Hagfish phylogeny and taxonomy, with description of the new genus Rubicundus (Craniata, Myxinidae)
Abstract
A recent phylogenetic analysis of the Myxinidae based on the 16S rRNA gene resulted in synonymization of Paramyxine with Eptatretus. This created homonymy of Paramyxine fernholmi with Eptatretus fernholmi and Paramyxine wisneri with Eptatretus wisneri. In order to resolve this nomenclatural dilemma, we made a more extensive phylogenetic assessment of the Myxinidae and examined the nomenclature of the family. We used 75 sequences (37 of which new for this study) of a 561 bp fragment of the 16S rRNA gene, representing 33 species, and 72 sequences (37 of which new for this study) of a 687 bp fragment of the cytochrome c oxidase subunit I (COI) gene, representing 23 species, to reconstruct the phylogeny of Myxinidae. The monophyly of the subfamily Myxininae, traditionally characterized by having a single pair of external gill openings, was rejected (0.50 Bayesian posterior probability) by the 16S analysis, but supported by the COI and combined COI+16S analyses (0.99 and 0.81 Bpp, respectively). The monophyly of the subfamily Eptatretinae, characterized by having several pairs of external gill openings, was not supported by the 16S analysis and rejected by the COI and combined COI+16S analysis due to the placement of Eptatretus lopheliae as the earliest branch of Myxinidae (0.71 and 0.57 Bpp, respectively). Eptatretus lopheliae and Eptatretus rubicundus formed a monophyletic group and were allocated to a new genus, Rubicundus, characterized by the presence of an elongated tubular nostril and reddish coloration. A new monotypic subfamily, Rubicundinae, was proposed for Rubicundus. The synonymy of the genera Paramyxine and Quadratus with Eptatretus was confirmed. E. fernholmi is renamed Eptatretus luzonicus. Eptatretus wisneri was renamed Eptatretus bobwisneri. Petromyzon cirrhatus Forster, 1801, Homea banksii Fleming, 1822, and Bdellostoma forsteri Müller, 1836 are synonyms, but no type specimens are known to exist. Petromyzon cirrhatus was designated as type species of Eptatretus, conserving present usage. Gastrobranchus dombeyi Shaw, 1804 has priority over other names for Chilean myxinids. Bdellostoma stoutii was designated as type species of Polistotrema Gill. The validity of the Western Atlantic Myxine limosa as distinct from the Eastern Atlantic Myxine glutinosa was confirmed.
Introduction
The Myxinidae are a family of basal marine craniates distributed in all oceans except the Arctic and Southern Ocean and the Red Sea (Froese and Pauly 2012). 78 species are currently recognized as valid (Froese and Pauly 2012). Myxinids are eel-like benthic scavengers and predators on small invertebrates and fish (Zintzen et al. 2011) with reduced eyes, no paired fins, a terminal complex jaw apparatus and slime pores along the lower side of their body. The taxonomy of the Myxinidae has been based mainly on variation in the teeth and gill structures, as well as a limited set of meristic, morphological and colour characteristics, with the latest review of the family by Fernholm (1998). Two subfamilies are recognized. The Myxininae Linnæus (1758) are characterized by having one pair of external gill openings while the Eptatretinae Bonaparte, 1850, have several (5–16) pairs of external gill openings (Nelson 2006).
Up to seven genera are currently recognized (Nelson 2006), but a number of them have been disputed (Strahan 1975; Fernholm 1998; Møller and Jones 2007; Kuo et al. 2010). The Myxininae include Myxine Linnæus, 1758, with 23 species, Nemamyxine Richardson, 1958, with two species, and the monotypic genera Notomyxine Nani & Gneri, 1951, and Neomyxine Richardson, 1953. A second, undescribed species of Neomyxine was reported by Zintzen et al. (2011). The Eptatretinae, with 51 species, include the genera Eptatretus Cloquet, 1819; Paramyxine Dean, 1904 and Quadratus Wisner, 1999.
Until recently, myxinids have been caught mostly over soft bottoms. However, recent opportunities to collect and observe in the deep-sea using submersibles, deep potting and submerged cameras, have enabled sampling and observations in a more diverse range of habitats. Eptatretus lakeside Mincarone and McCosker (2004) from the Galápagos Islands was collected from a submersible in 711 m depth. Eptatretus goliath Mincarone and Stewart (2006) from northern New Zealand, the largest (1275 mm long) and heaviest (6.2 kg) hagfish, was collected from 811 m depth using baited traps. Eptatretus strickrotti Møller and Jones (2007) was collected with a submersible from a hydrothermal vent habitat at 2211 m depth. An undescribed species of Eptatretus was observed on video and collected from a deep coral sponge garden at 300–700 m depth off northern New Zealand (Zintzen et al. 2011). Eptatretus lopheliae Fernholm and Quattrini (2008), collected with submersible from cold-water coral habitat in 430–442 m depth, and Eptatretus rubicundus Kuo et al. (2010), trawled in 800 m depth north-east of Taiwan, have been found in remarkable new biotopes for hagfishes and also show some interesting and novel morphologies.
The taxonomy of the Myxinidae currently suffers from several unresolved issues. The validity of the genera Paramyxine and Quadratus is uncertain. Synonymization of Paramyxine with Eptatretus was suggested already by Strahan (1975) and Fernholm (1998). Møller and Jones (2007), in an analysis of a 16S rDNA nucleotide data set from 17 species of the Myxinidae, provided molecular support for the synonymization of Paramyxine and Quadratus with Eptatretus. As a consequence of synonymizing Paramyxine with Eptatretus resulting junior homonyms require new names. Eptatretus wisneri McMillan (1999) is predated by Paramyxine wisneri Kuo et al. (1994), and Eptatretus fernholmi McMillan and Wisner (2004) by Paramyxine fernholmi Kuo et al. (1994). In addition, the taxonomy and nomenclature of Eptatretus is complicated by the dispute around the type species of Eptatretus (Gill 1901; Eschmeyer 2012), and also by the synonymization of Quadratus and Paramyxine by Møller and Jones (2007) which remains tentative because the type species of these genera, as well as the putative type species of Eptatretus, were not included in their analysis.
The present study aims first to provide an improved phylogenetic hypothesis and classification of the Myxinidae, then solve the resulting homonymy problems, and also resolve nomenclatural problems associated with Eptatretus.
Material and Methods
Selection of gene fragment for molecular phylogenetic analysis
This study uses mitochondrial 16S rDNA (16S, also known as the small subunit, ssu) and cytochrome oxidase subunit I (COI, also known as mt-CO1 and COX1) DNA fragments because these are the only genes with a significant number of published sequences, permitting a better sampling of myxinid diversity, and with variation and rate of evolution suitable for study of myxinid interrelationships. The 33 published 16S sequences mostly come from earlier phylogenetic studies (Kuo et al. 2003, 2010; Chen et al. 2005; Møller and Jones 2007), whereas the 30 published COI sequences are mostly products of the Barcode of Life consortium. Preliminary analysis showed that four published COI sequences (JF493943 and JF493945–493947) were obtained from misidentified specimens, and GenBank was notified.
DNA extraction and PCR
Mitochondrial DNA was extracted using a GeneMole (Mole Genetics) fully automated liquid-handling instrument, with the MoleStrips (Mole Genetics) kit and recommended protocol. The 16S fragment was amplified using the primers 16SARL and 16SBRH (Palumbi et al. 1991), the COI fragment was amplified using the standard barcoding primers Fish-F1 and Fish-R1 (Ward et al. 2005). PCRs (PCR cycling: 94°C 4′; 35* (94°C 30″; 52°C 30″; 72°C 30″); 72°C 8′) were performed with the puReTaq Ready-To-Go PCR kit (Amersham Biosciences AB, Uppsala, Sweden). PCR products were checked on minigel and purified using the FastAP Thermosensitive Alkaline Phosphatase (Thermo Fischer Scientific, Göteborg, Sweden) purification kit.
Sequencing
Sequencing of both strands of all fragments was carried out by Macrogen Europe (Amstelveen, Holland). All sequences were proofread and assembled using the software Geneious v. 6.1.2 (Drummond et al. 2012).
Voucher specimens
Voucher specimens were deposited at the Swedish Museum of Natural History, NRM (Stockholm, Sweden), Scripps Institute of Oceanography, SIO (San Diego, USA), Icelandic Museum of Natural History, NMSI (Reykjavik, Iceland) and Museum of New Zealand Te Papa Tongarewa, NMNZ (Wellington, New Zealand). Data on voucher specimens are available from the Global Biodiversity Information Facility (GBIF) portal (data.gbif.org).
Analysis
Sequences used in this study are summarized in (Table 1). All available myxinid 16S sequences from GenBank corresponding to the studied fragment and longer than 530 base pairs (bp) were included. All myxiniform COI sequences available from GenBank on the 10th of March 2013 corresponding to the studied fragment and longer than 610 bp have been included, except JF493943 which was excluded after analysis showed that the sequence is not of a myxiniform, but identical to published COI sequences of the congrid eel Bassanago bulbiceps. The sequences were aligned using the MUSCLE (Edgar 2004) plug-in for Geneious. Alignment of the 16S sequences was problematic, and alternative alignments were constructed using Geneious Alignment and the MAFFT (Katoh et al. 2002, 2005) plug-in for Geneious in order to test sensitivity to alignment.
| Species | GenBank# 16S | GenBank# COI | Voucher | Origin |
|---|---|---|---|---|
| Eptatretus burgeri | – | AJ278504 | – | Japan |
| Eptatretus burgeri | AF364616 | – | – | Taiwan |
| Eptatretus burgeri | AF364617 | – | – | Taiwan |
| Eptatretus burgeri | AY619579 | – | – | ‘China’ (Taiwan) |
| Eptatretus burgeri | JX442457 | KC807331 | NRM 50265-T3326 | Japan |
| Eptatretus burgeri | JX442458 | KC807331 | NRM 50265-T3325 | Japan |
| Eptatretus burgeri | JX442459 | KC807321 | NRM 50265-T3327 | Japan |
| Eptatretus burgeri | JX442460 | KC807323 | NRM 50265-T3329 | Japan |
| Eptatretus burgeri | JX442461 | KC807322 | NRM 50265-T3328 | Japan |
| Eptatretus burgeri | NC_002807 | NC_002807 | – | Japan |
| Eptatretus cf fernholmi | JX442463 | KC807333 | – | Taiwan |
| Eptatretus cheni | AF364620 | – | – | Taiwan |
| Eptatretus cheni | AF364621 | – | – | Taiwan |
| Eptatretus chinensis | AY619580 | – | – | ‘China’ (Taiwan) |
| Eptatretus cirrhatus | AF364619 | – | – | New Zealand |
| Eptatretus cirrhatus | JX442450 | KC807340 | NMNZ P.037107 | New Zealand |
| Eptatretus cirrhatus | JX442451 | KC807345 | NMNZ P.049408 | New Zealand |
| Eptatretus cirrhatus | JX442452 | KC807341 | NMNZ P.044178 | New Zealand |
| Eptatretus cirrhatus | JX442453 | – | NMNZ P.037133 | New Zealand |
| Eptatretus deani | – | FJ164594 | TZ05-FROSTI-325 | Canada, British Columbia |
| Eptatretus deani | – | FJ164595 | TZ05-FROSTI-330 | Canada, British Columbia |
| Eptatretus deani | – | FJ164596 | TZ05-FROSTI-331 | Canada, British Columbia |
| Eptatretus deani | – | FJ164597 | TZ05-FROSTI-302 | Canada, British Columbia |
| Eptatretus deani | – | FJ164598 | TZ05-FROSTI-215 | Canada, British Columbia |
| Eptatretus deani | – | GU440316 | MFC317 | USA |
| Eptatretus deani | EF014477 | – | KU 28249 | USA, California |
| Eptatretus deani | EU099514 | – | SIO 05-95 | USA, California |
| Eptatretus longipinnis | EF014476 | – | SAMA F07540 | Australia, South |
| Eptatretus minor | JX442454 | KC807329 | – | USA, Gulf of Mexico |
| Eptatretus minor | JX442455 | KC807331 | – | USA, Gulf of Mexico |
| Eptatretus minor | JX442456 | KC807331 | – | USA, Gulf of Mexico |
| Eptatretus nelsoni | AF364607 | – | – | Taiwan? |
| Eptatretus nelsoni | AF364608 | – | – | Taiwan |
| Eptatretus nelsoni | AF364609 | – | – | Taiwan? |
| Eptatretus sheni | AF364610 | – | – | Taiwan |
| Eptatretus sp. ‘Korea’ | JX442462 | KC807324 | NRM 50590 | South Korea |
| Eptatretus stoutii | – | FJ164599 | NEOCAL07-0004 | Canada, British Columbia |
| Eptatretus stoutii | – | FJ164600 | NEOCAL07-0003 | Canada, British Columbia |
| Eptatretus stoutii | – | FJ164601 | NEOCAL07-0002 | Canada, British Columbia |
| Eptatretus stoutii | – | FJ164602 | NEOCAL07-0001 | Canada, British Columbia |
| Eptatretus stoutii | – | FJ164603 | TZ-06-RICKER-573 | Canada, British Columbia |
| Eptatretus stoutii | – | FJ164604 | TZ-06-RICKER-574 | Canada, British Columbia |
| Eptatretus stoutii | – | FJ164605 | TZ-06-RICKER-607 | Canada, British Columbia |
| Eptatretus stoutii | – | FJ164606 | TZ-06-RICKER-608 | Canada, British Columbia |
| Eptatretus stoutii | – | FJ164607 | TZ-06-RICKER-490 | Canada, British Columbia |
| Eptatretus stoutii | – | GU440317 | MFC124 | USA, San Diego |
| Eptatretus stoutii | AF364618 | – | – | USA, Oregon |
| Eptatretus stoutii | EU099456 | – | SIO 95-2 | USA, California |
| Eptatretus strickrotti | EF014478 | – | CAS 223480 | East Pacific Rise |
| Eptatretus taiwanae | AF364611 | – | – | Taiwan |
| Eptatretus yangi | AF364612 | – | – | Taiwan |
| Eptatretus yangi | AF364613 | – | – | Taiwan |
| Eptatretus yangi | AF364614 | – | – | Taiwan |
| Eptatretus yangi | AF364615 | – | – | Taiwan |
| Myxine affinis | JX442466 | KC807353 | UNMDP-T 0919 | Argentina, Plataforma Patagonia |
| Myxine affinis | JX442467 | KC807352 | UNMDP-T 0041 | Argentina, Plataforma Patagonia |
| Myxine affinis | JX442471 | KC807349 | UNMDP-T 0025 | Argentina, Plataforma Patagonia |
| Myxine australis | – | EU074490 | INIDEP-T 0132 | Argentina |
| Myxine australis | JX442468 | KC807354 | UNMDP-T 0935 | Argentina, Ushuaia, Mar del Plata |
| Myxine australis | JX442469 | KC807351 | UNMDP-T 0035 | Argentina, Plataforma Patagonia |
| Myxine australis | JX442470 | KC807355 | UNMDP-T 0936 | Argentina, Ushuaia, Mar del Plata |
| Myxine capensis | – | JF493944 | ADC09_1.4#4 | South Africa, Agulhas Bank |
| Myxine capensis | – | JF493945 | ADC09_1.4 #3 | South Africa, Agulhas Bank |
| Myxine capensis | – | JF493946 | ADC09_1.4#2 | South Africa, Agulhas Bank |
| Myxine capensis | – | JF493947 | ACD07_1.4 #1 | South Africa |
| Myxine circifrons | AF364628 | – | – | USA |
| Myxine circifrons | EU099504 | – | SIO 05-104 | USA, California |
| Myxine fernholmi | JX442465 | KC807350 | UNMDP-T 0026 | Argentina, Plataforma Patagonia |
| Myxine formosana | – | JN027323 | NAFF 6195 | Taiwan? |
| Myxine formosana | AF364625 | – | – | Taiwan |
| Myxine glutinosa | – | KC807339 | NRM 47489 | |
| Myxine glutinosa | – | Y15182 | – | Sweden? |
| Myxine glutinosa | AJ404477 | AJ404477 | – | Sweden, ‘Baltic Sea’ |
| Myxine glutinosa | JX442475 | KC807338 | NRM 47485 | Sweden, Skagerrak |
| Myxine glutinosa | JX442476 | KC807337 | NRM 47487 | Sweden, Skagerrak |
| Myxine jespersenae | JX442472 | KC807336 | NMSI 760 | Iceland |
| Myxine jespersenae | JX442473 | KC807334 | NMSI 761 | Iceland |
| Myxine jespersenae | JX442474 | KC807335 | NMSI 759 | Iceland |
| Myxine limosa | – | KC015742 | SIO 06-781 | Canada, St. Lawrence |
| Myxine limosa | – | KC015744 | SIO 06-913 | Canada, St. Lawrence |
| Myxine limosa | JX442477 | KC807327 | – | USA, North Carolina |
| Myxine limosa | JX442478 | KC807326 | – | USA, North Carolina |
| Myxine limosa | JX442479 | KC807332 | – | USA, North Carolina |
| Myxine limosa | JX442480 | KC807328 | – | |
| Myxine sp. ‘1-CHK-2001’ | AF364622 | – | – | Taiwan |
| Myxine sp. ‘1-CHK-2001’ | AF364623 | – | – | Taiwan |
| Myxine sp. ‘1-CHK-2001’ | AF364624 | – | – | Taiwan |
| Myxine sp. ‘2-CHK-2001’ | AF364626 | – | – | Taiwan |
| Myxine sp. ‘3-CHK-2001’ | AF364627 | – | – | Taiwan |
| Neomyxine biniplicata | JX442447 | KC807347 | NMNZ P.052442/TS2 | New Zealand |
| Neomyxine biniplicata | JX442448 | KC807348 | NMNZ P.052443/TS2 | New Zealand |
| Neomyxine biniplicata | JX442449 | KC807346 | NMNZ P.052441/TS2 | New Zealand |
| Neomyxine sp. | JX442444 | KC807343 | NMNZ P.047438 | New Zealand |
| Neomyxine sp. | JX442445 | KC807342 | NMNZ P.044215 | New Zealand |
| Neomyxine sp. | JX442446 | KC807344 | NMNZ P.047862 | New Zealand |
| Notomyxine tridentiger | – | EU074500 | INIDEP-T 0131 | Argentina |
| Rubicundus lopheliae | JX442464 | KC807325 | – | USA, North Carolina |
| Rubicundus rubicundus | AY033088 | – | ASIZP 60660 | Taiwan |
| * Branchiostoma lanceolatum | NC_001912 | NC_001912 | ||
| * Lampetra fluviatilis | NC_001131 | NC_001131 | ||
| * Petromyzon marinus | U11880 | U11880 | ||
| * Protopterus dolloi | NC_001708 | NC_001708 |
The combined data set was produced with all taxa for which both 16S and COI data were available. The 16S alignment comprises 75 sequences representing 33 species and is 561 bp long, after insertions unique to out-groups had been deleted. The COI alignment comprises 72 sequences representing 23 species and is 687 bp long. The combined alignment comprises 42 sequences representing 18 species and is 1240 bp long. All alignments are available from the authors.
Phylogenetic analysis was performed using the software MrBayes v. 3.2 (Huelsenbeck and Ronquist 2001; Ronquist et al. 2012). COI data was partitioned according to codon position (first, second, third), and parameters estimated separately for each partition. In the combined analysis, the 16S data comprised a fourth partition. The GTR + Γ + I model was used as suggested by ModelTest (Posada and Crandall 1998). The analysis was run for two million generations, at which time average standard deviation of split frequencies reported by MrBayes was ≤0.01. Samples were taken every 1000 generations, and the first 25% of samples were discarded as ‘burn-in’.
Parsimony bootstrap estimates indicate the amount of conflicting signals in a data set. Parsimony bootstrap analysis was performed using the software TNT v1.1 (Goloboff et al. 2008), with 1000 replicates, 10 random additions per replicate, and using ‘classic search’ and TBR branch swapping. End gaps were coded as missing data, other gaps as a fifth state.
In all analyses, the trees were rooted with four non-myxinid chordates: Branchiostoma lanceolatum (Cephalochordata: Branchiostomatidae), Protopterus dolloi (Sarcopterygii: Lepidosirenidae), Petromyzon marinus (Petromyzontomorphi: Petromyzontidae) and Lampetra fluviatilis (Petromyzontomorphi: Petromyzontidae).
In all presented trees, (Figs 1-3) species represented by several sequences and recovered in our analyses as monophyletic have been reduced to a single terminal. This is because intraspecific population structure is irrelevant to the questions this study seeks to answer, and smaller trees are easier to read. However, the full, unreduced, versions of the presented trees are instead provided as supplementary information (Figures S4–S6).
Taxonomic revision
The taxonomic and nomenclatural analysis was performed studying publications and comparing with the International Code of Zoological Nomenclature (International Commission on Zoological Nomenclature 1999), abbreviated ‘Code’ below. Entries in Eschmeyer (2012) were used as seed for collecting names and relevant taxonomic literature.
Results
Molecular phylogenetic analysis
16S data analysis
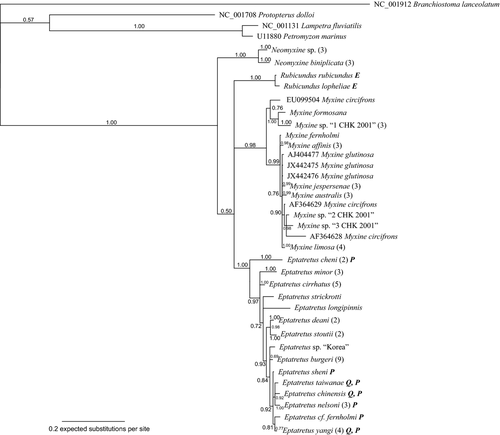
The Bayesian phylogenetic analysis of the 16S data is summarized in Fig. 1. The base of the tree separated into two sister groups, the first one composed of Neomyxine sp. and Neomyxine biniplicata (Bayesian posterior probability 1.0), the second composed of all other species of Myxinidae (Bpp 0.50). This second group is further divided into three groups corresponding to Rubicundus (Bpp 1.0), Myxininae excluding Neomyxine (Bpp 0.98), and Eptatretinae excluding Rubicundus (Bpp 1.0). Within the Myxininae, Myxine sp. ‘1-CHK-2001’, Myxine formosana, and one of the two Myxine circifrons formed a cohesive group (Bpp 0.76), whereas remaining Myxininae formed another supported (Bpp 0.99) group with little internal structure. The monophyly of N. sp., N. biniplicata, M. sp. ‘1-CHK-2001’, Myxine limosa and Myxine jespersenae was supported, (Bpp ≥ 0.99), but the monophyly of Myxine glutinosa was not supported (Bpp < 0.5), and the monophyly of M. circifrons was rejected.
In the Eptatretinae, Eptatretus cheni branched first. Eptatretus minor, E. cirrhatus and a group comprising the remaining Eptatretinae formed an unresolved polytomy (Bpp 0.97). Within this group of Eptatretinae, E. strickrotti, E. longipinnis and a group comprising the remaining Eptatretinae formed another unresolved polytomy (Bpp 0.93). Within this last group, E. deani + E. stoutii (Bpp 0.98) was the sister group of a clade comprising all remaining Eptatretinae (Bpp 0.84). The Bayesian analysis supported the monophyly of E. cheni, E. cirrhatus, E. minor, E. deani, E. stoutii and Eptatretus nelsoni (Bpp 1.0), and the monophyly of E. burgeri (0.69) and E. yangi (Bpp 0.77). All included members of the proposed genus Paramyxine except E. cheni formed a largely unresolved group (Bpp 0.72).
The majority-rule parsimony bootstrap tree inferred by TNT (not shown; provided as supporting information, Figure S1) was compatible with the majority-rule Bayesian tree inferred using MrBayes (Fig. 1). The parsimony bootstrap tree differs by having lower resolution in Eptatretinae and Myxininae.
The 16S tree was sensitive to alignment, with different methods and parameters of alignment resulting in different relative branching order of Myxininae, Eptatretinae, Rubicundus and Neomyxine. However, the tree was robust to the method of analysis, with parsimony bootstrap and Bayesian analysis producing compatible trees when analysing the same alignment.
COI data analysis
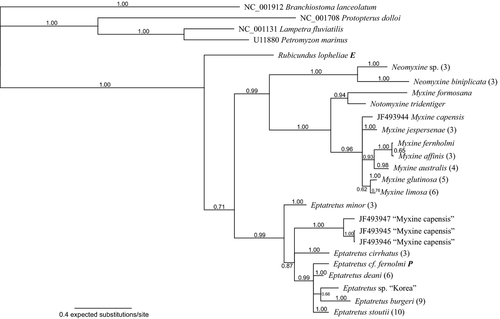
The Bayesian phylogenetic analysis of the COI data is summarized in Fig. 2. The base of the tree separated into two sister groups, the first one composed of Rubicundus lopheliae, the second one with all other species of Myxinidae (Bpp 0.71). This second group was further divided into two groups corresponding to Myxininae (Bpp 0.99) and Eptatretinae excluding Rubicundus (Bpp 0.99). Within the Myxininae, the earliest branch is Neomyxine sp. + Neomyxine biniplicata (Bpp 1.0). The next group to branch comprised Myxine formosana + Notomyxine tridentiger (Bpp 0.94), while the remaining Myxininae formed a polytomy in which Myxine fernholmi + Myxine affinis + Myxine australis formed a cohesive group (Bpp 0.93) and Myxine glutinosa + Myxine limosa another (Bpp 0.62). The monophyly of Neomyxine sp., N. biniplicata, M. affinis, M. australis, M. glutinosa, M. limosa and M. jespersenae was supported (Bpp 1.0 for all except limosa, Bpp 0.76) but the monophyly of M. capensis was rejected.
In the Eptatretinae, E. minor branched first. ‘M. capensis’, E. cirrhatus, and a group comprising the remaining Eptatretinae formed an unresolved polytomy (Bpp 0.99). Within this group of Eptatretinae, E. sp. ‘Korea’ formed a monophyletic group with E. burgeri (Bpp 0.66). The monophyly of E. minor, ‘M. capensis’, E. cirrhatus, E. deani, E. burgeri and E. stoutii was supported (Bpp 1.0 for all).
The majority-rule parsimony bootstrap tree inferred by TNT (not shown; provided as supporting information, Figure S2) was similar, but conflicted with the majority-rule Bayesian tree (Fig. 1) by reconstructing Neomyxine as the earliest branch within Myxinidae, while Rubicundus, Eptatretinae and Myxininae formed an unresolved trichotomy, and by having M. formosana as the earliest branch within Myxininae.
Combined 16S+COI data analysis
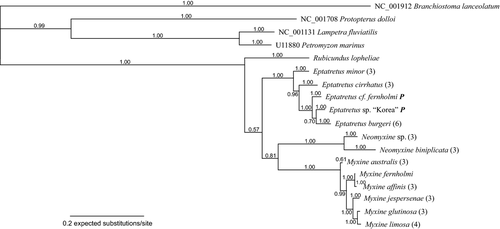
The Bayesian phylogenetic analysis of all myxinids for which both 16S and COI data were available is summarized in Fig. 3. The tree topology is compatible with the COI tree, differing only in support values. Excluding intraspecific nodes, all nodes have Bpp ≥ 0.99, except for the placement of Rubicundus lopheliae as first branch in Myxinidae (Bpp 0.57), the placement of Neomyxine as the sister group of all other Myxininae (0.81) and the sister group relationship of Eptatretus sp. and E. burgeri (Bpp 0.70). The monophyly of all included species is recovered (Bpp 1.00; M. australis Bpp 0.61).
The majority-rule parsimony bootstrap tree inferred by TNT (not shown; provided as supporting information, Figure S3) conflicted with the majority-rule Bayesian tree (Fig. 3) by reconstructing Neomyxine as the earliest branch within Myxinidae and Rubicundus as the sister group to Eptatretinae.
Taxonomy
Type species and validity of Eptatretus
The original description of Eptatretus (Cloquet, 1819) was based on two species, both identified as Eptatretus Dombeii (= Gasterobranchus dombeyi Shaw, 1804). With reference to Code article 70.3, we designate here as type species of Eptatretus the species corresponding to the diagnostic characters reported by Cloquet, viz., Homea banksii Fleming (1822). Homea banksii is a junior synonym of Petromyzon cirrhatus Forster (in Schneider 1801). Bdellostoma heptatrema Müller (1836) is an unjustified replacement name for P. cirrhatus and thus an objective junior synonym. Gasterobranchus dombeyi Shaw, 1804, described from Chile, is an available name for a different species, most likely a senior synonym of Eptatretus polytrema (Girard, 1855a), E. nanii Wisner and McMillan (1988), or E. bischhoffi (Schneider, 1880), but the type specimen was lost.
No types are known to exist of H. banksii or P. cirrhatus. A neotype from the type locality area of P. cirrhatus has been selected and is being formally designated by Zintzen et al. (in prep.) covering H. banksii, B. heptatrema and P. cirrhatus. This neotype designation is necessary because of the prevailing uncertainty concerning the type species of Eptatretus. Previous authors have confused H. banksii and Gasterobranchus dombeyi, and it has not been clear if H. banksii and P. cirrhatus even refer to the same specimen, as discussed further below. The type locality of P. cirrhatus is Dusky Bay and Queen Charlotte Sound in New Zealand.
New taxa resulting from the present phylogenetic analysis
The tree obtained from the molecular phylogenetic analyses suggested two major clades, one with the traditionally recognized Myxininae and Eptatretinae, and a second one composed of two highly distinctive species. These two species are recognized as belonging to a new genus. In order to preserve the recognition of the subfamilies Myxininae and Eptatretinae, a new subfamily for their sister clade is proposed.
Rubicundus, new genus.
Type species: Eptatretus rubicundus Kuo et al. (2010).
Referred species: Eptatretus lopheliae, Eptatretus eos Fernholm (1991), Eptatretus lakeside Mincarone and McCosker (2004).
Etymology: Rubicundus repeats the specific epithet of the type species, emphasizing the reddish coloration of the included species. It is a Latin adjective meaning red, here treated as a masculine noun in the nominative singular.
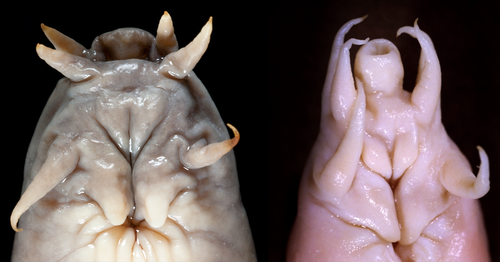
Diagnosis: Myxinids with five gill openings, uniquely distinguished by elongated, tubular nostril (versus short in all other Myxinidae; Fig. 4) and reddish colour (versus grey or brown, never reddish, in all other Myxinidae; Fig. 4).
Rubicundinae, new subfamily of Myxinidae.
Type-genus: Rubicundus, present paper, only included genus.
Diagnosis: Same as for the genus Rubicundus.
Replacement of scientific names resulting from the present phylogenetic analysis
Synonymization of species from Paramyxine with Eptatretus resulted in two cases of homonymy that requires replacement names. Eptatretus wisneri McMillan (1999) from the Galápagos Archipelago becomes a junior homonym of Paramyxine wisneri Kuo et al. (1994) from Taiwan. Eptatretus wisneri McMillan is herewith given the new name Eptatretus bobwisneri to maintain its affiliation to Robert L. Wisner (1921–2005) of the Scripps Institute of Oceanography. Eptatretus fernholmi McMillan and Wisner (2004) from the Philippines becomes a junior homonym of Paramyxine fernholmi Kuo et al. (1994) from Taiwan and is herewith renamed E. luzonicus. This specific name was mentioned by McMillan and Wisner (2004) in the description as the name initially intended for the species. The single known specimen was caught east of the southern end of the Luzon Island in the Philippines. The species epithet luzonicus, alluding to the type locality, is an adjective with alternative endings -a, and -um.
Discussion
Molecular phylogenetic analysis
The COI results were robust to methods of alignment, but in the 16S analysis, the position of Neomyxine, and to a lesser degree Rubicundus, was not. The tree depicted in (Fig. 1) was based on the alignment obtained from MUSCLE, but analysing alignments produced by Geneious and MAFFT variously recovered Neomyxine as either (1) the earliest branch within the Myxinidae (in which case Rubicundus either forms an unresolved trichotomy with the Eptatretinae and Myxininae, or is the sister taxon of the Myxininae), (2) the sister taxon of the Eptatretinae or (3) as a member of an unresolved polytomy encompassing Myxininae, Eptatretinae and Rubicundus. From this, we infer that there is insufficient information regarding the relative branching order of Neomyxine, Rubicundus, Eptatretinae and Myxininae in our 16S data and that resolution at that level in the tree is due to differences in alignment.
Given alignment, the Bayesian and the parsimony bootstrap majority-rule trees were largely compatible, differing mostly in resolution, with the exception of the placement of Neomyxine. In all parsimony bootstrap analyses, Neomyxine was the earliest branch in Myxinidae, whereas in Bayesian analyses, Neomyxine was the earliest branch in Myxinidae only in the 16S analysis, while Rubicundus was the earliest branch in the COI and combined analyses, as well as with some alternative alignments of 16S. This discrepancy may be a result of signal erosion (dubbed a Class II Long Branch artefact by Wägele and Mayer 2007), as all out-groups are distant, with branches roughly one order of magnitude longer than branches inside Myxinidae, except the long branches of Rubicundus and Neomyxine and Bayesian analysis is more resistant to long branch artefacts than parsimony analysis.
There exists no more closely related out-group taxa that could be added to the analysis, and performing the parsimony bootstrap analysis without out-groups results in Neomyxine, Rubincundinae, Myxininae and Eptatretinae forming a polytomy. The relative branching order of these taxa remains uncertain.
Our reconstructions of the phylogeny of the Myxinidae (Figs 1-3) are similar to what Kuo et al. (2010) proposed based on an analysis of 16S data, differing mainly in the placement of Rubicundus rubicundus. In their tree, R. rubicundus was the sister group to all other Eptatretinae, but with weak support (0.63 Bpp), and as previously discussed, the position of Rubicundus in analyses of 16S data is sensitive to alignment. Møller and Jones (2007) presented a tree with a topology similar to ours for the Myxininae, but different from ours for the Eptatretinae. In their tree, E. strickrotti was the first branch within the Eptatretinae. To test if this difference was due to the increased number of specimens and species included in our study, we performed an analysis with just the 27 sequences used in Møller and Jones (2007). We could not reproduce their results, and the tree obtained was similar to the results of the 16S analysis of this study (Fig. 1). The discrepancy may be explained by differences in method of analysis: Møller and Jones (2007) did not provide sufficient information on the parameters used in their analysis, such as method of alignment, data partitioning or model used in their Bayesian analysis, preventing us from repeating their results.
The rejected monophyly of Myxine circifrons suggests that one or several of the sequences EU099504, AF364628 and AF364629 deposited in GenBank were obtained from misidentified specimens. Voucher specimens do not exist for these sequences, leaving their identity unresolved. The long branch of AF364628 could be an artefact caused by an apparent sequencing error at positions 72–83.
The rejected monophyly of M. capensis also seems to be caused by misidentifications. One of the four M. capensis sequences (JF493944) groups inside Myxininae and is presumably correct, but the three remaining ‘M. capensis’ (JF493945–JF493947) group inside Eptatretinae, indicating that these sequences represent species of Eptatretus. The sequences JF493944–JF493947 were also included in an analysis by Kuraku (2013). Their analysis (their fig. 5) also recovered E. capensis as non-monophyletic. GenBank has been notified of the error. The ‘M. capensis’ sequences originate from the Barcode of Life initiative, preserved voucher specimens exist, and the sequence identity will hopefully be corrected.
The sequences KC015742 and KC015744 are listed in GenBank as M. glutinosa from Canada, but in this study represent M. limosa. This is not a result of misidentification: the north-west Atlantic M. limosa is often considered a synonym of the north-east Atlantic M. glutinosa.
Taxonomy
Twenty nominal genera have been recognized in the Myxinidae: Bdellostoma Müller, 1836; Dodecatrema Fowler, 1947, Heptatrema Duméril, 1832, Heptatremus Swainson, 1839, Heptatretus Regan, 1912, Hexabranchus Schultze, 1835, Homea Fleming, 1822; Polistotrema Gill, 1881 and Polytrema Girard, 1855b; are generally considered as junior synonyms of Eptatretus Cloquet, 1819; when not simply unjustified emendations. Paramyxine Dean (1904) and Quadratus Wisner (1999) are recognized as valid in recent publications (Nelson 2006), but also often considered synonyms of Eptatretus (Møller and Jones 2007). Myxine (Linnæus 1758) has well established junior synonyms Anopsis Agassiz, 1846, Anopsus Rafinesque, 1815, Gastrobranchus Bloch, 1791 and Muraenoblenna La Cepède, 1803. Neomyxine Richardson, 1953 and Nemamyxine Richardson, 1958, both with two species, and the monotypic Notomyxine Nani & Gneri, 1951, are three little studied genera, universally considered valid.
Traditionally, the Myxinidae has been organized in two subfamilies, Eptatretinae for the hagfishes with several pairs of gill openings and Myxininae for species with only one pair of gill openings (Fernholm 1998; Nelson 2006). Our molecular results are not compatible with that of dichotomous division. The new genus Rubicundus has several pairs of gill openings, but does not belong to the traditional Eptatretinae. Clearly, a more complicated taxonomy of the Myxinidae is emerging. We believe that having several pairs of external gill openings is the plesiomorphic character state within Myxinidae, while having one pair of external gill openings is a synapomorphy of the Myxininae. Neomyxine, with a single pair of gill openings, has traditionally been included in the Myxininae, a position challenged by the 16S phylogeny (Fig. 1) and parsimony bootstrap analysis. However, the placement of Neomyxine as the first branch in Myxinidae is dependent on alignment, and in the Bayesian COI (Fig. 2) and combined analyses (Fig. 3), Myxininae is monophyletic. Also, Nemamyxine and Notomyxine have one pair of gill openings and are consequently traditionally assigned to the Myxininae. Pending further studies, we choose to continue the prevailing praxis of assigning these three genera to the Myxininae.
Eptatretus
Any classification of myxinid species is problematic because of uncertainty about the type species of Eptatretus, a genus traditionally diagnosed by the multiple gill openings. The problem was already recognized by Gill (1901) who presented a detailed analysis of the status of Eptatretus and its synonym Bdellostoma. The first description of a hagfish with multiple gill openings may have been by Home (1815), who described it as an animal ‘intermediate between the lamprey and the myxine’, and considered it to be a new genus. Duméril (1812) established the family of ‘Cyclostomes’ for Myxine, lampreys and ammocetes (not known at the time to be larvae of lampreys). He described the anatomy of the cyclostomes in some detail, but only mentioning a single pair of gill openings in Myxine. Cloquet (1818) reviewed the cyclostomes, basing his work mainly on Duméril (1812). However, at the end of his text, Cloquet referred to Home's (1815) paper as having about the same content as that of Duméril (1812). Cloquet also provided a key to cyclostomes, which distinguishes between Myxine with two ventral gill openings, and Eptatrème with seven lateral gill openings. No species name was associated with Eptatrème, but the name doubtlessly refers to the description by Home (1815) and should be regarded as a French vernacular name.
Cloquet (1819) listed three generic names that apparently all refer to the Eptatrème of 1818. Eptacitrète is listed only with a reference to Eptatrème, for which the entry reads ‘Eptatrème ou Eptatrète’. In the paragraph explaining the etymology, eptatrème is explained followed by the remark that Duméril had first proposed the word eptacitrète. Below that are two uses of Eptatrète, and one of Eptatretus, and a mention that Duméril established his genus Eptatrème in Paris. The entry is signed by Cloquet, but it is evident that he considers Duméril as author of Eptatrème and the proposed Eptacitrète. No real distinction is made between Eptatretus, Eptatrète and Eptatrème, but only Eptatretus appears in italics together with a specific epithet (Eptatretus Dombeii). The position of later workers, adopted also by us, has obviously been to consider Eptatrète and Eptatrème as vernacular names, and Eptatretus as the intended scientific name, although we share Gill's (1901) resignation over Cloquet's confused text.
Cloquet (1819) based the diagnosis of Eptatretus on information about two species, viz., Gastrobranche dombey, described by La Cepède (1798), and the subsequently described H. banksii Fleming (1822). La Cepède's description and figure (1798: pl. 25, fig. 1) were based on a single dry skin. The characters described and the drawing are clearly of a hagfish, but the number of gill openings is not given, and gill openings are not shown on the drawing. In the introductory text, La Cepède (1798: 526) makes clear that he did not count the number of gill openings, stating that ‘All pétromyzons have seven gills on each side, the blind gastrobranche [Myxine glutinosa] only has six on the left and six on the right, and one should assume that the Gastrobranche dombey does not have more’ (‘Tous les pétromyzons ont sept branchies de chaque côté; le gastrobranche aveugle [Myxine glutinosa] n'en a que six à droite et six à gauche, et il est a présumer que le gastrobranche dombey n'en a pas un plus grand nombre.’). This description does not contain any information that may enable identification as to genus or species, and the specimen is apparently lost (Wisner and McMillan 1988). The type locality is given as the sea near Chili [= Chile]. The collector, Joseph Dombey, collected plants in Chile 1782–1784 (Ruiz 1940).
The status of Gastrobranche dombey remains in limbo. There is no character known by which it can be identified to genus or species. Putnam (1874) listed Bdellostoma polytrema as a valid species and added in a footnote that ‘This is unquestionably the species described by Lacepède [sic], under the name of Le Gastrobranche Dombey, and figured from a stuffed skin of a specimen collected by Dombey in Chili’. Later, many authors have cited Gastrobranche dombey as a scientific name, although it was consistently presented by La Cepède as a French vernacular name. The earliest scientific name for La Cepède's species appeared in 1804, but priority among the two descriptions is uncertain. Gasterobranchus dombeyi Shaw (1804: 265, pl. 134) is based entirely on La Cepède's description, and the figure is a copy of La Cepède's illustration. Latreille (1804: 75) lists the genus Gastrobanchus [sic] and provides a Latinized scientific name for La Cepède's species as ‘gastrobanc. dombey’, with clear indication to La Cepède's description. Dates of publication for those two works are not known to us, but we follow Eschmeyer (2012) who gives Shaw as author. Eptatretus dombeii Duméril (in Cloquet 1819) is a later independent Latinization. Cuvier (1816: 121) writes Gasterobranche dombey in italics, but apparently just cites the French name used by La Cepède.
Several post-1899 usages of Polistotrema dombey and Bdellostoma dombeyi as valid names were listed among unassignable records of Chilean hagfishes by Wisner and McMillan (1988), and therefore Gasterobranchus dombeyi is available as potential senior synonym for one of several species of hagfish known from the Pacific coast of Chile. Girard (1855a) described B. polytrema from Chile and reviewed La Cepède's description of Gastrobranche dombey, coming to the conclusion that the characters did not support inclusion in Bdellostoma, diagnosed by presence of eyes and multiple gill openings. Putnam (1874) nevertheless considered Gasterobranche dombey and B. polytrema to be the same species, but dismissed the first as a vernacular name. This was obviously not accepted by Jordan and Evermann (1896: 6) who used Putnam's (1874) diagnosis of B. polytrema as a diagnosis of Polistotrema dombey, to differentiate between this species and E. stoutii (Lockington, 1878). Putnam's B. polytrema was re-identified as E. bischhoffi (Schneider, 1880) by Wisner and McMillan (1988). Wisner and McMillan (1988) reported three species of Eptatretus from the Chilean coast (E. bischhoffi, E. polytrema, and E. nanii Wisner and McMillan, 1988). One of them is likely to be identical with La Cepède's species, but we leave the synonymization to a future revision of the concerned species.
Homea banksii was known to Cloquet (1819) from the description by Home (1815) based on a dissected specimen ‘brought from the South Seas by Sir Joseph Banks’. The description by Fleming (1822) is apparently based only on Home's description. Müller (1836) described the species as Bdellostoma heptatrema, again based apparently only on Home's description, making B. heptatrema a junior synonym of H. banksii. There is no record of extant specimens for the description of Petromyzon cirrhatus Forster, 1801 (in Schneider 1801: 532), which was based on a manuscript description by Forster [Johann Reinhold Forster]. Schneider (1801) gave the locality as the coastal sea in New Zealand. Müller (1836) renamed it Bdellostoma forsteri, and gave the locality as Queen Charlotte Sound, based on Forster's manuscript. Forster's manuscript was edited and published by Lichtenstein (1844), including a long, detailed description of P. cirrhatus, with the locality Dusky Bay and Queen Charlotte Sound in New Zealand. Banks's specimen(s) may have come from the first of Cook's journeys to the South Sea, in which Banks participated. Reinhold and Georg Forster participated in the second of Cook's South Sea journeys, during which the expedition anchored in Dusky Bay in March and April 1773 and in Queen Charlotte Sound in May and November 1773, and in October 1774 (Lichtenstein 1844; Forster 2007). It remains possible, however, that there was a specimen passed from Forster to Banks, as some objects and drawings were sold to Banks (Harpprecht 2007:19). It seems that the majority or all of the animals brought back by Cook's three journeys were in the possession of Banks for longer or shorter time, but became widely dispersed (Whitehead 1969). The specimen(s) examined by Home may have come from any of the journeys and would have been available to him at the Hunter museum, operated by his brother-in-law, or at the Royal College of Surgeons, both of which, along with other institutions in London possessed specimens from Banks (Whitehead 1969). Fish specimens from Cook's journey still exist in the Natural History Museum in London (Whitehead 1969), but no myxinid specimens coming for sure from Forster or Banks are present today in the Natural History Museum (James Maclaine, pers. comm., Sept. 2012). An unpublished drawing of a P. cirrhatus, made by Georg Forster is, however, preserved in the Natural History Museum (Whitehead 1978; Wheeler 1981). Müller (1836) considered Forster's and Home's species to be different. Later authors, however, have synonymized H. banksii with P. cirrhatus, and it seems not impossible that the descriptions were based on the same specimen. If this is the case, P. cirrhatus would be an objective junior synonym of H. banksii.
There are thus no type specimens known for H. banksii or P. cirrhatus, as would be desirable for establishing the relative status of these names, and for a stable nomenclature of the genus Eptatretus. It also turns out that two similar species of Eptatretus, including E. cirrhatus occur sympatrically around the South Island, probably including the type locality of P. cirrhatus. The taxonomy of these taxa will be addressed by Zintzen et al. (in prep.) who will designate a common neotype for H. banksii, B. heptatrema and P. cirrhatus. By being a replacement name, this neotype also becomes neotype of Bdellostoma forsteri.
Cloquet's description of Eptatretus is based on a diagnostic character (number of gill openings) not known from Gastrobranche dombey, but only from Home's description, which he cites at length, and apparently considers to be the same species. There is no doubt that Gastrobranche dombey, formally established as Gasterobranchus dombeyi Shaw, is the type species of Eptatretus by monotypy (Code articles 67.2.1 and 68.3). However, as already put forth by Gill (1901), it is also clear that Cloquet did not base the genus on the characters of this species, but on the misidentified specimen later named H. banksii. Consequently, Code article 70.3 applies, and we designate H. banksii as type species of Eptatretus. We consider H. banksii and P. cirrhatus to be synonyms. The older name, P. cirrhatus, takes priority.
Gastrobranche dombey is also the nominal type species of Polistotrema Gill (in Jordan & Gilbert, 1880). According to Eschmeyer (2012), the genus was based on misidentified E. stoutii and B. polytrema Girard in the synonymy of Gasterobranche dombey. The original description lists ‘Polistrotrema dombey (Muller) Gill’ as a species occurring on the Pacific Coast of the United States, with reference to a footnote with the two names Bdellostoma polytrema and B. stoutii. In Jordan and Gilbert (1881), it is obvious that the species they call Polistotrema dombeyi is rather, by a taxonomic conclusion, E. stoutii. Consequently Code article 70.3 applies, and we select here B. stoutii as the type species of Polistotrema, observing Code article 70.3.2.
Paramyxine and Quadratus
Paramyxine was introduced by Dean (1904) to distinguish the Japanese species Paramyxine atami Dean (1904) with crowded gill openings from species of Eptatretus, in which the gill openings are more spaced. Subsequently, another 13 species have been referred to Paramyxine and four of them eventually to Quadratus. Molecular data failed to support monophyly of either Paramyxine or Quadratus (Møller and Jones 2007; Kuo et al. 2010; this study). The type species of Quadratus is Paramyxine taiwanae (Shen & Tao, 1975), which is included in the 16S analyses, and nested among species of Eptatretus. Of the seven species of Eptatretus, labelled ‘P’ in Fig. 1, which possess crowded gill openings and are referable to Paramyxine, one species positions as sister group of remaining Eptatretus and the remainder form a monophyletic group within a clade composed of species with spaced gill openings and mixed with species that have been referred to Quadratus. Although Quadratus is clearly not distinct from Paramyxine, and all putative Paramyxine in this analysis should be referred to Eptatretus, P. atami was not included, and there remains a possibility that it is genetically distinct. If that was the case, Paramyxine would remain a valid genus, but with a different composition and in need of a revised diagnosis. Recognizing the six-species cluster of Paramyxine as a valid genus based on the tree topology in Fig. 1 would require recognition of seven or eight separate genera for the other clusters as well. There is no morphological support for such extensive splitting.
Rubicundus and Rubicundinae
Rubicundus lopheliae formed the sister clade to the remaining Myxinidae in the COI and combined analyses, while R. lopheliae and R. rubicundus formed a monophyletic group in a polytomy with Myxininae and Eptatretinae in the 16S analysis. Both species were placed in Eptatretus because of their multiple gill openings, and it was expected that they would group within or with the Eptatretinae. However, if — as has been suggested by Fernholm (1998) — having multiple pairs of gill openings with a gill opening adjacent to each gill pouch is the plesiomorphic state, then that character is not useful for informing phylogeny.
Rubicundus lopheliae was sampled in the western North Atlantic off the south-eastern United States, and R. rubicundus in the western Pacific, off the north-eastern coast of Taiwan (Fig. 3). Although they come from very distant sites, R. lopheliae and R. rubicundus formed a monophyletic group in this study. Both species were found in deep waters (400 and 800 m), and have five pairs of gills. They share two unique morphological character states, currently considered as synapomorphies: an elongated nostril (Fig. 2) and the reddish colour. Another possible synapomorphy is the localization of the slime pores in the gill area in a straight line with the gill openings (Fernholm and Quattrini 2008; Kuo et al. 2010).
Based on the results of the molecular phylogenetic analyses and the presence of distinctive morphological characters, we consider R. lopheliae and R. rubicundus to represent a distinct lineage of the Myxinidae, and place them in a separate genus and separate subfamily. Two more species share the same snout shape, reddish colour and five pairs of gills, and are also referred to Rubicundus. Both are known only from a single specimen each, and no tissue suitable for molecular phylogenetic analysis is available. Rubicundus eos (Fernholm, 1991) was described from one specimen caught in trawl at about 1000 m of depth in the southern hemisphere in the Tasman Sea, west of New Zealand. Rubicundus lakeside was based on one specimen caught in 762 m of depth off the Galápagos Archipelago (Mincarone and McCosker 2004).
Fernholm and Quattrini (2008) discussed the possibility of erecting a new genus for the long-snouted, reddish deep-water forms but refrained mainly because of the long geographic distances between the type localities. Kuo et al. (2010) also discussed the striking similarities between these deep-living hagfish species occurring at long distances from each other.
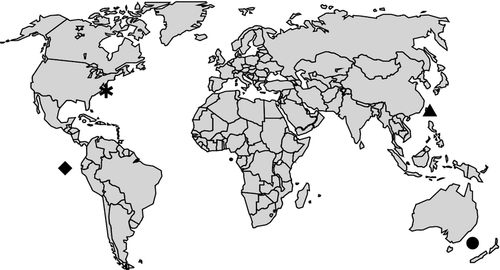
The extremely wide and patchy distribution of Rubicundus (Fig. 5) is likely due to insufficient sampling. Rubicundus eos, R. lakeside and R. rubicundus are all described from single specimens, and R. lopheliae is known from only a few individuals. The latter was collected by manned submersibles in a cold-water coral reef, and observations of live specimens within the reef indicate that this is their natural habitat (Fernholm and Quattrini 2008). Additional specimens have later been collected from deep coral areas off the south-east United States (S. W. Ross, unpublished data). Also R. lakeside was collected using manned submersibles. R. eos and R. rubicundus on the other hand were caught by trawl, which is not the best method to collect deep-living benthic hagfishes in complex habitats. We consider it likely that what we see here is the emerging pattern of high degree of endemism and speciation within complex deep-sea habitats (cf. Hubbs 1959; Hart and Pearson 2011). It is expected that with the on-going exploration of the deep sea using appropriate new collecting methods, we could find more hagfish species in these habitats. New technology to access deep-sea habitats was identified by Eschmeyer et al. (2010) as the principal trigger for the present overall increase in descriptions of new marine fish species.
Eptatretus strickrotti was also caught in a deep-sea habitat from a hydrothermal vent (Møller and Jones 2007). However, although the live specimen was described as bright pink, its morphology is different from the species discussed in connection with Rubicundus, being a very slender species with 12 pairs of gills and lacking an elongated nostril. Our molecular phylogenetic analysis places it in Eptatretus (Fig. 1).
Myxine
The genus Myxine was erected by Linnæus (1758) with inclusion of the single species M. glutinosa. Four syntypes (NRM 89) are preserved in the Swedish Museum of Natural History and provide a stable reference for taxonomic work on the Myxininae.
When describing M. jespersenae, Møller et al. (2005) included a comprehensive comparison and detailed discussion of all species of Myxine. They discussed whether M. glutinosa, from the north-east Atlantic, and M. limosa, from the north-west Atlantic, are conspecific or distinct species. Putnam (1874) synonymized M. limosa with M. glutinosa. Following that work, recognition of M. limosa has varied. Jordan and Evermann (1896) followed Putnam. Regan (1913) considered M. limosa to be valid, but also described another Western Atlantic species, Myxine atlantica. Wisner and McMillan (1995) synonymized M. atlantica with M. limosa, which they considered to be distinct from M. glutinosa based on colour characters and larger size. Møller et al. (2005) stated that the holotype of M. limosa, from Bay of Fundy, was in poor condition and could not be used to investigate useful characters, concluding that input from molecular data would be necessary to assess the validity of M. limosa. In all analyses, sequences from our specimens identified as M. limosa from North Carolina form a monophyletic group that is clearly separate from M. glutinosa, supporting the validity of M. limosa and the diagnosis provided by Wisner and McMillan (1995). Our results supported the monophyly of M. jespersenae, but only the COI and combined analyses supported the monophyly of M. glutinosa.
Acknowledgements
We thank Bodil Kajrup (Swedish Museum of Natural History) for taking photographs of hagfishes, Georg B. Friðriksson (Icelandic Museum of Natural History) and Juan M. Díaz de Astarloa (Universidad Nacional de Mar del Plata) for graciously making tissue samples available to us, Nicolas Lartillot (Université de Montreal) for advice on analytical interpretation and Nicolas Bailly (FishBase) for his forceful encouragement. We also thank Steve W. Ross (University North Carolina Wilmington) for valuable contributions. FishBase Sweden provided financial support.




