1826) | TTAGG + | Frydrychová et al. (2004) | FISH, SBH |
| Leptynia attenuata algarvica Scali, Milani & Passamonti, 2012 | TTAGG + | Scali et al. (2016) | FISH |
| L. a. attenuata Pantel, 1890 | TTAGG + | Scali et al. (2016) | FISH |
| L. a. iberica Scali, Milani & Passamonti, 2012 | TTAGG + | Scali et al. (2016) | FISH |
| L. montana Scali, 1996 | TTAGG + | Scali et al. (2016) | FISH |
| Medauroidea extradentata (Brunner von Wattenwyl, 1907) | TTAGG + | Liehr et al. (2017) | FISH |
| Peruphasma schultei Conle & Hennemann, 2005 | TTAGG + | Liehr et al. (2017) | FISH |
| Phaenopharos khaoyaiensis Zompro, 2000 | TTAGG + | Liehr et al. (2017) | FISH |
| Sipyloidea sipylus (Westwood, 1859) | TTAGG + | Liehr et al. (2017) | FISH |
| Sungaya inexpectata Zompro, 1996 | TTAGG + | Liehr et al. (2017) | FISH |
| Mantophasmatodea | |||
| Austrophasmatidae | |||
| Karoophasma biedouwense Klass, Picker, Damgaard, van Noort & Tojo, 2003 | TTAGG + | Lachowska-Cierlik et al. (2015) | FISH |
| Mantodea | |||
| Hymenopodidae | |||
| Creobroter pictipennis Wood-Mason, 1878 | TTAGG + | Frydrychová et al. (2004) | SBH |
| Mantidae | |||
| Hierodula sp. | TTAGG + | Frydrychová et al. (2004) | FISH |
| Blattodea | |||
| Blaberidae | |||
| Blaberus craniifer Burmeister, 1838 | TTAGG + | Marziliano (1999) | FISH |
| Blattellidae | |||
| Blattella germanica (Linnaeus, 1767) | TTAGG + | Marziliano (1999) | FISH |
| Blattidae | |||
| Periplaneta americana (Linnaeus, 1758) | TTAGG + | Sasaki and Fujiwara (2000) | TRAP-Seq |
| TTAGG + | Frydrychová et al. (2004) | FISH, SBH | |
| P. fuliginosa Serville, 1839 | TTAGG + | Okazaki et al. (1993) | SBH |
| Isoptera | |||
| Rhinotermitidae | |||
| Reticulitermes santonensis Feytaud, 1924 | TTAGG + | Frydrychová et al. (2004) | FISH, SBH |
| Termopsidae | |||
| Hodotermopsis japonicus Holmgren, 1912 | TTAGG + | Okazaki et al. (1993) | SBH |
| Hemiptera | |||
| Auchenorrhyncha | |||
| Aphrophoridae | |||
| Philaenus arslani Abdul-Nour & Lahoud, 1996 | TTAGG + | Maryańska-Nadachowska et al. (2013) | FISH |
| Ph. italosignus Drosopoulos & Remane, 2000 | TTAGG + | Maryańska-Nadachowska et al. (2013) | FISH |
| Ph. loukasi Drosopoulos & Asche, 1991 | TTAGG + | Maryańska-Nadachowska et al. (2013) | FISH |
| Ph. maghresignus Drosopoulos & Remane, 2000 | TTAGG + | Maryańska-Nadachowska et al. (2013) | FISH |
| Ph. signatus Melichar, 1896 | TTAGG + | Maryańska-Nadachowska et al. (2013) | FISH |
| Ph. spumarius (Linnaeus, 1758) | TTAGG + | Maryańska-Nadachowska et al. (2013) | FISH |
| Ph. tarifa Remane & Drosopoulos, 2001 | TTAGG + | Maryańska-Nadachowska et al. (2013) | FISH |
| Ph. tesselatus Melichar, 1899 | TTAGG + | Maryańska-Nadachowska et al. (2013) | FISH |
| Cercopidae | |||
| Mahanarva fimbriolata (Stål, 1854) | TTAGG + | Anjos et al. (2016) | FISH |
| M. liturata (Le Peletier & Serville, 1825) | TTAGG + | Anjos et al. (2016) | FISH |
| M. quadripunctata (Walker, 1858) | TTAGG + | Anjos et al. (2016) | FISH |
| M. spectabilis Distant, 1909 | TTAGG + | Anjos et al. (2016) | FISH |
| M. tristis (Fabricius, 1803) | TTAGG + | Anjos et al. (2016) | FISH |
| M. vittata (Walker, 1851) | TTAGG + | Anjos et al. (2016) | FISH |
| Cicadellidae | |||
| Alebra albostriella (Fallén, 1826) | TTAGG + | Kuznetsova, Maryańska-Nadachowska, et al. (2015) | FISH |
| A. coryli Le Quesne, 1977 | TTAGG + | Kuznetsova, Maryańska-Nadachowska, et al. (2015) | FISH |
| A. viridis Rey, 1894 | TTAGG + | Kuznetsova, Maryańska-Nadachowska, et al. (2015) | FISH |
| A. wahlbergi (Boheman, 1845) | TTAGG + | Kuznetsova, Maryańska-Nadachowska, et al. (2015) | FISH |
| Alebra sp. | TTAGG + | Kuznetsova, Maryańska-Nadachowska, et al. (2015) | FISH |
| Cicadidae | |||
| Yezoterpnosia nigricosta (Motschulsky, 1866) | TTAGG + | Okazaki et al. (1993) (as Terpnosia) | SBH |
| Delphacidae | |||
| Javesella pellucida (Fabricius, 1794) | TTAGG + | Frydrychová et al. (2004) (as Calligypona) | SBH |
| Issidae | |||
| Agalmatium bilobum (Fieber, 1877) | TTAGG + | Maryańska-Nadachowska et al. (2016) | FISH |
| A. flavescens (Olivier, 1791) | TTAGG + | Maryańska-Nadachowska et al. (2016) | FISH |
| Hemisphaerius sp. | TTAGG + | Maryańska-Nadachowska et al. (2016) | FISH |
| Issus lauri Ahrens, 1814 | TTAGG + | Maryańska-Nadachowska et al. (2016) | FISH |
| Kervillea basiniger (Dlabola, 1982) | TTAGG + | Maryańska-Nadachowska et al. (2016) | FISH |
| Mycterodus pallens (Stål, 1861) | TTAGG + | Maryańska-Nadachowska et al. (2016) | FISH |
| Palaeolithium distinguendum (Kirschbaum, 1868) | TTAGG + | Maryańska-Nadachowska et al. (2016) | FISH |
| Scorlupella discolor (Germar, 1821) | TTAGG + | Maryańska-Nadachowska et al. (2016) | FISH |
| Thabena sp. | TTAGG + | Maryańska-Nadachowska et al. (2016) | FISH |
| Zopherisca penelopae (Dlabola, 1974) | TTAGG + | Maryańska-Nadachowska et al. (2016) | FISH |
| Z. tendinosa (Spinola, 1839) | TTAGG + | Maryańska-Nadachowska et al. (2016) | FISH |
| Myerslopiidae | |||
| Mapuchea chilensis (Nielson, 1996) | TTAGG + | Golub et al. (2014) | FISH |
| Sternorrhyncha | |||
| Aphidoidea | |||
| Aphididae | |||
| Acyrthosiphon pisum (Harris, 1776) | TTAGG + | Okazaki et al. (1993) | SBH |
| TTAGG + | Bizzaro et al. (2000) | FISH | |
| TTAGG + | Monti, Giusti, et al. (2011) | FISH | |
| Amphorophora tuberculata Brown & Blackman, 1985 | TTAGG – | Spence et al. (1998) | FISH |
| Diuraphis noxia (Mordvilko, 1913) | TTAGG – | Novotná et al. (2011) | FISH, SBH |
| Myzus antirrhinii (Macchiati, 1883) | TTAGG + | Spence et al. (1998) | FISH |
| M. persicae (Sulzer, 1776) | TTAGG +/TSRT | IAGC (2010) | FISH |
| TTAGG + | Monti, Giusti, et al. (2011) | DBH, SBH | |
| TTAGG +/TSRT | Monti et al. (2013) | FISH | |
| TTAGG + | Mandrioli et al. (2014) | FISH | |
| TTAGG + | Spence et al. (1998) | FISH | |
| Macrosiphum euphorbiae (Thomas, 1878) | TTAGG + | Monti, Manicardi, et al. (2011) | FISH, SBH |
| Megoura viciae Buckton, 1876 | TTAGG + | Monti, Giusti, et al. (2011) | FISH, SBH |
| Rhopalosiphum padi (Linnaeus, 1758) | TTAGG + | Monti, Giusti, et al. (2011) | FISH, SBH |
| Coccoidea | |||
| Coccidae | |||
| Planococcus lilacinus (Cockerell, 1905) | TTAGG + | Mohan et al. (2011) | FISH, SBH |
| Psylloidea | |||
| Aphalaridae | |||
| Rhinocola aceris (Linnaeus, 1758) | TTAGG + | Maryańska-Nadachowska et al. (2018) | FISH |
| Psyllidae | |||
| Baeopelma foersteri (Flor, 1861) | TTAGG + | Maryańska-Nadachowska et al. (2018) | FISH |
| Cacopsylla mali (Schmidberger, 1836) | TTAGG + | Maryańska-Nadachowska et al. (2018) | FISH |
| C. sorbi (Linnaeus, 1767) | TTAGG + | Maryańska-Nadachowska et al. (2018) | FISH |
| Psylla alni (Linnaeus, 1758) | TTAGG + | Maryańska-Nadachowska et al. (2018) | FISH |
| Aleyrodoidea | |||
| Aleyrodidae | |||
| Bemisia tabaci (Gennadius, 1889) | TTAGG + | Luan et al. (2018) | DNA-Seq, MA, NGS |
| Trialeurodes vaporariorum (Westwood, 1856) | TTAGG + | Frydrychová et al. (2004) | SBH |
| Coleorrhyncha | |||
| Peloridiidae | |||
| Peloridium pomponorum Shcherbakov, 2014 | TTAGG + | Kuznetsova, Grozeva, et al. (2015) | FISH |
| Heteroptera | |||
| Nepomorpha | |||
| Belostomatidae | |||
| Belostoma candidum Montandon, 1903 | TTAGG + | Chirino et al. (2017) | FISH |
| B. dentatum (Mayr, 1863) | TTAGG + | Chirino et al. (2017) | FISH |
| B. elegans (Mayr, 1871) | TTAGG + | Chirino et al. (2017) | FISH |
| B. elongatum Montandon, 1908 | TTAGG + | Chirino et al. (2017) | FISH |
| B. micantulum (Stål, 1858) | TTAGG + | Chirino et al. (2017) | FISH |
| B. oxyurum (Dufour, 1863) | TTAGG + | Chirino et al. (2017) | FISH |
| Lethocerus patruelis (Stål, 1855) | TTAGG + | Kuznetsova et al. (2012) | FISH |
| Nepidae | |||
| Nepa cinerea Linnaeus, 1758 | TTAGG + | Angus et al. (2017) | FISH |
| Ranatra linearis (Linnaeus, 1758) | TTAGG + | Angus et al. (2017) | FISH |
| Cimicomorpha | |||
| Cimicidae | |||
| Cimex lectularius Linnaeus, 1758 | TTAGG – | Grozeva et al. (2011) | FISH, DBH |
| TTTTGGGG – | Grozeva et al. (2011) | DBH | |
| TTGGGG – | Grozeva et al. (2011) | DBH | |
| TTAGGC – | Grozeva et al. (2011) | DBH | |
| TAACC– | Grozeva et al. (2011) | DBH | |
| TTAGGG – | Grozeva et al. (2011) | DBH | |
| TTTAGGG – | Grozeva et al. (2011) | DBH | |
| Miridae | |||
| Deraeocoris ruber (Linnaeus, 1758) | TTAGG – | Grozeva et al. (2011) | FISH |
| D. rutilus (Herrich-Schaffer, 1838) | TTAGG – | Grozeva et al. (2011) | FISH |
| Megaloceroea recticornis (Geoffroy, 1785) | TTAGG – | Grozeva et al. (2011) | FISH |
| Nabidae | |||
| Nabis sp. | TTAGG – | Grozeva et al. (2011) | DBH |
| TTTTGGGG – | Grozeva et al. (2011) | DBH | |
| TTGGGG – | Grozeva et al. (2011) | DBH | |
| TTAGGC – | Grozeva et al. (2011) | DBH | |
| TAACC – | Grozeva et al. (2011) | DBH | |
| TTAGGG – | Grozeva et al. (2011) | DBH | |
| TTTAGGG – | Grozeva et al. (2011) | DBH | |
| Reduviidae | |||
| Dipetalogaster maxima (Uhler, 1894) | TTAGG + | Pita et al. (2016) | FISH |
| Rhodnius prolixus Stål, 1859 | TTAGG + | Pita et al. (2016) | FISH |
| Triatoma dimidiata (Latreille, 1811) | TTAGG + | Pita et al. (2016) | FISH |
| T. infestans Klug, 1834 | TTAGG + | Pita et al. (2016) | NGS, FISH |
| Tingidae | |||
| Acalypta marginata Wolff, 1804 | TTAGG – | Golub et al. (2018) | FISH |
| Agramma atricapillum Spinola, 1837 | TTAGG – | Golub et al. (2018) | FISH |
| A. fallax (Horváth, 1906) | TTAGG – | Golub et al. (2017) | FISH |
| A. femorale (Thompson, 1871) | TTAGG – | Golub et al. (2015) | FISH |
| Catoplatus carthusianus Goeze, 1778 | TTAGG – | Golub et al. (2018) | FISH |
| Copium teucrii (Host, 1788) | TTAGG – | Golub et al. (2017) | FISH |
| Derephysia longispina Golub, 1974 | TTAGG – | Golub et al. (2018) | FISH |
| Dictyla platyoma Fieber, 1861 | TTAGG – | Golub et al. (2018) | FISH |
| D. humuli (Fabricius, 1794) | TTAGG – | Golub et al. (2017) | FISH |
| Dictyonota stichnocera Fieber, 1844 | TTAGG – | Golub et al. (2018) | FISH |
| Elasmotropis testacea (Herrich-Schäffer, 1830) | TTAGG – | Golub et al. (2015) | FISH |
| Galeatus sinuatus (Herrich-Schäffer, 1838) | TTAGG – | Golub et al. (2017) | FISH |
| Lasiacantha hermani Vásárhelyi, 1977 | TTAGG – | Golub et al. (2018) | FISH |
| Oncochila simplex Herrich-Schaeffer, 1830 | TTAGG – | Golub et al. (2018) | FISH |
| Tingis cardui (Linnaeus, 1758) | TTAGG – | Golub et al. (2015) | FISH |
| T. crispata (Herrich-Schäffer, 1838) | TTAGG – | Golub et al. (2015) | FISH |
| T. pilosa Hummel, 1825 | TTAGG – | Golub et al. (2018) | FISH |
| T. reticulata Herrich-Schäffer, 1835 | TTAGG – | Golub et al. (2018) | FISH |
| Pentatomomorpha | |||
| Lygaeidae | |||
| Oxycarenus lavaterae (Fabricius, 1787) | TTAGG – | Grozeva et al. (2011) | FISH, DBH |
| TTTTGGGG – | Grozeva et al. (2011) | DBH | |
| TTGGGG – | Grozeva et al. (2011) | DBH | |
| TTAGGC – | Grozeva et al. (2011) | DBH | |
| TAACC – | Grozeva et al. (2011) | DBH | |
| TTAGGG – | Grozeva et al. (2011) | DBH | |
| TTTAGGG – | Grozeva et al. (2011) | DBH | |
| Pentatomidae | |||
| Eurydema oleracea (Linnaeus, 1758) | TTAGG – | Grozeva et al. (2011) | FISH |
| Graphosoma lineatum (Linnaeus, 1758) | TTAGG – | Grozeva et al. (2011) | FISH |
| Halyomorpha halys (Stål, 1855) | TTAGG – | Okazaki et al. (1993) (as H. mista) | SBH |
| Pyrrhocoridae | |||
| Pyrrhocoris apterus (Linnaeus, 1758) | TTAGG – | Sahara et al. (1999) | FISH, SBH |
| TTAGG – | Grozeva et al. (2011) | FISH | |
| Thysanoptera | |||
| Thripidae | |||
| Parthenothrips dracaenae (Heeger, 1854) | TTAGG + | Frydrychová et al. (2004) | SBH |
| Psocoptera | |||
| Stenopsocidae | |||
| Stenopsocus lachlani Kolbe, 1880 | TTAGG + | Frydrychová et al. (2004) | FISH, SBH |
| Phthiraptera | |||
| Pediculidae | |||
| Pediculus humanus humanus Linnaeus, 1758 | TTAGG +/TSRT | Kirkness et al. (2010) | NGS |
| Raphidioptera | |||
| Raphidiidae | |||
| Phaeostigma major (Burmeister, 1839) | TTAGG – | Frydrychová et al. (2004) (as Magnoraphidia) | SBH |
| Megaloptera | |||
| Sialidae | |||
| Sialis lutaria (Linnaeus, 1758) | TTAGG + | Frydrychová et al. (2004) | FISH, SBH |
| Neuroptera | |||
| Ascalaphidae | |||
| Libelloides macaronius (Scopoli, 1763) | TTAGG + | Kuznetsova et al. (2016) | FISH |
| Protidricerus japonicus (McLachlan, 1891) | TTAGG + | Okazaki et al. (1993) (as Mecoptera) | SBH |
| Chrysopidae | |||
| Chrysoperla carnea (Stephens, 1836) | TTAGG – | Frydrychová and Marec (2002) | SBH |
| Myrmeleontidae | |||
| Acanthaclisis occitanica (Villers, 1789) | TTAGG + | Kuznetsova et al. (2016) | FISH |
| Distoleon tetragrammicus (Fabricius, 1798) | TTAGG + | Kuznetsova et al. (2016) | FISH |
| Palpares libelluloides (Linnaeus, 1764) | TTAGG + | Kuznetsova et al. (2016) | FISH |
| Coleoptera | |||
| Adephaga | |||
| Carabidae | |||
| Pterostichus oblongopunctatus (Fabricius, 1787) | TTAGG – | Frydrychová and Marec (2002) | FISH, SBH |
| Dytiscidae | |||
| Graphoderus cinereus (Linnaeus, 1758) | TTAGG + | Frydrychová and Marec (2002) | FISH, SBH |
| Gyrinidae | |||
| Orectochilus villosus (Müller, 1776) | TTAGG – | Frydrychová and Marec (2002) | FISH, SBH |
| Polyphaga | |||
| Anobiidae | |||
| Stegobium paniceum (Linnaeus, 1758) | TTAGG + | Frydrychová and Marec (2002) | FISH, SBH |
| Buprestidae | |||
| Agrilus viridis (Linnaeus, 1758) | TTAGG + | Frydrychová and Marec (2002) | FISH, SBH |
| Cerambycidae | |||
| Arhopalus coreanus (Sharp, 1905) | TTAGG + | Okazaki et al. (1993) | SBH |
| Rhagium inquisitor (Linnaeus, 1758) | TTAGG + | Mravinac et al. (2011) | SBH |
| Spondylis buprestoides (Linnaeus, 1758) | TTAGG + | Okazaki et al. (1993) | SBH |
| Chrysomelidae | |||
| Acanthoscelides obtectus (Stay, 1831) | TTAGG + | Mravinac et al. (2011) | SBH |
| Chrysolina americana (Linnaeus, 1758) | TTAGG + | Mravinac et al. (2011) | FISH, SBH |
| Ch. herbacea (Duftschmid, 1825) | TTAGG + | Mravinac et al. (2011) | SBH |
| Ch. polita (Linnaeus, 1758) | TTAGG + | Mravinac et al. (2011) | SBH |
| Leptinotarsa decemlineata (Say, 1824) | TTAGG + | Frydrychová and Marec (2002) | FISH |
| TTAGG + | Mravinac et al. (2011) | SBH | |
| TCAGG – | Mravinac et al. (2011) | SBH | |
| Timarcha balearica Gory, 1829 | TTAGG + | Mravinac et al. (2011) | SBH, FISH |
| TCAGG – | Mravinac et al. (2011) | SBH | |
| TTAGGG – | Mravinac et al. (2011) | SBH | |
| CTGGG – | Mravinac et al. (2011) | SBH | |
| TTGGG – | Mravinac et al. (2011) | SBH | |
| CTAGG – | Mravinac et al. (2011) | SBH | |
| Cleridae | |||
| Thanasimus formicarius (Linnaeus, 1758) | TTAGG – | Frydrychová and Marec (2002) | FISH, SBH |
| Coccinelidae | |||
| Henosepilachna argus (Geoffroy, 1762) | TTAGG + | Mora et al. (2015) | FISH |
| Curculionidae | |||
| Cionus ganglbaueri Wingelmüller, 1914 | TTAGG – | Mravinac et al. (2011) | SBH |
| TCAGG – | Mravinac et al. (2011) | SBH | |
| TTAGGG – | Mravinac et al. (2011) | SBH | |
| CTGGG – | Mravinac et al. (2011) | SBH | |
| TTGGG – | Mravinac et al. (2011) | SBH | |
| CTAGG – | Mravinac et al. (2011) | SBH | |
| C. nigritarsis Reitter, 1904 | TTAGG – | Mravinac et al. (2011) | SBH |
| TCAGG – | Mravinac et al. (2011) | SBH | |
| TTAGGG – | Mravinac et al. (2011) | SBH | |
| CTGGG – | Mravinac et al. (2011) | SBH | |
| TTGGG – | Mravinac et al. (2011) | SBH | |
| CTAGG – | Mravinac et al. (2011) | SBH | |
| Ips typographus Linnaeus, 1758 | TTAGG + | Sahara et al. (1999) | FISH, SBH |
| Sitophilus granarius (Linnaeus, 1758) | TTAGG – | Frydrychová and Marec (2002) | FISH, SBH |
| TTAGG – | Mravinac et al. (2011) | SBH | |
| TCAGG – | Mravinac et al. (2011) | SBH | |
| TTAGGG – | Mravinac et al. (2011) | SBH | |
| CTGGG – | Mravinac et al. (2011) | SBH | |
| TTGGG – | Mravinac et al. (2011) | SBH | |
| CTAGG – | Mravinac et al. (2011) | SBH | |
| Elateridae | |||
| Ampedus sanguineus (Linnaeus, 1758) | TTAGG + | Frydrychová and Marec (2002) | FISH, SBH |
| Diacanthous undosus (Lewis, 1894) | TTAGG + | Okazaki et al. (1993) | SBH |
| Melanotus legatus Candèze, 1860 | TTAGG + | Okazaki et al. (1993) | SBH |
| Geotrupidae | |||
| Geotrupes stercorarius (Linnaeus, 1758) | TTAGG – | Frydrychová and Marec (2002) | FISH, SBH |
| Hydrophilidae | |||
| Hydrochara affinis (Sharp, 1873) | TTAGG + | Okazaki et al. (1993) | SBH |
| Laemophloeidae | |||
| Cryptolestes sp. | TTAGG – | Mravinac et al. (2011) | SBH |
| TCAGG – | Mravinac et al. (2011) | SBH | |
| TTAGGG – | Mravinac et al. (2011) | FISH | |
| CTGGG – | Mravinac et al. (2011) | FISH | |
| TTGGG – | Mravinac et al. (2011) | FISH | |
| CTAGG – | Mravinac et al. (2011) | FISH | |
| Mycetophagidae | |||
| Typhaea stercorea (Linnaeus, 1758) | TTAGG – | Mravinac et al. (2011) | SBH |
| TCAGG + | Mravinac et al. (2011) | SBH | |
| Meloidae | |||
| Mylabris sp. | TTAGG – | Mravinac et al. (2011) | SBH |
| TCAGG + | Mravinac et al. (2011) | SBH | |
| Scarabeidae | |||
| Anomala cuprea (Hope, 1839) | TCAGG – | Okazaki et al. (1993) | SBH |
| Silvanidae | |||
| Oryzaephilus surinamensis (Linnaeus, 1758) | TTAGG + | Frydrychová and Marec (2002) | FISH, SBH |
| Silphidae | |||
| Silpha obscura Linnaeus, 1758 | TTAGG + | Frydrychová and Marec (2002) | FISH, SBH |
| Tenebrionidae | |||
| Gonocephalum sexuale (Marseul, 1876) | TTAGG – | Okazaki et al. (1993) | SBH |
| Palorus ficicola (Wollaston, 1867) | TTAGG – | Mravinac et al. (2011) | SBH |
| TCAGG + | Mravinac et al. (2011) | SBH | |
| P. genalis Blair, 1930 | TTAGG – | Mravinac et al. (2011) | SBH |
| TCAGG + | Mravinac et al. (2011) | SBH | |
| P. ratzeburgii (Wissmann, 1848) | TTAGG – | Mravinac et al. (2011) | SBH |
| TCAGG + | Mravinac et al. (2011) | SBH | |
| P. subdepressus (Wollaston, 1864) | TTAGG – | Mravinac et al. (2011) | SBH |
| TCAGG + | Mravinac et al. (2011) | SBH | |
| Pimelia cribra Solier, 1836 | TTAGG – | Mravinac et al. (2011) | SBH |
| TCAGG + | Mravinac et al. (2011) | SBH | |
| P. elevata Sénac, 1887 | TTAGG – | Mravinac et al. (2011) | SBH |
| TCAGG + | Mravinac et al. (2011) | SBH | |
| P. monticola Rosenhauer, 1856 | TTAGG – | Mravinac et al. (2011) | SBH |
| TCAGG + | Mravinac et al. (2011) | SBH | |
| Tenebrio molitor Linnaeus, 1758 | TTAGG – | Sahara et al. (1999) | FISH |
| TTAGG – | Mravinac et al. (2011) | SBH | |
| TCAGG + | Mravinac et al. (2011) | FISH, SBH | |
| T. obscurus Fabricius, 1792 | TTAGG – | Mravinac et al. (2011) | SBH |
| TCAGG + | Mravinac et al. (2011) | SBH | |
| Tribolium anaphe Hinton, 1948 | TTAGG – | Mravinac et al. (2011) | SBH |
| TCAGG + | Mravinac et al. (2011) | SBH | |
| T. audax Halstead, 1969 | TTAGG – | Mravinac et al. (2011) | SBH |
| TCAGG + | Mravinac et al. (2011) | SBH | |
| T. brevicornis (LeConte, 1859) | TTAGG – | Mravinac et al. (2011) | SBH |
| TCAGG + | Mravinac et al. (2011) | SBH | |
| T. castaneum (Herbst, 1797) | TTAGG – | Osanai et al. (2006) | SBH |
| TCAGG + | Osanai et al. (2006) | SBH | |
| TCAGG +/TSRT | Richards et al. (2008) | NGS | |
| TTAGG – | Mravinac et al. (2011) | SBH | |
| TCAGG + | Mravinac et al. (2011) | FISH, SBH | |
| T. confusum Jacquelin du Val, 1868 | TTAGG – | Mravinac et al. (2011) | SBH |
| TCAGG + | Mravinac et al. (2011) | SBH | |
| T. destructor Uyttenboogard, 1933 | TTAGG – | Mravinac et al. (2011) | SBH |
| TCAGG + | Mravinac et al. (2011) | SBH | |
| T. freemani Hinton, 1948 | TTAGG – | Mravinac et al. (2011) | SBH |
| TCAGG + | Mravinac et al. (2011) | SBH | |
| T. madens (Charpentier, 1825) | TTAGG – | Mravinac et al. (2011) | SBH |
| TCAGG + | Mravinac et al. (2011) | SBH | |
| Hymenoptera | |||
| Aculeata | |||
| Ampulicidae | |||
| Ampulex sp. | TTAGG – | Menezes et al. (2017) | DBH |
| TTAGGG – | Menezes et al. (2017) | DBH | |
| Apidae | |||
| Apis mellifera Linnaeus, 1758 | TTAGG + | Meyne et al. (1995) | FISH, SBH |
| TTAGG + | Robertson and Gordon (2006) | NGS, Seq+GA | |
| Berhylidae | |||
| Epyris sp. | TTAGG – | Menezes et al. (2017) | DBH |
| TTAGGG – | Menezes et al. (2017) | DBH | |
| Chrysididae | |||
| Caenochrysis paraca (Linsenmaier, 1984) | TTAGG – | Menezes et al. (2017) | DBH |
| TTAGGG – | Menezes et al. (2017) | DBH | |
| Ipsiura tropicalis Bohart, 1985 | TTAGG – | Menezes et al. (2017) | DBH |
| TTAGGG – | Menezes et al. (2017) | DBH | |
| Crabronidae | |||
| Bicyrtes cingulata Burmeister, 1874 | TTAGG – | Menezes et al. (2017) | DBH |
| TTAGGG – | Menezes et al. (2017) | DBH | |
| Formicidae | |||
| Acromyrmex striatus (Roger, 1863) | TTAGG + | Pereira et al. (2018) | FISH |
| Aphaenogaster gibbosa (Latreille, 1798) | TTAGG + | Lorite et al. (2002) | SBH |
| TTAGGG – | Lorite et al. (2002) | SBH | |
| A. senilis Mayr, 1853 | TTAGG + | Lorite et al. (2002) | SBH |
| TTAGGG – | Lorite et al. (2002) | SBH | |
| Bothriomyrmex corsicus Santschi, 1923 | TTAGG + | Lorite et al. (2002) (as B. gibbus) | SBH |
| TTAGGG – | Lorite et al. (2002) | SBH | |
| Camponotus micans (Nylander, 1856) | TTAGG + | Lorite et al. (2002) | SBH |
| TTAGGG – | Lorite et al. (2002) | SBH | |
| C. pilicornis (Roger, 1859) | TTAGG + | Lorite et al. (2002) | SBH |
| TTAGGG – | Lorite et al. (2002) | SBH | |
| Cataglyphis velox Santschi, 1929 | TTAGG + | Lorite et al. (2002) | SBH |
| TTAGGG – | Lorite et al. (2002) | SBH | |
| Crematogaster auberti Emery, 1869 | TTAGG + | Lorite et al. (2002) | SBH |
| TTAGGG – | Lorite et al. (2002) | SBH | |
| C. laestrygon Emery, 1869 | TTAGG + | Lorite et al. (2002) | SBH |
| TTAGGG – | Lorite et al. (2002) | SBH | |
| Formica cunicularia Latreille, 1798 | TTAGG + | Lorite et al. (2002) | SBH |
| TTAGGG – | Lorite et al. (2002) | SBH | |
| Iberoformica subrufa (Roger, 1859) | TTAGG + | Lorite et al. (2002) (as Formica) | SBH |
| TTAGGG – | Lorite et al. (2002) | SBH | |
| Lasius niger (Linnaeus, 1758) | TTAGG + | Lorite et al. (2002) | SBH |
| TTAGGG – | Lorite et al. (2002) | SBH | |
| Linepithema humile Mayr, 1868 | TTAGG + | Lorite et al. (2002) | SBH |
| TTAGGG – | Lorite et al. (2002) | SBH | |
| Manica yessensis Azuma, 1955 | TTAGG + | Okazaki et al. (1993) | SBH |
| Messor bouvieri Bondroit, 1918 | TTAGG + | Lorite et al. (2002) | SBH |
| TTAGGG – | Lorite et al. (2002) | SBH | |
| M. structor (Latreille, 1798) | TTAGG + | Lorite et al. (2002) | SBH |
| TTAGGG – | Lorite et al. (2002) | SBH | |
| Monomorium subopacum (Smith, 1858) | TTAGG + | Lorite et al. (2002) | SBH |
| TTAGGG – | Lorite et al. (2002) | SBH | |
| Myrmecia arnoldi Clarck, 1951 | TTAGG + | Meyne et al. (1995) | FISH |
| TTAGGG + | Meyne et al. (1995) | FISH | |
| M. chasei Forel, 1894 | TTAGG + | Meyne et al. (1995) | FISH |
| TTAGGG + | Meyne et al. (1995) | FISH | |
| M. croslandi Taylor, 1991 | TTAGG + | Meyne et al. (1995) | FISH |
| TTAGGG + | Meyne et al. (1995) | FISH | |
| M. forficata (Fabricius, 1787) | TTAGG + | Meyne et al. (1995) | FISH |
| TTAGGG + | Meyne et al. (1995) | FISH | |
| M. fulvipes Roger, 1861 | TTAGG + | Meyne et al. (1995) | FISH |
| TTAGGG + | Meyne et al. (1995) | FISH | |
| M. gulosa (Fabricius, 1775) | TTAGG + | Meyne et al. (1995) | FISH |
| TTAGGG + | Meyne et al. (1995) | FISH | |
| M. haskinsorum Taylor, 2015 | TTAGG + | Meyne et al. (1995) | FISH |
| TTAGGG + | Meyne et al. (1995) | FISH | |
| M. pilosula Smith, 1858 | TTAGG + | Meyne et al. (1995) | FISH |
| TTAGGG+ | Meyne et al. (1995) | FISH | |
| Pheidole pallidula (Nylander, 1849) | TTAGG + | Lorite et al. (2002) | SBH |
| TTAGGG – | Lorite et al. (2002) | SBH | |
| Solenopsis invicta Buren, 1972 | TTAGG + | Wurm et al. (2011) | FISH |
| Tapinoma nigerrimum Nylander, 1856 | TTAGG + | Lorite et al. (2002) | FISH, SBH |
| TTAGGG – | Lorite et al. (2002) | FISH, SBH | |
| Tetramorium forte Forel, 1904 | TTAGG + | Lorite et al. (2002) (as T. hispanicum) | SBH |
| TTAGGG – | Lorite et al. (2002) | SBH | |
| T. semilaeve André, 1883 | TTAGG + | Lorite et al. (2002) | SBH |
| TTAGGG– | Lorite et al. (2002) | SBH | |
| Pompilidae | |||
| Pepsis sp. | TTAGG – | Menezes et al. (2017) | DBH |
| TTAGGG – | Menezes et al. (2017) | DBH | |
| Scoliidae | |||
| Campsomeris sp. 1 | TTAGG – | Menezes et al. (2017) | DBH |
| TTAGGG – | Menezes et al. (2017) | DBH | |
| Campsomeris sp. 2 | TTAGG – | Menezes et al. (2017) | DBH |
| TTAGGG – | Menezes et al. (2017) | DBH | |
| Priochilus sp. | TTAGG – | Menezes et al. (2017) | DBH |
| TTAGGG – | Menezes et al. (2017) | DBH | |
| Sphecidae | |||
| Sceliphron sp. | TTAGG – | Menezes et al. (2017) | DBH |
| TTAGGG – | Menezes et al. (2017) | DBH | |
| Vespidae | |||
| Angiopolybia pallens (Lepeletier, 1836) | TTAGG – | Menezes et al. (2017) | DBH |
| TTAGGG – | Menezes et al. (2017) | DBH | |
| Apoica sp. | TTAGG – | Menezes et al. (2017) | DBH |
| TTAGGG – | Menezes et al. (2017) | DBH | |
| Chartergus sp. | TTAGG – | Menezes et al. (2017) | DBH |
| TTAGGG – | Menezes et al. (2017) | DBH | |
| Epipona media Cooper (2002) | TTAGG – | Menezes et al. (2017) | DBH |
| TTAGGG – | Menezes et al. (2017) | DBH | |
| Metapolybia decorata Gribodo, 1896 | TTAGG – | Menezes et al. (2013) | FISH |
| TTAGGG – | Menezes et al. (2017) | DBH | |
| Mischocyttarus cerberus Ducke, 1918 | TTAGG – | Menezes et al. (2017) | DBH |
| TTAGGG – | Menezes et al. (2017) | DBH | |
| M. consimilis Zikan, 1949 | TTAGG – | Menezes et al. (2017) | DBH |
| TTAGGG – | Menezes et al. (2017) | DBH | |
| M. drewseni Saussure, 1954 | TTAGG – | Menezes et al. (2017) | DBH |
| TTAGGG – | Menezes et al. (2017) | DBH | |
| Pachymenes ater (Saussure, 1852) | TTAGG – | Menezes et al. (2017) | DBH |
| TTAGGG – | Menezes et al. (2017) | DBH | |
| Parachartergus pseudapicalis Willink, 1859 | TTAGG – | Menezes et al. (2017) | DBH |
| TTAGGG – | Menezes et al. (2017) | DBH | |
| Paragia vespiformes Smith, 1865 | TTAGG – | Menezes et al. (2017) | DBH |
| TTAGGG – | Menezes et al. (2017) | DBH | |
| Polistes versicolor (Olivier, 1792) | TTAGG – | Menezes et al. (2017) | DBH |
| TTAGGG – | Menezes et al. (2017) | DBH | |
| Polybia ruficeps Schrottky, 1902 | TTAGG – | Menezes et al. (2017) | DBH |
| TTAGGG – | Menezes et al. (2017) | DBH | |
| Synoeca ilheensis Lopes & Menezes, 2017 | TTAGG – | Menezes et al. (2017) | DBH |
| TTAGGG – | Menezes et al. (2017) | DBH | |
| Zethus cylindricus Fox, 1899 | TTAGG – | Menezes et al. (2017) | DBH |
| TTAGGG – | Menezes et al. (2017) | DBH | |
| Parasitica | |||
| Pteromalidae | |||
| Nasonia giraulti Darling, 1990 | TTAGG – | NGWG (2010) | NGS |
| N. longicornis Darling, 1990 | TTAGG – | NGWG (2010) | NGS |
| N. vitripennis (Walker, 1836) | TTAGG – | NGWG (2010) | NGS |
| Eurytomidae | |||
| Eurytoma compressa (Fabricius, 1794) | TTAGG – | Gokhman et al. (2014) | FISH |
| Eu. robusta Mayr, 1878 | TTAGG – | Gokhman et al. (2014) | FISH |
| Eu. serratulae (Fabricius, 1798) | TTAGG – | Gokhman et al. (2014) | FISH |
| Torymidae | |||
| Torymus bedeguaris (Linnaeus, 1758) | TTAGG – | Gokhman et al. (2014) | FISH |
| Ichneumonidae | |||
| Ichneumon amphibolus Kriechbaumer, 1888 | TTAGG – | Gokhman et al. (2014) | FISH |
| Cynipidae | |||
| Diplolepis rosae (Linnaeus, 1758) | TTAGG – | Gokhman et al. (2014) | FISH |
| Symphyta | |||
| Tenthredinidae | |||
| Taxonus agrorum (Fallén, 1808) | TTAGG + | Gokhman and Kuznetsova (2018) | FISH |
| Tentheredo omissa (Förster, 1844) | TTAGG + | Gokhman and Kuznetsova (2018) | FISH |
| Trichoptera | |||
| Limnephilidae | |||
| Limnephilus decipiens (Kolenati, 1848) | TTAGG + | Frydrychová et al. (2004) | FISH, SBH |
| Stenopsychidae | |||
| Stenopsyche marmorata Navas, 1920 | TTAGG + | Okazaki et al. (1993) (as S. japonica) | SBH |
| Lepidoptera | |||
| Bombycidae | |||
| Bombyx mandarina Moore, 1872 | TTAGG + | Okazaki et al. (1993) | SBH |
| B. mori Linnaeus, 1758 | TTAGG + | Okazaki et al. (1993) | FISH, SBH |
| TTAGG + | Okazaki et al. (1995) | FISH, SBH | |
| TTAGG+/TSRT | Sahara et al. (1999) | FISH, SBH | |
| Lycaenidae | |||
| Polyommatus caeruleus (Staudinger, 1871) | TTAGG + | Vershinina et al. (2015) | FISH |
| P. hamadanensis (de Lesse, 1959) | TTAGG + | Vershinina et al. (2015) | FISH |
| P. karindus (Riley, 1921) | TTAGG + | Vershinina et al. (2015) | FISH |
| P. morgani (Le Cerf, 1909) | TTAGG + | Vershinina et al. (2015) | FISH |
| P. peilei (Bethune-Baker, 1921) | TTAGG + | Vershinina et al. (2015) | FISH |
| P. pfeifferi (Brandt, 1938) | TTAGG + | Vershinina et al. (2015) | FISH |
| P. sennanensis (de Lesse, 1959) | TTAGG + | Vershinina et al. (2015) | FISH |
| Erebidae | |||
| Orgyia antiqua (Linnaeus, 1758) | TTAGG + | Rego and Marec (2003) | FISH |
| TTAGG + | Yoshido et al. (2005) | FISH | |
| O. thyellina Butler, 1881 | TTAGG + | Yoshido et al. (2005) | FISH |
| Samia cynthia ricini (Anderson, 1788) | TTAGG + | Okazaki et al. (1993) | SBH |
| TTAGG + | Yoshido et al. (2005) | FISH | |
| S. cynthia walkeri (Felder & Felder, 1862) | TTAGG + | Yoshido et al. (2005) | FISH |
| S. cynthia subsp. indet. | TTAGG + | Yoshido et al. (2005) | FISH |
| Noctuidae | |||
| Mamestra brassicae (Linnaeus, 1758) | TTAGG + | Mandrioli (2002) | FISH, SBH |
| Spodoptera exigua (Hübner, 1808) | TTAGG + | Gong et al. (2015) | TRAP-Seq |
| Nymphalidae | |||
| Danaus plexippus (Linnaeus, 1758) | TTAGG + | Traut et al. (2007) | FISH |
| Melitaea cinxia (Linnaeus, 1758) | TTAGG + | Traut et al. (2007) | FISH |
| Pyralidae | |||
| Ephestia kuehniella Zeller, 1879 | TTAGG + | Sahara et al. (1999) | FISH, SBH |
| TTAGG + | Rego and Marec (2003) | FISH | |
| Galleria mellonella (Linnaeus, 1758) | TTAGG + | Sahara et al. (1999) | FISH, SBH |
| Saturniidae | |||
| Antheraea pernyi (Guérin-Méneville, 1855) | TTAGG + | Okazaki et al. (1993) | SBH |
| TTAGG + | Sasaki and Fujiwara (2000) | FISH | |
| A. yamamai (Guérin-Méneville, 1861) | TTAGG + | Okazaki et al. (1993) | SBH |
| TTAGG + | Sasaki and Fujiwara (2000) | FISH | |
| Mecoptera | |||
| Panorpidae | |||
| Panorpa communis Linnaeus, 1758 | TTAGG – | Frydrychová et al. (2004) | FISH, SBH |
| Siphonaptera | |||
| Pulicidae | |||
| Ctenocephalides canis (Curtis, 1826) | TTAGG – | Frydrychová et al. (2004) | SBH |
| Diptera | |||
| Nematocera | |||
| Chironomidae | |||
| Chironomus dilutus Shobanov, Kiknadze & Butler, 1999 | LCTTR | Rosén and Edström (2000) | FISH, DNA-Seq |
| Ch. pallidivittatus Malloch, 1915 | LCTTR | Saiga and Edström (1985) | FISH, SBH, DNA-Seq |
| LCTTR | Zhang et al. (1994) | FISH, SBH, PFGE | |
| Ch. riparius (Meigen, 1804) | LCTTR | Martínez-Guitarte et al. (2014) | RT-PCR |
| LCTTR | Carmona et al. (1985) (as Ch. thummi) | FISH, NBH, DNA-Seq | |
| Ch. tentans Fabricius, 1805 | LCTTR | Nielsen and Edström (1993) | DAN-Seq |
| Rosén and Edström (2002) | PFGE, IGH | ||
| LCTTR | Rosén et al. (2002) | NBH, IPC | |
| LCTTR | |||
| Culicidae | |||
| Anopheles gambiae Giles, 1902 | LCTTR | Roth et al. (1997) | SBH, DNA-Seq, FIGE |
| LCTTR | Biessmann et al. (1998) | SBH, DAN-Seq | |
| LCTTR | Walter et al. (2001) | SBH | |
| Sciaridae | |||
| Rhynchosciara americana (Wiedemann, 1821) | TSRT | Madalena, Amabis, et al. (2010) | stSTR, DNA-Seq, FISH, SBH |
| Madalena, Fernandes, et al. (2010) | FISH, SBH, DNA-Seq | ||
| stSTR | Fernandes et al. (2012) | FISH, SBH, DAN-Seq | |
| Brachycera | |||
| Asilidae | |||
| Neoitamus angusticornis (Loew, 1858) | TTAGG – | Okazaki et al. (1993) | SBH |
| Drosophilidae | |||
| Drosophila americana Spencer, 1938 | TSRT | Biessmann et al. (2000) | FISH, SBH, FIGE, DNA-Seq |
| TSRT | Casacuberta and Pardue (2003) | FISH, NBH, SBH, DNA-Seq | |
| D ezoana Takada & Okada, 1958 | TSRT | Biessmann et al. (2000) | FISH, SBH, FIGE |
| D. littoralis Meigen, 1830 | TSRT | Biessmann et al. (2000) | FISH, SBH, FIGE |
| D. lummei Hackman, 1972 | TSRT | Biessmann et al. (2000) | FISH, SBH, FIGE, DNA-Seq |
| D. mauritiana Tsacas & David, 1974 | TSRT | Danilevskaya et al. (1998) | FISH, SBH, DNA-Seq |
| D. melanogaster (Meigen, 1830) | TTAGG – | Okazaki et al. (1993) | SBH |
| TSRT | Levis et al. (1993) | SBH, FISH, DNA-Seq | |
| TTAGG – | Sahara et al. (1999) | FISH, SBH | |
| TSRT | Danilevskaya et al. (1998) | FISH, SBH, DNA-Seq | |
| D. montana Stone, Griffen & Patterson, 1941 | TSRT | Biessmann et al. (2000) | FISH, SBH, FIGE,DNA-Seq |
| D. novamexicana Patterson, 1941 | TSRT | Biessmann et al. (2000) | FISH, SBH, FIGE, DNA-Seq |
| D. simulans Sturtevant, 1919 | TSRT | Danilevskaya et al. (1998) | FISH, SBH, DNA-Seq |
| D. teissieri Tsacas, 1971 | TSRT | Danilevskaya et al. (1998) | FISH, SBH, DNA-Seq |
| D. virilis Sturtevant, 1916 | TSRT | Biessmann et al. (2000) | FISH, SBH, FIGE, DNA-Seq |
| TSRT | Casacuberta and Pardue (2003) | FISH, NBH, SBH, DNA-Seq | |
| D. yakuba Burla, 1954 | TSRT | Danilevskaya et al. (1998) | FISH, SBH, DNA-Seq |
| TSRT+ | Casacuberta and Pardue (2003) | FISH, NBH, SBH, DNA-Seq | |
| Phoridae | |||
| Megaselia scalaris (Loew, 1866) | TTAGG – | Sahara et al. (1999) | FISH, SBH |
| Syrphidae | |||
| Eristalomya tenax (Linnaeus, 1758) | TTAGG – | Okazaki et al. (1993) | SBH |
| Monoceromyia pleuralis (Coquillett, 1898) | TTAGG – | Okazaki et al. (1993) (as Sphyximorphoides) | SBH |
| Tabanidae | |||
| Tabanus trigonus Coquillett, 1898 | TTAGG – | Okazaki et al. (1993) | SBH |
Abbreviations: DBH, Dot Blot Hybridization; DNA Seq, DNA sequencing; FIGE, Field-Inversion Gel Electrophoresis; FISH, Fluorescence In Situ Hybridization; IAGC, International Aphid Genomics Consortium; IGH, In-Gel Hybridization; IPC, Isopycnotic Centrofugation; LCTTR, Long Complex Terminal Tandem Repeats; MA, Microsatellite Analysis; NGS, New Generation Sequencing; NGWG, Nasonia Genome Working Group; PFGE, Pulsed-Field Gel Electrophoresis; RT-PCR, Reverse Transcription PCR; SBH, Southern Blot Hybridization; Seq+GA, Sequencing + Genome Assemblage; stSTR, special type of Short Tandem Repeats; TRAP-Seq, Sequencing of TRAP products; TSRT, Telomere Specific Retrotransposons are inserted into the simple telomeric repeats.




