Effect of mineral oil as a lubricant to collect feces from cats for microbiome studies
Funding information: Alabama Agriculture Experiment Station; Auburn University College of Veterinary Medicine Animal Health and Disease Research; Auburn University Intramural Grant Program, Grant/Award Number: 180254; EveryCat Health Foundation, Grant/Award Number: EC22-023; National Science Foundation, Grant/Award Number: 1928770; USDA National Institute of Food and Agriculture, Grant/Award Number: 1018100
Abstract
Background
Fecal specimens are critical for disease screening, diagnosis, and gut microbiome research. For domestic cats, lubricants are often necessary to obtain a sufficient quantity of sample. However, the effect of lubrication on feline microbiome analysis has not been assessed.
Objectives
To evaluate if lubrication using mineral oil during cat feces sample collection affects the DNA extraction, metagenomic sequencing yield, and the microbial composition and diversity in subsequent gut microbiome analyses.
Animals
Eight 6-year-old male, neutered, domestic short-haired cats housed in a research facility.
Methods
Cohort study. The gut microbiomes were investigated for fecal sample collection with and without lubrication using whole-genome shotgun metagenomic sequencing.
Results
Fecal specimens were collected using a fecal loop under sedation without lubrication and with mineral oil lubrication. There were no significant differences between the 2 groups in the microbial DNA yield in ng/mg fecal sample (75.75 [25.8-125.7] vs 60.72 [33.49-87.95], P = .95), metagenomic sequencing yield in Gbp (10.31 [6.29-14.32] vs 13.53 [12.04-15.02], P = .2), proportion of host contamination (0.1 [0.02-0.18] vs 0.15 [0-0.3], P = .84), relative taxonomy abundance (P > .8), or the number of microbial genes covered (408 132 [341 556-474 708] vs 425 697 [358 505-492 889], P = .31).
Conclusions and Clinical Importance
Fecal sampling with mineral oil lubrication did not change the microbial DNA extraction yield, metagenomic sequencing yield, level of host contamination, the microbial composition and diversity in subsequent gut microbiome analyses. Here we reported a proven cat-friendly protocol for fecal sample collection in clinical and research setting for gut microbiome analyses.
Abbreviations
-
- bp
-
- base pair
-
- BWA
-
- Burrows-Wheeler Aligner
-
- CPM
-
- counts per million mapped reads
-
- DNA
-
- deoxyribonucleic acid
-
- IACUC
-
- Institutional Animal Care and Use Committee
-
- LefSe
-
- linear discriminant analysis Effect Size
-
- miOil
-
- lubrication using mineral oil
-
- NCBI
-
- National Center for Biotechnology Information
-
- NIH
-
- National Institutes of Health
-
- noLub
-
- no lubricant applied
-
- PCoA
-
- principal coordinates analysis
-
- PE
-
- paired-end
-
- PERMANOVA
-
- permutational multivariate analysis of variance
-
- WGS
-
- whole-genome shotgun
1 INTRODUCTION
The gut microbiota is the collection of microorganisms that inhabit the host's gastrointestinal tract. There is clinical and physiological importance to understanding the gut microbiome based on its interactions with the host in healthy states and its involvement in the etiology of many diseases, including rheumatoid arthritis,1 colorectal cancer,2, 3 cardiovascular disease,4 and inflammatory bowel disease.5, 6
Low-stress handling and fear reduction techniques are the new standard of care for veterinary patients.7, 8 Using a lubricant can minimize stress, discomfort, and pain and prevent escalating fear in cats. An additional challenge of the dry collection (nonlubricated) approach using a fecal loop is that, in many cases, little to no fecal material is obtained, resulting in variable and insufficient amounts of material for metagenomic analysis, which leads to missing data and an imbalanced experimental design. During sample collection with a fecal loop, lubrication can consistently guarantee sufficient specimens for analysis. In addition to the previously mentioned welfare benefits, the lubrication approach will reduce potential host contamination. Fecal loop use could cause abrasion to the intestinal wall, leading to the shedding of host cells and bleeding, which increases host DNA contamination. Therefore, using lubricant will allow sufficient fecal sample collection with reduced incidence of bleeding, pain, discomfort, and potential infection and less chance for host contamination in the fecal samples. However, using lubricant might alter the microbial composition and abundance in the gut microbiome. Currently, no research has addressed this issue.
Reproducibility and stability of microbial profiles recovered from fecal samples are vital to the reliability of the analytical results. In this study, we evaluated the WGS metagenomic data consistency from cat fecal samples collected using mineral oil lubrication vs no lubrication, to provide information for veterinarians and researchers regarding appropriate methods to evaluate the cat gut microbiome.
2 MATERIALS AND METHODS
2.1 Animals
The study was approved by the Auburn University Institutional Animal Care and Use Committee (IACUC) with protocol number PRN 2019-3482. Eight obese, 6-year-old, neutered male cats were enrolled. All the animals were maintained at the Scott-Ritchey Research Center, College of Veterinary Medicine, Auburn University (Auburn, Alabama), and cared for according to the principles outlined in the NIH Guide for the Care and Use of Laboratory Animals.
2.2 Study design
The sample size of 8 was determined based on the rarefaction plots in our previous research on the feline microbiome,9 to ensure at least 90% of bacterial species and microbial genes in the reference assembly (NCBI Assembly ID JAIZPE000000000) were covered. Two fecal samples were collected from each of the 8 enrolled cats under sedation. The first batch of collections was performed without any lubrication (noLub sample group), and the second batch of collections was performed on the same day with mineral oil lubricant (miOil sample group). All fecal specimens were placed into 1.5 mL sterile Eppendorf tubes, flash-frozen, and stored immediately in a −80°C freezer (CryoCube F570, Eppendorf North America, Enfield, Connecticut).
2.3 Sedation and rectum fecal sample collection procedure
Adopting low-stress handling methods,8 cats were sedated to effect using a cocktail of intramuscularly administered medetomidine, ketamine, and butorphanol. For each participant, the small end of a plastic fecal loop (cat. no. 7500, Covetrus, Dublin, Ohio) was inserted into the rectum and descending colon to obtain an adequate amount (>200 mg) of feces. For the miOil samples, the fecal loop was coated with mineral oil (Equate, Bentonville, Arkansas), and the same sampling procedure was repeated.
2.4 Microbial DNA extraction and quality control
Genomic DNA samples were extracted from 200 mg fecal samples using the Allprep PowerFecal DNA/RNA kit (Qiagen, Redwood City, California) according to the manufacturer's protocols. To reduce technical variability, the homogenization step was performed for all fecal samples in the same batch using a PowerLyzer24 instrument (Qiagen, Redwood City, California). DNA concentrations were measured by a Qubit 3.0 Fluorometer (Thermo Fisher Scientific, Waltham, Massachusetts), and the A260/A280 absorption ratios were assessed by a NanoDrop One C Microvolume Spectrophotometer (Thermo Scientific, Waltham, Massachusetts).
2.5 Whole-genome shotgun metagenomic library construction and sequencing
For each sample, 1 μg of DNA was sheared into fragments of 500 bp by an M220 Focused-ultrasonicator (Covaris, Woburn, Massachusetts). Whole-genome shotgun metagenomic sequencing libraries were constructed using NEBNext Ultra II DNA Library Prep Kit for Illumina (New England BioLabs, Ipswich, Massachusetts). LabChip GX Touch HT Nucleic Acid Analyzer (PerkinElmer, Hopkinton, Massachusetts) was used to determine the library concentrations and size distributions, which were 600 bp on average, including sequencing adapters. The final libraries were measured by qPCR before being sequenced on an Illumina NovaSeq6000 sequencing machine in 150-bp paired-end mode at Novogene (Novogene Corporation Inc., Sacramento, California).
2.6 Processing and bioinformatic analysis of metagenomic data
The 16 metagenomes yielded a total of 1.28 billion raw sequencing reads (192 Gbp). After trimming the adapter sequences and low-quality bases using Trimmomatic (version 0.36),10 the high-quality reads were aligned to the feline reference genome Felis_catus_9.011 using Burrows-Wheeler Aligner (BWA; version 0.7.17-r1188)12 to detect host contaminations. The feline reads were extracted using SAMtools (version 1.6)13 and BEDTools (version 2.30.0).14 The remaining microbial reads were aligned to the feline gut metagenomic reference contigs (GCA_022675345.1)15 with comprehensive taxonomy assignments and microbial gene annotations. Taxonomic abundances were determined in the form of read counts, which were normalized by the total amount of sequences to obtain counts per million mapped reads (CPM) for subsequent metagenomic analyses.
2.7 Microbial diversity analyses
Alpha- and beta-diversity analyses were performed with the R package vegan (version 2.5.7).16 The alpha diversity measures of microbial profiles at different taxonomic levels and the microbial gene level were calculated using the Shannon index.17 Beta diversity was calculated based on the Bray-Curtis dissimilarity matrices18 generated from the composition of the microbial profiles. Visualization of the beta diversity was in the format of PCoA (Principal Coordinates Analysis) plot using the R software.19
2.8 Statistical analysis
The statistical analyses were performed using the statistical software package R, version 4.0.2.19 Differences in DNA yield (ng/mg fecal specimen), library yield (Gbp), percentage of host contamination, number of microbial genes, the relative abundance of specific taxa, and Shannon index from α-diversity analyses of microbial profiles at each taxonomic levels between groups, were analyzed using the Wilcoxon signed-rank test in the R software.20, 21 Permutational multivariate analysis of variance (PERMANOVA) test was performed using Adonis function in the R package vegan,22-26 to estimate the percentage of variability (R2) explained by different fecal sample collection methods based on Bray-Curtis distance matrices. When P-value was less than .05, the null hypothesis was rejected. To estimate the correlation of taxonomy composition of fecal samples collected using mineral oil lubrication or without lubrication, Spearman's rank-order correlation coefficients were calculated using the R software. The P-value was determined using Spearman's correlation test (a permutation test). To determine the differences in the variance, nonparametric Levene's test of equality of variances was performed using the R software.
3 RESULTS
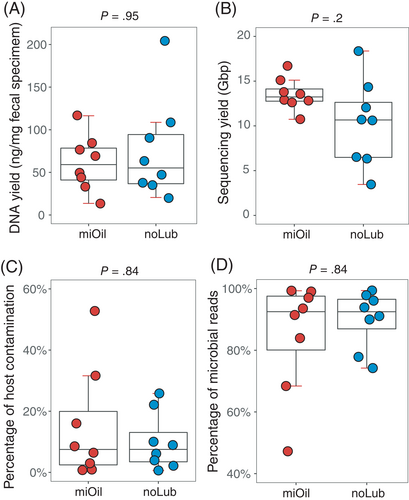
No significant differences were observed in DNA yield per mg fecal specimen between miOil and noLub groups (75.75 ng [25.80-125.70] vs 60.72 ng [33.49-87.95], P = .95, Wilcoxon signed-rank test; Figure 1A), suggesting a lack of effect on microbial DNA yield using mineral oil lubricant. In total, 1.28 billion 150-bp reads (or 192 Gbp reads) were generated in the WGS metagenomic sequencing of 16 fecal DNA samples. Of these, 1.2% were adapter sequences or low-quality bases, and 12.5% were cat sequences. The yield from the miOil group (13.5 Gbp on average) was not statistically different from the noLub group (10.3 Gbp; P = .2, Wilcoxon signed-rank test; Figure 1B). We did observe an increased variation in sequencing yield in the noLub group (23.1) compared to the miOil group (3.2), which is statistically significant (P = .04, Levene's test of homogeneity of variance). The percentage of host contamination was not significantly different between the miOil group and the noLub group (P = .84, Wilcoxon signed-rank test; Figure 1C,D).
In the WGS metagenomic data from the noLub group, on average, we discovered 84.5 phyla [75.1-93.9], 73.8 classes [69.1-78.4], 157.8 orders [146.4-169.1], 330.5 families [303.7-357.3], 1141.6 genera [1002.5-1280.7], and 4985.9 species [4300.3-5671.5]. Under mineral oil lubrication, there was no significant change in the number of phyla (86.6, [77.2-96]; P = .96), classes (75.1, [71-79.2]; P = .96), orders (162.4, [153-171.8]; P = .83), families (342.8, [323.1-362.4]; P = .83), genera (1196.6, [1076.2-1317]; P = .67), and species (5300.6, [4679.3-5921.9]; P = .67) detected in the rectum microbiome (Figure 2).
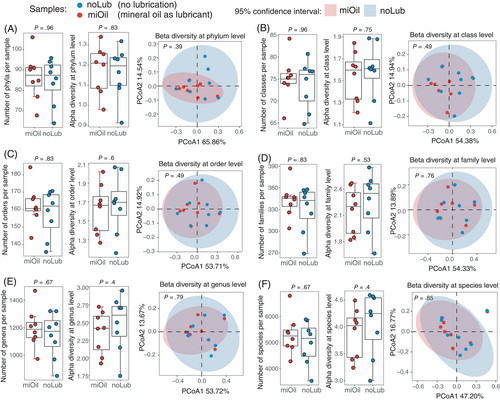
Alpha-diversity was measured for the noLub and miOil metagenomes using the Shannon index (Figure 2). No significant differences in α-diversity were detected between the noLub and miOil samples at phylum (1.16 vs 1.18, [1.06-1.27] vs [1.08-1.28]; P = .83), class (1.59 vs 1.54, [1.37-1.82] vs [1.35-1.73]; P = .75), order (1.67 vs 1.62, [1.41-1.93] vs [1.4-1.84]; P = .6), family (2.25 vs 2.15, [1.97-2.52] vs [1.93-2.37]; P = .53), genus (2.48 vs 2.35, [2.16-2.81] vs [2.1-2.6]; P = .4), and species level (4.06 vs 3.88, [3.56-4.55] vs [3.49-4.26]; P = .4; Figure 2). When β-diversity was examined using Bray-Curtis dissimilarity in PCoA analyses, no significant changes were detected either (P > .39 for comparisons at all 6 taxonomical levels; PERMANOVA test; Figure 2). The overlapping confidence intervals indicated the microbial compositions could not be distinguished between the 2 fecal sample collection methods.
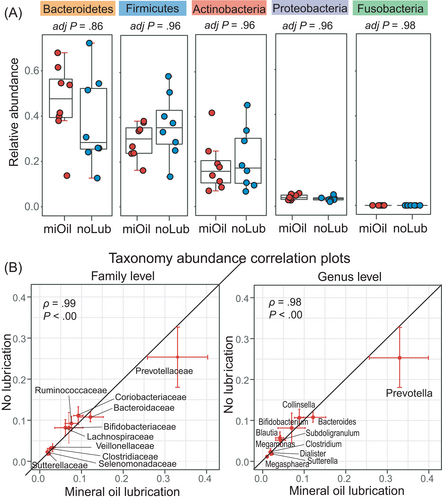
In our WGS metagenomic data, none of the 5 major phyla (Bacteroidetes, Firmicutes, Actinobacteria, Proteobacteria, and Fusobacteria) had significant differences in their relative abundance between miOil and noLub groups (P > .05, adjusted P > .85; Wilcoxon signed-rank tests; Figure 3A). In both the miOil and noLub groups, 99.72% of the metagenomic sequences belonged to bacteria reads, 0.11% were viral reads, 0.01% were archaeal reads, and the rest (0.16%) remained unknown. Considering the biological importance, we examined the microbial taxa with high relative abundance (>1.00%) at different taxonomical levels, which included 7 classes, 7 orders, 10 families, 11 genera, and 14 species. No significant changes were detected between miOil and noLub groups in the relative abundance at any taxonomic level (adjusted P > .8, Wilcoxon signed-rank test). The Spearman's rank-order correlation coefficients were extremely high between the noLub and miOil groups at the family (ρ = .99, P < .000001) and the genus levels (ρ = .98, P < .0000001; Spearman's rank-order correlation test; Figure 3B).
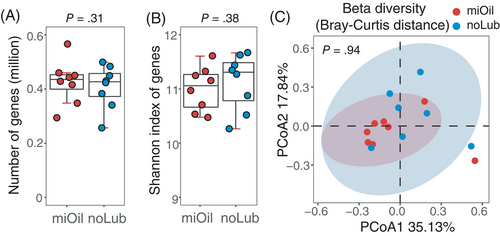
A total of 834 014 nonredundant microbial genes were identified in the 16 metagenomes. Of these, 798 430 genes were identified in 8 miOil metagenomes, and 769 476 genes were identified in 8 noLub metagenomes (Figure 4A). There was no significant difference in the number of observed genes between fecal samples collected with mineral oil and samples collected without lubrication (P = .31, Wilcoxon signed-rank test; Figure 4A). The alpha diversity based on the Shannon index of the observed genes from the 2 groups showed no significant difference either (P = .38, Wilcoxon signed-rank test; Figure 4B). Based on the Bray-Curtis distance matrix, no significant dissimilarity was detected between miOil and noLub groups in the PCoA analysis, and 95% confidence interval ellipses were overlapped (P = .94, PERMANOVA test; Figure 4C).
4 DISCUSSION
Fecal specimen collection is a commonly used approach in veterinary clinics and research to detect zoonotic parasites and diagnose pathogenic infections. Common methods for fecal sample collection in veterinary medicine include collection from the litter box and collection from the rectum using a fecal loop. Studies using cat fecal samples from litter boxes have 2 potential shortcomings. First, it is difficult to determine the freshness of the specimen, and excessive time at room temperature will shift the microbial composition. Second, it is likely the sample is contaminated by the environment. For projects designed to accurately represent the gut microbiome, fecal samples are collected from the rectum using a fecal loop, which can cause increased distress compared to litterbox collections. Lubrication is necessary to increase sample quality and to improve animal welfare during sample collection. In this group of cats, our results suggest that if adequately lubricated, the mineral oil applied will not affect the fecal microbial DNA extraction or gut microbiome analysis. The benefits of collection with lubrication include improved animal welfare and reduced variability in sequencing yield.
Fecal sample collection using a fecal loop in cats is challenging. It can cause discomfort, pain, and bleeding in the cat and result in little to no samples if it is done incorrectly. In an earlier attempt to collect fecal samples from the 8 cats enrolled in this research using a dry collection approach, we were only able to obtain a sufficient quantity of feces from 6/8 cats, and bleeding was observed in 5/8 cats. This failure led us to explore modifications to the fecal sample collection methods. Lubrication can ease the fecal sample collection process and ensure a sufficient amount of samples, but the lubricant could introduce materials that could prevent DNA extraction and sequencing or alter the microbiome composition. To determine if using lubricants during fecal sample collection has potential effects on the gut microbiome, we performed fecal collections with lubrication and without lubrication on the same 8 cats. The sample size was determined from the rarefaction plots from a previous study, in which a sample size of n = 6 will detect >90% of the bacterial species and microbial genes in the cat reference microbiome.15 In our metagenomic data analyses, we did not observe any significant changes in alpha-diversity, beta-diversity, the number of taxa discovered at each taxonomy level, and the relative abundances of taxonomic units are also highly correlated, indicating that the microbial composition was not affected by the use of lubricant.
We expected to see less host sequence contamination in the samples collected with mineral oil lubrication because the use of the fecal loop without lubrication is more likely to damage the intestinal wall, resulting in a higher proportion of host contamination. However, we did not observe any significant differences in host contamination, which could be because we sampled the same cat on the same day, and the more readily sloughed host cells had already exfoliated after the fecal loop collection without lubrication. The level of host DNA contamination (12.5%) is acceptable in both groups, given that intestinal cells are constantly sloughed off into the gut lumen.
Lubrication did not cause any issues in microbial DNA extraction, and a similar yield was observed in both groups, which is sufficient for subsequent research purposes. Interestingly, we observed a lower variability of the metagenomic sequencing yield, and this homogenous yield across the samples is beneficial for achieving an even level of metagenome coverage.
Whole-genome shotgun (WGS) metagenomic sequencing was performed in this study to assess the feline microbiome, as this method is rapidly becoming the new standard for assessing microbiomes across all species. We anticipate that the results will be applicable to 16S rDNA ampliconic sequencing, because a high correlation in taxonomic composition was observed in the feline microbiome using these 2 approaches.15
One limitation of this study is that our results only applied to fecal samples stored immediately in an ultracold freezer after collection. The same results might not hold in other storage conditions (room temperature, 4°C refrigeration, DNA stabilizing solution, etc.).
ACKNOWLEDGMENT
Funding provided by Alabama Agriculture Experiment Station, Auburn University College of Veterinary Medicine Animal Health and Disease Research, Auburn University Intramural Grant Program (number 180254), EveryCat Health Foundation (number EC22-023), National Science Foundation (number 1928770), and United States Department of Agriculture National Institute of Food and Agriculture (number 1018100).
CONFLICT OF INTEREST DECLARATION
Authors declare no conflict of interest.
OFF-LABEL ANTIMICROBIAL DECLARATION
Authors declare no off-label use of antimicrobials.
INSTITUTIONAL ANIMAL CARE AND USE COMMITTEE (IACUC) OR OTHER APPROVAL DECLARATION
The study was reviewed and approved by the Auburn University IACUC, project number 2019-3482.
HUMAN ETHICS APPROVAL DECLARATION
Authors declare human ethics approval was not needed for this study.
Open Research
DATA AVAILABILITY STATEMENT
The whole-genome shotgun metagenomic sequencing data is available at NCBI SRA under accession number PRJNA821230.




