A review on protein utilization and its interactions with carbohydrate and lipid from a molecular perspective in aquaculture: An implication beyond growth
Abstract
The world's increasing need for protein faces challenges in aquaculture production. New applications and tools will need to be added at every stage of the manufacturing line to attain this expansion sustainably, safely, and effectively. Utilizing experimental methods to increase aquatic animal production has become more common as aquatic biotechnology has advanced. High-throughput omics technologies have been introduced to address these issues, including transcriptomic, metabolomic, proteomic, and genomes. But it also faces many difficulties, like other food manufacturing industries. One of the best and most durable approaches to address these issues is probably to understand nutritional requirements and modify diet based on need. Molecular approaches are a subset of multiomics technology. Previously, most of the published work was devoted to the biochemical aspects of protein–lipid interactions in biological systems. In this review, we explore this idea and highlight various works that fall under the umbrella of nutrigenomics, with a particular emphasis on protein utilization and its interactions with carbohydrates and lipids.
1 INTRODUCTION
Proteins constitute the major feed components in aquaculture. Its contributions to the functional and nutritional properties of feeds are well established. Protein, the most expensive ingredient in fish feed, is also deemed the most important because its regular intake requires the fish to utilize amino acids to build new proteins during growth and reproduction or to replace existing ones during maintenance (Abasubong et al., 2018). Inadequate protein in feed results in growth reduction, but when it is overloaded in a diet, the excess protein is converted to energy through the direct oxidation of amino acids. However, the immune response is highly influenced by the nutritional background (Oliva-Teles, 2012), and the use of plant-based diets is often accompanied by intestinal infiltration of inflammatory cells and impaired disease outcomes (Estensoro et al., 2016; Piazzon et al., 2017; Romarheim et al., 2013; Urán et al., 2008). This drawback effect is remediated, at least partially, using different dietary supplementation strategies (Table 1).
| Protein sources | Advantages | Disadvantages | References |
|---|---|---|---|
| Soybean meal | The essential amino acid (EAA) profile is favourable, and the protein level is high. Possibility of making soy protein concentration (low in antinutritional factors, soluble carbohydrates) | Contains saponins, phytic acid, and some processing techniques (solvent extraction) that leave antinutrient molecules in the food | Bandara (2018) |
| Canola/Rapeseed | Ability to produce canola protein concentrate by aqueous extraction of fibre (higher protein content). Good source of linolenic acid and low levels of linoleic acid in rapeseed oil | It contains a higher amount of fibre and glucosinolates. Higher content of phytic acids and glucosinolates | Bandara (2018) |
| Sunflower meal | Dehulling and other preprocessing processes remove a greater amount of fibre | Higher content of fibre. Higher in protease inhibitors, arginase inhibitors, and phytic acids | Bandara (2018) |
| Wheat gluten meals | Most fish have higher digestibility. Fish distal intestine tissues show no morphological alterations | Deficiency in lysine, methionine, and arginine | Bandara (2018) |
| Rice protein concentrate | High protein and lipid content | Deficiency in lysine | Abasubong et al. (2018) |
| Poultry by-products | Free from antinutrient | Expensive feed ingredients in aqua diets and low levels of lysine, methionine, and histidine | Bandara (2018) |
| Feather meal (hydrolysed) | They are rich in cystine (74%–61%) and protein content | Very hard to digest, low in lysine and methionine content. During processing with application heat typically deteriorates haemoglobin and causes low palatability | Grazziotin et al. (2008); Yu et al. (2020) |
| Blood meal (Cow blood) | Higher protein content and relatively rice-in lysine content | The deficiency of Methionine content heat sensitivity and drying conditions have a big impact on protein digestion | Aladetohun and Sogbesan (2013); Hussain et al. (2011) |
| Guar meal | Guar meal can be used in place of soy meal without affecting the growth of some fish | Antinutritional and antidigestive chemicals, such as phytate, protease, residual gum, saponin, and inhibitor tannin, are present. Gastrointestinal evacuation takes time and indigestible amino acids | Nidhina and Muthukumar (2015); Ullah et al. (2016) |
| Lupins | Arginine and glutamic acid levels are high, but antinutritional factors are low | Deficiency in methionine and lysine and contain alkaloid compound | Bandara (2018) |
To achieve zero waste in the agro-food value chain and feed cost-effectiveness, however, substantial research is required on innovative fish feed formulations that follow the circularity principles and optimize resource efficiency (Campos et al., 2020). As a result, some of the most promising substitutes for producing aquafeed are single-cell biomasses and oils, insect meal products, and aquaculture by-products (Figure 1). Of course, microbial biomass production is more intriguing than other animal or plant protein sources due to its quick production timeframes, low land needs, and weather independence (Piazzon de Haro et al., 2022). The effectiveness of using them in place of marine feedstuffs has been evaluated in fish and shrimp, with varying degrees of success depending on the species, amount of replacement, and kind and source of microbial biomass (Davies & Wareham, 1988; Delamare-Deboutteville et al., 2019; Hardy et al., 2018).
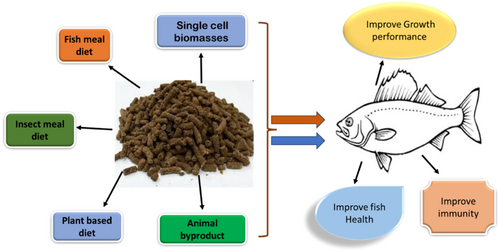
In addition, fish require energy satiation to feed; therefore, the diet should have the right amount of protein and nonprotein energy-supplying components. This will allow the fish to consume all the necessary nutrients within the allotted feed intake amount (Guillaume, 2001). Since lipids are the most bioavailable component in fish, they can preserve protein for growth more successfully than carbohydrates. Many biological structures in biological cells and tissues are organized in a desired way due to interactions between proteins, lipids, and carbohydrates. For instance, protein–lipid interactions influence the morphology and physiology of membrane cells and cell organelles. Complexes of proteins, lipids, and carbohydrates have a variety of uses in cellular membranes, lipid transport, and metabolism. Because of how these elements naturally interact to support aquaculture methods, they also impact different feed qualities. Therefore, the protein, fat/lipid, and carbohydrate combination must be appropriate for encouraging growth, immunity, metabolic response, and biological pathways to achieve optimal growth without undue nutritional loss. The assessment of the impact of various nutrient formulations on fish species has been made easier in these areas because of recent advancements in biotechnology. Hence, ‘molecular’ techniques represent one of the biotechnology's most promising fields.
Of these ‘molecular’ approaches, genomics (the study of genomes) has been used in genetic improvement, nutrition health, and pathogen characterization (Jun et al., 2020; Tekedar et al., 2020) to improve feed production. Additional subfields of research include proteomics, which studies changes in protein expression, dynamics, and posttranslational modifications, and transcriptomics, which profiles gene expression in a cell, tissue, or organism (Whitfield & Kirwan, 2010). Transcriptomics, metabolomics, and proteomics are increasingly used in aquaculture (Raposo de Magalhães et al., 2020). This information gives insight into what appears to be occurring within the body at any moment (Figure 2). The speed at which these fields are developing and expanding is largely due to recent developments in analytical platforms) and bioinformatics, which access and analyze vast amounts of previously unattainable data (Berger et al., 2013; Mutz et al., 2013). This work intends to review the impacts of protein from a molecular perspective comprehensively. Also, the molecular interactions with lipids and carbohydrates will be reviewed.
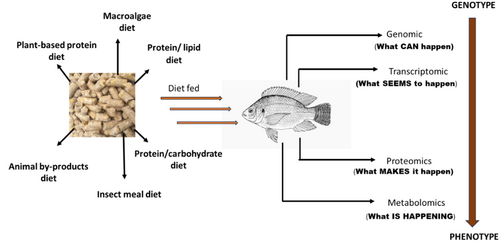
2 OVERVIEW OF MOLECULAR TOOLS IN AQUACULTURE
Gene is defined as a sequence of nucleotides/segments of chromosome/unit of heredity. Genes encode proteins, and proteins dictate cell function. Transcription and translation are mechanisms used to express the genetic information contained in DNA (Lockhart & Winzeler, 2000). DNA's genetic information is expressed through the processes of transcription and translation (Lockhart & Winzeler, 2000). The cell's nucleus is where transcription, also known as DNA to messenger RNA (mRNA), occurs. This process changes genetic information from DNA to RNA (Birnstiel et al., 1985). This data is sent via RNA from the nucleus into the cytoplasm, where translation takes place and directly aids in the synthesis of proteins (Birnstiel et al., 1985). Molecular techniques and tools have been used to address biological issues in potential aquaculture species. Recent advances in computational tools and high-throughput sequencing technology have opened the door to a greater understanding of biological processes, including development, immunity, physiology, evolution, stress, and adaptations. Several techniques, including genomics, molecular biology, and biochemistry, have been used to study these fish at the molecular level. They provide thorough biomolecule characterization and quantification that replicates the responses of an organism to environmental and internal stimuli (Hickok & Maslej, 2023). Genomics (Snyder & Gerstein, 2003) has been employed in several fields, including genetic improvement (Jun et al., 2020), nutrition (Skugor et al., 2011), health and pathogen characterization (Tekedar et al., 2020), immunization, and therapeutic development (Mweemba Munang'andu et al., 2018). Proteomics (Graves & Haystead, 2002) and transcriptomics (Chandhini & Rejish Kumar, 2019) are two more subdisciplines.
Aquaculture genomic research's primary goals include higher growth and development rates, increased feeding efficiency, resistance to illness and infections, and molecular phenotypic selection for better broodstock traits (Yuvarajan et al., 2019). In commercially valuable fish species like Atlantic halibut (Hippglossus hippoglossus), common cap (Cyprinus carpio) (Abasubong et al., 2022), and so on, differential gene expression has been employed to alleviate production issues. Fish adaptive evolution is believed to be significantly influenced by gene expression and regulation (Cresko et al., 2004). Aquaculture and fisheries can benefit from the genetic mechanisms causing adaptive differences in critical physiological features (Garcia de Leaniz et al., 2007) by creating new technologies and fundamentally altering how genetic information is used in culture management.
Proteomics has become a potent instrument for understanding biological systems and their behaviour under many circumstances. As a result, this technology has been used increasingly to address various fish biology-related issues in recent years. Numerous studies have documented the impact on fish health and, consequently, on food quality of radiation exposure (Andersson et al., 2004), viral infection (Booy et al., 2005), and stress resulting from handling, anoxia, or density (Provan et al., 2006; Wulff et al., 2008). Research on pathogen infection using proteomics has also been conducted in families of farmed channel catfish (Ictalurus punctatus) with high and low susceptibilities to enteric septicaemia of catfish to find novel genes linked to disease resistance (Booth & Bilodeau-Bourgeois, 2009). The anoxia effect in crucian carp (Carassius carassius) has also been studied (Andersson et al., 2004). Quality of gametes, embryos, and nutrition, which are directly related to alevin growth, developmental competency, and fertility, respectively, is another problem in farmed fish that has been studied using proteomics. The proteome of the females' coelomic fluid was examined to make educated guesses about the proteins that either increase or vanish concurrently with the decline in egg viability during postovulatory aging in trout (Rime et al., 2004).
According to Lankadurai et al. (2013), metabolomics aims to assess the effects of physical, chemical, and biological stresses on the metabolisms of wild and farmed organisms, including fish. For instance, studies on pollution exposure have demonstrated that metabolomics was a more sensitive approach than conventional methods when evaluating the effects of pollutants on Japanese medaka (Oryzias latipes) and eggs (Viant et al., 2006). Several metabolomic studies have looked into the impact of cyanobacterial blooms (Sotton et al., 2017), heavy oils (Kokushi et al., 2012), pesticides (Viant et al., 2006), endocrine-disrupting chemicals (Samuelsson et al., 2006), and heavy metals (Santos et al., 2020). The primary uses of metabolomics for fish health include the study of disease characterization (Southam et al., 2008), host–pathogen interactions (Guo et al., 2014; Peng et al., 2015), and therapy efficacy (Zhao et al., 2015). It is interesting to note that metabolomics techniques are used to create feed compositions that improve treatment efficacy (Cheng et al., 2014) or nutraceutical feed that delays the onset of disease (Robles et al., 2013; Silva et al., 2014).
These days, aquaculture extensively uses transcriptome profiling to identify different gene expression patterns and create new choosing markers. Gene expression, molecular markers, and novel genes have all been found through large transcriptome research. Nuclear receptors, a biological marker for sex determination, were found to be exclusively expressed in female shrimps and RNAi genes, according to transcriptome sequencing of the common caridean shrimp Crangon crangon in five different phases. According to Christiaens et al. (2015), RNA interference genes are utilized as antiviral therapies. At the same time, the nuclear receptor superfamily is a family of transcription factors necessary for cell differentiation, growth, development, reproduction, and moulting. Numerous significant aquaculture species, such as Nile tilapia Oreochromis niloticus (Zhang et al., 2013), Barramundi Lates calcarifer (Xia & Yue, 2010), Orange-spotted grouper Epinephelus coioides (Huang et al., 2011), Giant freshwater prawn (Macrobrachium rosenbergii) (Jung et al., 2011), Chinese mitten crab Eriocheir sinensis (Li, Cui, et al., 2013), eastern oyster Crassostrea virginica (Zhang et al., 2014), grass carp Ctenopharyngodon idella (Chen et al., 2012), and Chinese fleshy prawn Fenneropenaeus chinensis (Li, Zhang, et al., 2013) are among the many significant aquaculture species now available for de novo transcriptome analysis. In aquaculture, transcriptome sequencing has proven useful for identifying the genes and metabolic pathways behind superior performance features in growth, immunity, and reproduction.
3 THE ROLE OF PROTEIN SOURCES ON FISH SPECIES FROM A MOLECULAR PERSPECTIVE
Protein typically has an inclusion level of between 30% and 50%, making it the most expensive ingredient in fish diets. In fish diets, fishmeal (FM) has been the primary source of protein for many years because of its high protein content and well-balanced amino acid profile (Glencross et al., 2020). Individual amino acid bonds form the structure of proteins. About 20 amino acids are prevalent, although more than 200 amino acids exist. Fish cannot produce 10 of these amino acids, making them essential. The 10 necessary amino acids—lysine, valine, phenylalanine, isoleucine, histidine, arginine, threonine, tryptophan, and histidine—must be obtained by diet. Methionine and lysine are frequently the first limiting amino acids among them. Furthermore, FM has been called the ‘golden standard’ due to its great palatability, high nutrient digestibility, and general lack of antinutritional elements (Gatlin et al., 2007; Hardy, 2010). Global demand for FM affects its availability for use as an ingredient in diets for various aquaculture species making plant-based proteins an ideal alternative. However, plant protein sources contain antinutritional components and nonstarch polysaccharides, fatty acid, and amino acid profiles, which are less suitable for fish (Abasubong et al., 2018). Due to this, there has been a recent surge in research on novel ingredients for fish feeds, including macroalgae, insect meals, microalgae, and other single-cell proteins as an alternative for FM replacement (Naylor et al., 2021). Macroalgae are multicellular, macroscopic algae with varying nutritional values depending on the species. Several studies have recently focused on using insect meal to replace plant protein and FM in aquaculture (Fontes et al., 2019; Mancuso et al., 2019). With their intriguing nutritional profile, insects are a sustainable ingredient.
At the molecular level, gilthead fed plant-based diet observed an upregulated interleukin 1 beta (il-1β), tumour necrosis factor alpha (tnfα), major histocompatibility complex-II (mhcII), and cyclooxygenase-2 (cox-2) expression than the control after eight weeks of feeding. Also, high mortality rates observed in gilthead seabream fed a diet consisting entirely of broad bean, soybean, pea, and sunflower meals in place of FM were consistent with lower expression of immune-related genes like immunoglobulin (igm) and proinflammatory response genes like il-1β, il-6, and cox-2 (Estruch et al., 2018). It has also been shown that high doses of soybean meal (SBM) included in the diets of some fish species might cause intestinal irritation with and display elevated expression of proinflammatory genes (tnfα, il-1β, il-8, il-17, and il-4), lowered expression of anti-inflammatory genes (transforming growth factor beta 1 [tgfβ1]), and immune parameters (lysozyme, complement factors C3 and C4, and IgM) in hybrid grouper after 10 days (Hedrera et al., 2013; Zhang et al., 2021). Molecular findings of different protein sources are presented in Table 2
| Fish species | Protein ingredients and Level of ingredients | Duration of feeding | Molecular findings | References |
|---|---|---|---|---|
| Red tilapia juveniles Oreochromis aureus × Oreochromis mossambicus | CP levels (24%, 30%, 36% and 42%) | 42 days | Procarboxypeptidase, lipase, and leptin expression did not differ significantly across any of the groups. The expression of ghrelin and insulin decreased, and that of cholecystokinin and peptide yy increased in response to the 42% CP diet | Santos et al. (2020) |
| Juvenile blunt snout bream Megalobrama amblycephala | Plant proteins supplemental essential amino acids | 8 weeks | nf-κb, il-8, tnf-α, il10, and their relative expressions were all markedly lower in the FM-reduced groups. The relative expression of signal transducer and transcription activator 4 (STAT4) was dramatically elevated by the addition of EAA | Mokrani et al. (2020) |
| Common carp (Cyprinus carpio) | Chicken intestinal hydrolysates 0% (CIH-0), 25% (CIH-25), 50% (CIH-50), 75% (CIH-75), and 100% (CIH-100) of the fishmeal with CIH | 8 weeks | tnf-α and il-1β expression were lower in the CIH-50 group, whereas the CIH-50 and CIH-75 groups had significantly higher expression levels of tgf-β2 and il-10. zo-1, occludin, and mlck were considerably upregulated compared with the CIH-0 group. In the CIH-75 and CIH-100 groups, there was a downregulation of claudin-1 expression | Wu et al. (2022) |
| Pearl gentian grouper (Epinephelus fuscoguttatus ♀ × Epinephelus lanceolatus ♂) | 0% (0 g kg−1), 10% (50 g kg−1), 20% (100 g kg−1) and 30% (150 g kg−1) fishmeal were replaced with BSF | 8 weeks | Significant upregulation of nf-κbem1, r-cel, and il-10 mRNA levels was seen in response to elevated dietary BSF levels. The mRNA levels of myd88 and tlr22 in fish fed the BSF30 diet were notably greater than the FM diet | Huang et al. (2022) |
| Largemouth bass, Micropterus salmoides | Composite mixture of shrimp hydrolysate and plant proteins 45 (FM45, the control), 40 (FM40), 35 (FM35), 30 (FM30) and 25% (FM25) | 8 weeks | The substitution of fishmeal resulted in a large drop in 4e-bp1 expression but increased tor and s6k expression, indicating that the tor pathway had been activated | Li et al. (2021) |
| Juvenile channel catfish, Ictalurus punctatus | Cricket meal (Gryllus bimaculatus) 0%, 25%, 50%,75% and 100% CM | 10 weeks | Immune-related gene expression levels of il-1β, il-8, il-10, and hif1α were upregulated in the intestine of fish fed CM75 group compared to others. Significantly higher tnf-α, il-22, and ifn-γ expression levels were seen in the fish fed CM100 group. Interestingly, fish fed CM50 to CM100 groups showed a downregulation of nf-κb expression compared to the other groups | Fan et al. (2023) |
| Juvenile large yellow Croaker (Larimichthys crocea) | Fishmeal protein with degossypolized cottonseed protein diets replaced fishmeal protein with 0%, 20%, 40%, 60%, and 80% DCP | 70 days | The DCP20 and DCP40 groups exhibited a considerable upregulation in the transcription of hepatic tnf-α, il-6, and ifn-γ compared to the control group. Hepatic tor and s6k transcription were markedly upregulated, but hepatic 4e-bp1 transcription showed the reverse pattern in the DCP group compared to the control group | Chen, Tang, et al. (2022) |
| Abalone, Haliotis discus hannai | CGM protein replacing 25%, 50%, 75% and 100% of dietary FM protein | 110 days | The expressions of s6k, mtor, and eif4e in the digestive gland were considerably downregulated as dietary CGM incorporation increased | Wu et al. (2022) |
| Gilthead seabream (Sparus aurata) | HFM replaced FM protein at 50% (HFM50) and 100% (HFM100) | 100 days | Increased levels of FM replacement decreased the gh/igf axis gene expression in the liver, which decreased growth and nutrition metabolism | Psofakis et al. (2020) |
| Largemouth Bass | Composite mixture of soybean protein hydrolysates and at 40% (FM40; control), 35% (FM35), 30% (FM30), or 25% (FM25) fish meal | 10 weeks | FM35 group exhibited a considerable increase in the expression of the lat1 gene, with no significant variation seen in lat2 and tor expression among treatment groups. On the other hand, s6k expression was much higher in the FM35 and FM30 groups compared to the other two groups, with reduced expression of 4ebp1 | Wang et al. (2023a) |
| Juvenile black sea bream (Acanthoparus schlegelii) | 0% (control diet), 10%, 20%, 30%, 40%, and 60% protein from FM with poultry by-product meal | 8 weeks | akt, s6k1, pi3k, tor, and igf1 expressions were significantly upregulated when FM replacement with PBM increased from 0% to 30%. fish fed a 30% replacement of FM with PBM exhibited the lowest expression of 4e-bp2 among all treatment groups | Irm et al. (2020) |
| Hybrid grouper (E. fuscoguttatus♀ × E. lanceolatus♂) | Rendered animal protein blend replacing 20% (FM56), 40% (FM42), 60% (FM28) and 80% (FM14) of FM protein | 8 weeks | Dietary APB inclusion decreased lpl levels while upregulating the pparα, cpt1, fas, and apo-ai mRNA levels. Also, upregulated tgf-β1, il-8, and il-10, p53, caspase-3, caspase-8, and caspase-9 in the liver | Ye et al. (2019) |
| Obscure pufferfish (Takifugu obscurus) | Clostridium autoethanogenum protein control diet (CAP0), and then 20% (CAP20), 40% (CAP40), and 80% (CAP80) dietary fishmeal were replaced by CAP | 63 days | Compared to the control group (CAP0), the CAP40 groups showed a considerable upregulation of the expression of tor in muscle and pept1 in the gut. However, graded amounts of CAP had no discernible effect on the relative expressions of tat1, y+ lat, and 4f2hc in the gut. In the liver and muscle, the expression of s6k1, 4e-bp1, and 4e-bp2 was not substantially impacted by the addition of CAP | Cui et al. (2022) |
| Hybrid groupers (female E. fuscoguttatus× male E. lanceolatus) | Replace 0% (FM), 25% (PSB25), 50% (PSB50), and 75% (PSB75) of the FM protein with PSB (peptides from swine blood} | 8 weeks | No notable distinctions were seen between the four groups regarding, tnf-α, il-2, il-8, and il-10 expression levels | He et al. (2021) |
| African catfish (Clarias gariepinus) | Fermented soy pulp FSP replacing D1 (0% FSP), D2 (25% FSP), D3 (50% FSP), D4 (75% FSP) and D5 (100% FSP) to FM component of the diets | 70 days | Gene expression (tgf-β1, lyzg, nf-kβ, and hsp90a) were significantly highest in the D3 diet | Kari et al. (2022) |
| Litopenaeus vannamei | The soybean meal was added to the diets at the rate of 20% (T20), 28% (T28), 35% (T35), 42% (T42), and 50% (T50 | 42 days | Growth hormone, myogenic regulatory factor 5, and target of rapamycin mRNA relative expression were all downregulated when the percentage of dietary soybean meal rose from 20% to 50% | Peng et al. (2022) |
| Hybrid grouper (E. fuscoguttatus × E. lanceolatus) juveniles | Formulated to replace 0%, 15%, 30%, 45%, 60%, 75% and 90% FMP with haemoglobin powder protein | 8 weeks | In the fish hypothalamus, large HPP substitutions (45–90%) to FM protein in diets dramatically reduced the relative mRNA expression levels of agrp, npy, and orexin genes | Yao et al. (2018) |
| Black sea bream (Acanthopagrus schlegelii | Fish soluble meal replaced 0% (control diet), 10%, 20%, 30%, 40% and 60% protein from FM | 8 weeks | The relative expression levels of, 1, and tor were upregulated in the liver when replacement levels of FM with FSM increased from 0% to 40%. Nevertheless, as the percentage of FM replaced by FSM rose from 0% to 40%, the relative expression 4e-bp2 was downregulated | Irm et al. (2020) |
| Turbot (Scophthalmus maximus L. | 400 g/kg FM replacement by plant protein blend | Intestine | The PP40 diet significantly decreased AA transporters (snat2 and b0, +at), but the gene expression of L-type and T-type AA transporters (lat2 and tat1), as well as the anabolism and AA catabolism enzymes (BCKDH-E2), was significantly elevated (asparagine synthetase) | Xu et al. (2017) |
| M. amblycephala | Graded replacing levels of fishmeal (replacing 0, 37.5, 75, 112.5, and 150 g/kg FM with canola meal | Liver | There was a considerable upregulation of pept1 mRNA in the gut. Hepatic expression of tor was reduced when FM was replaced with CM, with hepatic genes akt and 4e-bp2 showing an increasing expression | Zhou et al. (2018) |
| Pacific white shrimp L. vannamei | 25% of fishmeal (FM) and 10% (BSF10), 20% (BSF20), and 30% (BSF30) of FM protein were replaced with BSF black soldier fly larvae meal (BSF) | 7 weeks | Shrimp fed BSF30 had downregulated mRNA expressions of pk, hk, pepck, pfk, mcd, ampk cpt-1, and scd1 in their hepatopancreas but upregulated mRNA expressions of acc1. Shrimp fed BSF20 had upregulated hepatopancreatic mRNA expressions of fbp, cpt-1, fas, and 6pgd in contrast to BSF30 | Chen, Chi, et al. (2022) |
- Abbreviations: 4e-bp2, 4E-binding proteins (4E-BPs); 6pgd, 6-phosphogluconate dehydrogenase; agrp, agouti related neuropeptide; akt, serine-threonine protein kinase; ampk, AMP-activated protein kinase; apo-ai, apolipoprotein AI; cpt1, carnitine palmitoyltransferase 1; Fas, fatty acid synthase; fbp, fructose-1,6-bisphosphatase 1; Hk, hexokinase; igf-1, insulin-like growth factor 1; il10, interleukin 10; il-8, interleukin-8; irs-1, insulin receptor substrate 1; lat2, linker for activation of T-cells family member 2; lyzg, lysozyme; mcd, malonyl-CoA decarboxylase; mlck, myosin light-chain kinase; mtor, mechanistic target of rapamycin kinase; myd88, myeloid differentiation primary response 88; nf-κb, nuclear factor kappa-light-chain-enhancer of activated B; npy, neuropeptide Y; pepck, phosphoenolpyruvate carboxykinase; pfk, phosphofructokinase; pi3k, phosphoinositide 3-kinase; pk, protein kinase; pparα, peroxisome proliferator-activated receptor alpha; s6k, ribosomal protein S6 kinase beta-1; scd1, stearoyl-CoA desaturase; snat2, sodium-dependent neutral amino acid transporter-2; tat1, tyrosine aminotransferase; tgf-β2, transforming growth factor beta-2; tlr21, toll-like receptors; tnf-α, tumour necrosis factor alpha; zo-1, tight junction protein-1.
In proteomic, tropomyosins, beta enolase, and creatine kinase exhibited differential muscle protein expression. Also, the expression of multiple proteins involved in immune system function, cellular processes, stress, and inflammation response was observed to be modulated in the liver of a fish fed creatine supplementation diet in gilt-head bream Sparus aurata (Schrama et al., 2018). Atlantic salmon's immunological response and skin mucus proteome were not significantly affected by the dietary addition of black soldier fly larvae meal (Li et al., 2019). Enrichment analysis was used to identify immune-related proteins connected to cell chemotaxis cytokine production, bacterial infection, and pathogen detection, phosphorylation levels of the mitogen-activated protein kinase pathway, and leucocyte transendothelial migration in large yellow croaker Larimichthys crocea fed soybean diet. According to iTRAQ-based proteomics, Turbot (Scophthalmus maximus) fed lysine and leucine diet impacts the metabolism of amino acids, protein synthesis and degradation, and liver metabolism (Wei et al., 2021).
Metabolomics findings reveal that yellow catfish Pelteobagrus fulvidraco fed diet substituted with 50% of fish meal with peanut meal (PM), sesame meal (SEM), cottonseed meal (CM), corn gluten meal (CGM), and SBM and regulated biosynthesis of variations in muscle content of phenylalanine, proline, glutamic acid, vitamin B6, and tyrosine after 12 weeks of feeding (Li et al., 2023). Also, the metabolites of rainbow trout fed insect (Hermetia illucens) protein extract showed an alteration in plasma and muscle tissues. This was associated with an adverse impact on proteins and energy metabolism (Roques et al., 2020). Palma et al. (2021) noted a variation in the plasma and digesta metabolite profiles of the PM-fed ARS-Sel and CS groups, suggesting an early stage of enteritis development in the intestine of Rainbow trout strains. According to metabolomic studies in red drum (Sciaenops ocellatus) reveal potential dietary stress (N formimino l glutamate) in the liver, muscle, intestine, and plasma in fish fed soybean diet after 8 weeks of feeding (Casu et al., 2019). A 1H nuclear magnetic resonance (NMR) shows that the muscle and liver tissue demonstrated a significant decrease in metabolites associated with the energy shortage of juvenile turbot (Scophthalmus maximus) fed plant and animal protein sources (Hoerterer et al., 2023). When comparing Arctic charr (Salvelinus alpinus) fed zygomycetes and fish meal diet using the OPLS-DA fitted model, it revealed that fed fish meal exhibited a stronger signal for β-alanine, acetate, choline, formate, creatine, inosine, glucose, SN-glycero-3-phosphocholine, lysine, and two unidentified metabolites than fish fed diet zygomycete (Abro et al., 2014).
Transcriptomic analysis results also revealed that a diet of high-level/low-gossypol had a detrimental effect on hepatic gluconeogenesis and glycolysis pathways for cholesterol production, significantly influencing lipid synthesis and catabolism in the liver of Silver sillago (Sillago sihama) (Liu et al., 2023). A similar result was obtained in the liver and intestine of Gibel carp (Carassius gibelio) after feeding a FM and rapeseed meal (RM) diet for 10 weeks. Hybrid grouper (Epinephelus fuscoguttatus♀ × E. lanceolatus♂) fed Soy protein concentrate was enriched in metabolic processes, biological regulation, single-organism processes, and cellular processes. Also, several DEGs were involved in the digestive system's metabolism of lipids and amino acids in the intestine, liver, and muscle after 8 weeks of feeding. This was attributed to Soy protein concentrates (SPC's) modulatory effect on these metabolic processes (Chen et al., 2020). heat shock protein 90 kDa alpha (hsp90aa1), ornithine decarboxylase (odc1), collagen, type II, alpha 1 (col2a1a), and myosin light chain, phosphorylatable fast skeletal muscle (Mylpfb) was among the genes with differential expression in the skin and muscle of Zebrafish and Atlantic Salmon after receiving a pea, soy, and wheat diet for 16 weeks. According to the authors, these genes are crucial for controlling muscle growth, preserving muscle shape and function, and preserving muscle tissue homoeostasis (Dhanasiri et al., 2020). SPC and bean protein concentrate (BPC) caused less significant changes to the gut transcriptome than diets containing SPC or BPC alone. Compared to single-plant protein meals, mixed-plant protein diets were also linked to improved fish body composition in Atlantic salmon after 16 weeks of feeding (Król et al., 2016). A similar outcome was observed in the liver and intestine of rainbow trout (Oncorhynchus mykiss) after receiving a yeast diet (Frohn et al., 2024). The transcriptome study showed that the diet-based terrestrial animal by-products supplemented with 3% yeast extract positively affected the expressions of genes associated with the immune system, inflammation, combating pathogens, and coagulation in Oncorhynchus mykiss intestine and liver (Frohn et al., 2024). Fan et al. (2021) reported that over 28 DGEs are associated with lipid production, protein metabolism, absorption, or other metabolic pathways, immunological-related signalling pathways, encompassing genes linked to the immune system and pathogen resistance in the liver of pompano Trachinotus ovatus after being fed high plant protein-based diet.
Also, different hydrolysate products (Shrimps and Tilapia) supplemented with low fish meal diets affected more immune-related metabolic pathways. It was seen that combining the two hydrolysates boosted genes and metabolic pathways compared to when each hydrolysate was evaluated separately in European sea bass Dicentrarchus labrax intestinal mucosa (Kiron et al., 2020; Leduc et al., 2018). In contrast to the findings in the distal intestine of Atlantic Salmon fed soy derivative-linked diet, the transcriptome of the head kidney did not demonstrate any altered gene expression after 8 weeks of feeding. A plant-based diet was also shown to regulate biological processes linked to amino acid transamination, blood coagulation, protein catabolism, and RNA splicing in the liver of two European sea bass (Dicentrarchus labrax) half-sib families. In contrast, the half-sib families differed in the gene expression associated with adenosine triphosphate (ATP) synthesis and protein production (Geay et al., 2011). According to the result of transcriptomic analysis in the liver of two strains of gibel carp (Carassius gibelio) fed a FM-based diet and RM-based diet shows that the RM diet adversely impacted the digestive system, lipid metabolism, and amino acid metabolism as compared to FM diet (Xu et al., 2019). In juvenile yellow perch (Perca favescens) fed the SBM-based diet, transcriptome analysis in the intestine identified a few differentially expressed genes, most fundamental to the cholesterol production pathway (Kemski et al., 2020). Six dietary protein levels (25%, 30%, 35%, 40%, 45%, and 50%) were formulated by Lu et al. (2021) to fed Juvenile top-mouth culter (Erythroculter ilishaeformis) for 16 weeks; from their observation of five clusters were identified by trend analysis as the DEGs' substantial clustering: this include growth (insulin-like growth factor 1 [igf1]), lipid metabolism (fatty acid synthase (fasn) and acetyl-CoA carboxylases [accα]), glucose metabolism (phosphoenolpyruvate carboxykinase [pepck], pyruvate kinase [pk], glycerol kinase [gk], and glucose-6-phosphate dehydrogenase [g6pd]), aspartate biosynthesis (AST), and immune system function which were related to antioxidation, metabolism, immunology, and different signal pathways. Also, SBM50 and FM groups had 6390 differentially expressed genes (DEGs) and 92 DEmiRNAs. DEmiRNAs in the intestine and their DE target genes were implicated in phagosome-related immune responses, cytotoxicity mediated by natural killer cells, and phagocytosis mediated by IgA synthesis pathways within the intestinal immune network. In zebrafish, adding SBM changed the expression of several genes associated with intestinal barrier function. The intestinal tissue showed changes in several metabolic genes. In addition, genes linked to the DNA damage, cell cycle, and DNA repair, as well as immune-related genes such as PR domain zinc finger protein 1 (prdm1), tryptophan hydroxylase 1 (tph1a) NOD-like receptor 12 (nlrp12), NOD-like receptor 3 (nlrc3), and GTPase of immunity-associated proteins (gimap8), were expressed differently in the soya bean fed group (Rehman et al., 2021).
4 DIETARY PROTEIN INTERACTION WITH CARBOHYDRATES FROM A MOLECULAR PERSPECTIVE
Two realistic diets were developed for gilthead seabream: diet P50/CHO10, which has high protein (50%) and low starch (10%), and diet P40/CHO20, which is low protein (40%) and high starch (20%) in Sparus aurata juveniles Fish fed the P40/CHO20 diet showed reduced glutamate dehydrogenase expression and elevated glucokinase (gk) gene expression. Only the P/CHO ratio interaction was seen in the growth hormone receptor II (ghr-ii) and igf1 gene expression (Basto-Silva, Enes, et al., 2022). Based on the same dietary formulation, there was an impact on cox-2, which was more prevalent in fish fed the P50/CHO10 diets compared to the P40/CHO20 diets. Glutathione reductase and glutathione peroxidase were more expressed in fish fed FM-based diets, whereas superoxide dismutase was more expressed in fish fed P50/CHO10 diets (Basto-Silva, García-Meilán, et al., 2022). It was found that, following 8 weeks of feeding crayfish diets with different P/C ratios, a decrease in P/C ratio caused glucose transporter 1 (glut1) expression to significantly increase and hexokinase and pyruvate kinase activity and expression in the hepatopancreas to increase gradually. A higher P content in the diet was also associated with significant increases in the expression of the 4ebp1 and mechanistic mtor genes (Wen et al., 2022).
5 DIETARY PROTEIN INTERACTION WITH LIPIDS FROM A MOLECULAR PERSPECTIVE
For 16 weeks, lake whitefish (Coregonus clupeaformis) were found to have either high or low levels of protein (54% and 48%, respectively) and fat (18% and 12%, respectively). The relative expression of a gene involved in cellular repair, hsp70, was significantly downregulated in fish fed high-lipid diets. However, the expression of genes related to innate immunity and oxidative stress remained unchanged. Furthermore, the fish fed the high protein and lipid diet showed elevated (but not statistically significant) levels of il8 and catalase (Chen et al., 2023). Three lipid levels (60, 90, or 120 g kg−1) and three protein levels (430, 470, or 510 g kg−1) were measured in juvenile small Larimichthys polyactis during 51 days. The relative expression of igf1 and ghr increased significantly when dietary fat levels increased (igf-1). Dietary lipids markedly elevated and decreased the relative levels of FAS and hepatic lipoprotein lipase (lpl), respectively (Ma et al., 2020). The 60-day experiment examined the impact of lipids on protein sparing in the diet of genetically improved farmed tilapia fingerlings (Oreochromis niloticus L). Eight purified diets with four crude protein (CP) levels (30%, 35%, 40%, and 45%) and two lipid levels (6% and 10%) were used in the 4 × 2 factorial design experiment. The liver of fish fed 35% CP with 10% lipid showed the highest expression of the igf-i gene (Thirunavukkarasar et al., 2022). The effects of varying dietary CP (30%, 35%, and 40%) and ether extract (EE: 6%, 10%, and 14%) on P. dabryanus metabolism were investigated using nine different diets. Metabolome analysis revealed that the CP40EE10 group exhibited significantly enriched pathways in the glycerophospholipid metabolism, d-arginine, mtor signalling pathway, and d-ornithine metabolism compared to the other groups. Additionally, transcriptome results demonstrated a significant expression of arginase, or protein synthesis, in the CP40EE10 group, while the CP30EE6 group showed opposite expressions of fructose-1,6-bisphosphatase and secretory phospholipase A2 when compared to the CP40EE10 group. Compared to other groups, Kyoto Encyclopedia of Genes and Genomes showed considerably enriched pathways in the CP40EE10 pathway related to glycolysis/gluconeogenesis, glycerophospholipid metabolism, and arginine and proline metabolism (Wang et al., 2023b). Several important genes, including cysteine dioxygenase, ADP-ribosylation factor 1/2, sodium/potassium-transporting ATPase subunit alpha, carnitine/acylcarnitine translocase, and calcium/calmodulin-dependent protein kinase II (camk), were obtained by enrichment for the DEGs after an 8-week feeding regimen containing 10% fat and 1% taurine. These genes were found to be abundant in the pathways related to phospholipase D signalling, insulin secretion, bile secretion, taurine and hypotaurine metabolism, and thermogenesis, respectively (Chen, Bai, et al., 2022). Varying protein/lipid levels (LR, 28.8% protein/26.3% lipid; MR, 37.5% protein/20.9% lipid; and HR, 42.8% protein/17.3% lipid, for 8 weeks in Black Sea Bream (Acanthopagrus schlegelii). Numerous signalling pathways on lipid metabolism, energy metabolism, amino acid metabolism, carbohydrate metabolism, and immune system function have been found to exhibit varying degrees of gene expression. While the HR diet decreased the genes linked to glycolysis and protein catabolism, the LR diet inhibited the genes linked to lipid oxidation, protein metabolism, and glycolysis (Rehman et al., 2021).
6 CONCLUSIONS
The future of aquafeed formulation probably relies on the mixture of different protein sources in an attempt to replace expensive FM. In this way, it will be possible to take advantage of the nutritional properties of each ingredient, as well as to benefit from the functional properties that some of these alternative sources seem to present. However, by mixing different protein sources, synergetic or antagonistic effects might be observed in the growth performance and well-being of aquatic species. Therefore, more studies are needed to provide additional value to this new generation of aquafeeds.
Furthermore, molecular analysis can shed light on physiological processes by identifying the genes, sequences, and metabolites contributing to variations in fish growth performance. Over the past several decades, computational power and analytical platform advancements have transformed molecular analysis. Future research must use molecular-based platforms like genomic, transcriptomic, metabolomic, and proteomic to advance our understanding of nutrition formulation and boost industry benefits. More financing, cooperative agreements, and the development of more approachable molecular-based equipment are advised to improve industry adoption. Adopting dietary supplements will be highly beneficial for long-term and repeated-measures research. Also, researchers must follow standard reporting criteria and expedite presampling and sample processing methods. The mechanics of starvation, functional feeds, dietary supplements, and bioactive substances should also be focused on. Researchers in protein sources should focus on identifying welfare biomarkers and forecasting key production attributes by integrating genomes and epigenetics technologies. These will aid in improving the precision and interpretation of molecular results which could be useful in enhancing growth performance and well-being of aquatic species by aquatic researchers.
AUTHOR CONTRIBUTIONS
Hesham Eed Desouky: Conceptualization; writing—original draft. Nouran Mahmoud Sayed: Writing—review and editing. Kenneth Prudence Abasubong: writing—review and editing. Ziping Zhang: Supervision; funding acquisition; writing—review and editing.
ACKNOWLEDGEMENTS
This work was financially supported by the Research Project of Fuzhou Institute of Oceanography (2022Fll) and Fujian Innovation and Industrialization Development of the abalone seed industry (2021FJSCZY02). The authors wish to thank the Egyptian Central Missions Administration and the Ministry of Higher Education for their support.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
New data were not created nor analysed in this review, so data sharing is not applicable.




