Development and Validation of a High-Sensitivity Droplet Digital PCR Assay for Serum Hepatitis B Virus DNA Detection
ABSTRACT
Real-time polymerase chain reaction (PCR) is the current standard for serum HBV DNA measurement. However, conventional real-time PCR assays have technical limitations, and are not sensitive enough to detect low-level residual viremia in chronic hepatitis B (CHB) patients. We developed and validated a droplet digital PCR (ddPCR) assay for high-sensitivity detection of serum HBV DNA. A ddPCR assay was developed on the QX200 ddPCR System (Bio-Rad) for detection of serum HBV DNA in 200 μL of serum. The primers and probe were designed to target a highly-conserved region in the HBV X gene. The AcroMetrix HBV Panel (Thermo Fisher Scientific) and CHB patient samples were used for validation experiments to determine the assay sensitivity, specificity, linearity, intra-run variability, and inter-run variability. The ddPCR assay demonstrated lower limit of detection of 1.6 IU/mL and lower limit of quantification of 9.4 IU/mL for serum HBV DNA in probit regression. The assay also achieved excellent specificity (96.2%), linearity (R = 0.994, R2= 0.988, p < 0.001), intra-run variability (mean coefficient of variation [CV]: 0.69%, average intra-run difference: 0.026 log IU/mL), and inter-run variability (mean coefficient of variation [CV]: 4.54%, average inter-run difference: 0.18 log IU/mL). To conclude, we developed a robust ddPCR assay that achieved higher detection sensitivity with lower serum input volume than conventional real-time PCR assays. Our assay may be utilised for measuring residual viremia after nucleos(t)ide analogue therapy or for monitoring patients on novel HBV antivirals.
1 Introduction
Hepatitis B virus (HBV) DNA is an important biomarker in the management of chronic hepatitis B (CHB), as it reflects viral replication, guides treatment decisions, and predicts long-term risks of liver-related complications [1].
Real-time polymerase chain reaction (PCR) is the current standard for serum HBV DNA detection, with multiple commercial assays available. Despite the widespread use of real-time PCR, it is not without limitations. Real-time PCR requires external calibration for calculating the quantification cycle value, has high performance variation, and is prone to interference [2]. Conventional assays also generally have a serum HBV DNA lower limit of detection (LLOD) of 10–20 IU/mL. This detection sensitivity is insufficient to detect low-level residual viremia in CHB, which is important for HCC risk assessment in patients receiving nucleot(s)ide analogues (NUCs) [3]. Furthermore, most conventional assays require a serum input of 500 μL or more, which may necessitate additional bloodtaking from patients.
Droplet digital PCR (ddPCR) is a PCR technique that enables absolute quantification of target DNA with high sensitivity. ddPCR does not require external calibration for quantification and is also less prone to influence by PCR inhibitors, cross-contamination, or background noise [2]. ddPCR has been explored for serum HBV viral load detection, although the LLOD and performance of such ddPCR assays have not been well described. Given the limitations of prior studies, we developed and validated a ddPCR assay for high-sensitivity detection of serum HBV DNA.
2 Methods
The QX200 ddPCR System (Bio-Rad) was utilized for all experiments in this study.
2.1 Primer and Probe Design
The primers and probe for our ddPCR assay were designed in accordance with the manufacturer's (Bio-Rad) protocol. 37 reference HBV genomes (encompassing genotypes A–H) were used for primer and probe design. The primer and probe were designed to target a highly conserved region in the HBV X gene to amplify a 160-nucleotide length sequence. The sequences of the forward and reverse primers are 5′-CGTCTGTGCCTTCTCATCTG-3′ and 5′-TGAAGTATGCCTCAAGGTCG-3′, respectively. The sequence of the probe is 5′-ACCACCGTGAACGCCCACCAGGT-3′.
2.2 Droplet Digital PCR Workflow
2.2.1 DNA Extraction
Serum input volume of 200 μL was selected for our ddPCR assay, which is lower than that in most conventional real-time PCR assays. Serum DNA extraction was performed with the QIAamp MinElute Virus Spin kit (QIAGEN) in accordance with the manufacturer's protocol. The extracted DNA was eluted to a total volume of 16 μL.
2.2.2 Mastermix Preparation
The PCR Mastermix was prepared with 0.72 μL of the forward primer (concentration: 25 uM), 0.72 μL of the reverse primer (concentration: 25 uM), 0.5 μL of the probe (concentration: 10 uM), and 10.56 μL of the ddPCR Supermix (Bio-Rad). Approximately 8.5 μL of the extracted DNA was added to make up a reaction mix of 21 μL.
2.2.3 Droplet Generation
The reaction mix was then transferred to DG8 Cartridges for QX200 (Bio-Rad) and combined with 70 μL of Droplet Generation Oil for Probes (Bio-Rad) to form emulsified droplets by the QX200 Droplet Generator (Bio-Rad). The emulsified droplets were then transferred to a 96-well plate through slow pipetting (over 5 s) to avoid lysing the droplets.
2.2.4 Thermal Cycling
The sealed plate was taken to thermal cycling in the C1000 Touch Thermal Cycler (Bio-Rad). The cycling protocol involved enzyme activation at 95°C for 10 min, 50 cycles of denaturation at 94°C for 30 s, 50 cycles of annealing/extension at 60°C for 1 min, followed by deactivation at 98°C for 10 min.
2.2.5 Droplet Reading
After thermal cycling, the 96-well plate was taken for analysis within 24 h in the QX200 Droplet Reader (Bio-Rad). The reader detected fluorescence from the HEX dye and interpreted each droplet as positive (fluorescence detected) or negative (fluorescence not detected). The QuantaSoft software (Version 1.7, Bio-Rad) performed automatic thresholding based on the amplitude of fluorescence. Finally, the software utilised Poisson statistics to calculate HBV DNA concentration for each sample.
2.3 Testing Samples
The AcroMetrix HBV Panel (Thermo Fisher Scientific) was utilised in validation experiments for assay sensitivity and linearity. The AcroMetrix HBV Panel is calibrated against the WHO International Standard for HBV DNA and is frequently utilised for validation of serum HBV DNA assays [3]. The HBV DNA concentration in the AcroMetrix Panel ranges from 50 IU/mL to 50,000,000 IU/mL. For required HBV DNA concentrations < 50 IU/mL, serial dilutions of the AcroMetrix Panel were performed until the desired concentration was reached.
Archived serum samples from 10 CHB patients were also utilised in validation experiments for intra-run and inter-run variability. This study was performed in accordance with the 1975 Declaration of Helsinki and was approved by the University of Hong Kong/Hong Kong West Cluster Institutional Review Board (UW24-470). The consent for this study was waived by the Institutional Review Board as it involved the use of archived serum samples without identifiable information.
2.4 Validation Experiments
The ddPCR assay sensitivity was determined by probit regression on serial dilutions of the AcroMetrix HBV Panel. LLOD was defined as the lowest concentration at which 50% of positive samples were detected, while the lower limit of quantification (LLOQ) was defined as the lowest concentration at which 95% of positive samples were detected [4, 5]. Specificity of the ddPCR assay was determined by applying the ddPCR assay to 52 negative controls. Measurements with negative readings were assessed as true negatives, while positive readings were assessed as false positives. Specificity was calculated as (true negative)/ (true negative + false positive). Linearity of the ddPCR assay was determined by comparing the measured HBV DNA concentrations with expected HBV DNA concentrations in serial dilutions of the AcroMetrix HBV Panel in 6 repeat runs, with the Pearson correlation coefficient test used to assess the linear correlation. The intra-run variability was tested on 5 CHB patient samples with 4 repeated measurements in the same run, whereas the inter-run variability was tested on 5 CHB patient samples on 3 separate runs. The assay variability was presented as the coefficient of variation (CV), calculated by (standard deviation of DNA concentration)/(mean DNA concentration).
3 Results
3.1 Assay Sensitivity and Specificity
Serial dilutions of the AcroMetrix HBV Panel down to an HBV DNA concentration of 1 IU/mL were tested with the ddPCR assay. In probit regression, the LLOD of the ddPCR assay was determined to be 1.6 IU/mL (95% CI 0.4–2.8 IU/mL), and the LLOQ was determined to be 9.4 IU/mL (95% CI 4.5–14.2 IU/mL).
On measurement of 52 negative control samples, weak positive DNA readings at detectable but unquantifiable levels were noted in 2 samples only, giving an assay specificity of 96.2%. The false positive readings may be due to noise or interference in testing and were less likely to be due to contamination, as the readings were only at the weak positive range.
3.2 Assay Linearity
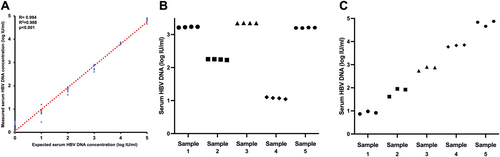
Serial dilutions of the AcroMetrix HBV Panel from 0 log IU/ml to 5 log IU/ml were tested with ddPCR in 6 repeat runs. The measured HBV DNA concentration significantly correlated with the expected concentrations (R = 0.994, R2 = 0.988, p < 0.001) (Figure 1A).
3.3 Intra-Run Variability
In 4 intra-run repeat measurements on 5 patient samples, the CV ranged from 0.09% to 2.26%, and a mean CV of 0.69% was achieved. The average intra-run difference in HBV DNA concentration was 0.026 log IU/mL (Figure 1B).
3.4 Inter-Run Variability
In 3 separate runs on five patient samples, the CV ranged from 1.10% to 10.15%, and a mean CV of 4.54% was achieved. The average inter-run difference in HBV DNA concentration was 0.18 log IU/mL (Figure 1C).
4 Discussion
We presented the development and validation experiments for a ddPCR serum HBV DNA assay. The ddPCR assay was designed based on reference genomes from all HBV genotypes and should have pangenotypic diagnostic capability through targeting a conserved sequence in the X gene. The ddPCR assay achieved HBV DNA LLOD and LLOQ of 1.6 IU/mL and 9.4 IU/mL, respectively, which are more sensitive than commercially available real-time PCR assays. Our assay also demonstrated excellent specificity, linearity and intra-run/inter-run repeatability. Furthermore, our assay can operate with 200 μL serum as the input volume, which minimises the consumption of patient samples and facilitates the testing of archived samples. Considering the assay sensitivity, assay performance and the minimal required sample volumes, we have developed a high-performing ddPCR assay for serum HBV DNA.
Conventional real-time PCR HBV DNA assays are not sensitive enough to detect low-level residual viremia in patients on NUCs or with hepatitis B surface antigen seroclearance. Residual viremia in CHB is associated with hepatocellular carcinoma (HCC) [3], yet research in the area is minimal due to current assay limitations. High-sensitivity HBV DNA detection hence presents an opportunity to measure residual viremia for risk prediction. ddPCR may be utilised to guide personalised follow-up and HCC surveillance strategies in CHB. Our novel assay may also have a role in monitoring patients on novel HBV antivirals—such as new-generation capsid assembly modulators, which induce deeper HBV DNA suppression than NUCs [6, 7]. Residual viremia detection will be essential for treatment response assessment as we enter the era of novel antivirals.
A limitation of ddPCR is its complex workflow, which involves special lab techniques and equipment. Nonetheless, if ddPCR is widely adopted, the testing protocols can be streamlined, and trained personnel should be able to perform the experiments efficiently. The cost of performing ddPCR is also expected to reduce as testing is scaled up.
To conclude, our data support the utility of ddPCR for serum HBV DNA detection. The importance of developing high-sensitivity HBV DNA assays was highlighted in the 2022 expert consensus on HBV treatment endpoints [8]. Our assay, hence, covers an important research gap and has the potential to alter clinical practice.
Author Contributions
R.W.-H.H. was involved in data acquisition, data interpretation and drafting of the manuscript. D.K.-H.W., L.-Y.M., J.F. and W.-K.S. were involved in the critical revision of the manuscript. M.-F.Y. was involved in the study concept, critical revision of the manuscript and overall study supervision. All authors have seen and approved the final version of the manuscript.
Acknowledgements
The authors have nothing to report.
Conflicts of Interest
L.-Y.M. is an advisory board member for Gilead Sciences. W.-K.S. received speaker's fees from AstraZeneca, is an advisory board member and received speaker's fees of Abbott, received research funding from Alexion Pharmaceuticals, Boehringer Ingelheim, Pfizer and Ribo Life Science and is an advisory board member, received speaker's fees and researching funding from Gilead Sciences. M.-F.Y. serves as advisor/consultant for AbbVie, Assembly Biosciences, Aligos Therapeutics, Arbutus Biopharma, Bristol Myer Squibb, Clear B Therapeutics, Dicerna Pharmaceuticals, Finch Therapeutics, GlaxoSmithKline, Gilead Sciences, Immunocore, Janssen, Merck Sharp and Dohme, Hoffmann-La Roche and Springbank Pharmaceuticals, Vir Biotechnology and receives grant/research support from Assembly Biosciences, Aligos Therapeutics, Arrowhead Pharmaceuticals, Bristol Myer Squibb, Fujirebio Incorporation, Gilead Sciences, Immunocore, Merck Sharp and Dohme, Hoffmann-La Roche, Springbank Pharmaceuticals and Sysmex Corporation. The remaining authors have no conflicts of interest.
Open Research
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.




