Analysis of pathogen distribution and antimicrobial resistance at infected sites in plastic surgery
Fengli Jiang and Xueshang Su have contributed equally to this work.
Abstract
Objective
By analyzing the distribution and drug resistance of common pathogen in different sites in plastic surgery to provide reference for clinicians to choose the best antibacterial treatment plan.
Methods
Pathogens of postoperative infection in plastic surgery from January 2011 to December 2021 were retrospectively analyzed to determine the species and quantity, and to access the trend of each pathogen's detection rate. The antibiotic sensitivity and distribution characteristics of common pathogens were studied in conjunction with the site of infection.
Results
A total of 1709 bacterial strains were detected, including 1244 gram-positive bacterial strains and 465 gram-negative bacterial strains. The main pathogen of perineum was Escherichia coli (E. coli) and Pseudomonas aeruginosa (P. aeruginosa), while Staphylococcus aureus (S. aureus) was the most common pathogen in the other infected sites. The detection rate of methicillin-resistant S. aureus (MRSA) and methicillin-resistant coagulase-negative staphylococcus (MRCNS) was on the rise from 2011 to 2021. No S. aureus and coagulase-negative staphylococcus (CoNS) strains were resistant to vancomycin. The sensitive rate of S. aureus from all parts and CoNS from all sites except lower limbs and mandible was higher than 80% to linezolid. The resistance rate of S. aureus and CoNS in all parts to penicillin, clindamycin, and erythromycin was high. The susceptibility rate of CoNS in lower mandible was high to gentamicin.
Conclusions
Staphylococcus aureus was the primary pathogen of gram-positive bacteria in all site of plastic surgery except perineum, followed by CoNS. The distribution and drug resistance of pathogen in different infection sites were different. We should formulate more accurate and reasonable antibacterial programs according to drug resistance results of various parts to reduce the emergence of resistant strains and effectively prevent and control infection.
1 INTRODUCTION
Infection is one of the common complications in plastic surgery and the leading cause of delaying wound recovery, an extended treatment time, poor cosmesis, and even letting deformities. Higher requirements were demanded in the healing quality and appearance of the wound in plastic surgery, which was the fusion of medicine and aesthetics. Various treatment methods such as flap transplantation, application of expander, foreign body implantation, and bacterial colonization in some treatment areas1 will increase the risk of infections. Therefore, the prevention of infections between the perioperative and treatment period is of utmost importance in plastic surgery.
In addition, the irregular abuse, misuse, and overuse of antibiotics lead to the emergence of antibiotics-resistant bacterial strains and high selective antibiotic pressure, which accompany with the increasing morbidity and mortality of infection. Numerous studies have revealed that reasonable antibiotic application of antibiotics can reduce medical costs, the incidence of infection, and the colonization prevalence of drug-resistant strains.2 Previous studies have mostly focused on the profile and the antibiotic resistance of microorganisms from the perspective of specimen type and bacterial classification, and few studies were conducted from the perspective of infected sites. In addition, the first use of antibiotics is usually performed without microbiological results, and mostly based on the epidemiology of microbiology. Therefore, it is crucial to investigate the distribution differences and drug resistance characteristics of common pathogenic bacteria in different parts of plastic surgery to provide useful and valuable scientific evidence and information for the prevention, control, and treatment of infections in plastic surgery.
2 MATERIALS AND METHODS
2.1 Clinical specimens
A retrospective study was performed to analyze the information of patients infected after plastic surgery in the plastic surgery hospital located in Beijing from January 2011 to December 2021. In this study, the presence of one of the following symptoms was considered as infection, including erythema, local warmth, swelling, purulent discharge, delayed wound healing beyond expectations, new or increasing pain, and increasing malodour.3 Routine smears, gram staining, inoculation and culture, isolation, and identification of all specimens were performed in accordance with the guidelines of National Clinical Laboratory Procedures (4th edition). A total of 1709 strains which were mainly from wound secretion and pus were isolated after the excluding of a repeated result of the same pathogen from the same patient. The location of the patient's wound mainly included ear, nose, oral cavity (including pharynx), lip (including angulus oris), lower mandible, face (including forehead, temporal region, zygomatic region, cheek, and chin), scalp, neck, upper limbs (including axillary region), lower limbs (including groin), trunk (including the chest, abdomen, back, waist, hip, sacrococcygeal region), breast, and perineum (including the perineum, genital, and urethra).
This study was approved by the Medical Ethics Review Board of Plastic Surgery Hospital of Chinese Academy of Medical Sciences (approval number: 2022170).
2.2 Microbe species identification and antimicrobial susceptibility test
Blood and MaikangKai dishes (Antu, Zhengzhou) were used for the culture and inoculation of microbial samples. Different types of samples were inoculated in the corresponding Petri dishes and incubated in the corresponding incubators for 18–24 h as required and then taken out for the observation of colony morphology. The pathogens including gram-negative bacteria and gram-positive bacteria were identified by API identification strip (bioMérieux, France) based on standard microbiological procedures. The sensitivity to drugs was tested by Kirby-Bauer Paper flakes diffusion method. The test judgment standard was according to the 2021 Clinical and Laboratory Standards Institute (CLSI) document M100.4 The antimicrobial susceptibility test in this study was only used for Staphylococcus aureus and CoNS; therefore, a total of 11 antibacterial drugs selected for gram-positive bacteria were used in this study, including cefoxitin, cotrimoxazole, penicillin, clindamycin, tetracycline, levofloxacin, erythromycin, vancomycin, chloramphenicol, gentamicin, and linezolid. Drug-sensitive piece of paper came from the British Oxoid company. The quality control strains were S. aureus (ATCC25923), Escherichia coli (ATCC25922), Pseudomonas aeruginosa (ATCC27853), and Enterococcus faecalis (ATCC29212). The identified strains were stored at −80°C.
2.3 Statistical analysis
Data were primarily input and processed using Microsoft Excel 2016, and percentages were calculated. The categorical data were expressed as frequencies and percentages. The statistical analysis was performed using SPSS 19.0 (IBM analytics) and WHONET 5.6 software. Trend chi-square test (linear-by-linear association) was used to analyze whether there was a linear relationship between the detection rates of pathogen and years. p < 0.05 indicated that there was a linear trend between the detection rates of pathogen and years. For pathogens with a linear trend with the years, use Microsoft Excel 2016 to make trend charts and determine trend direction.
3 RESULTS
3.1 The overall distribution of pathogens
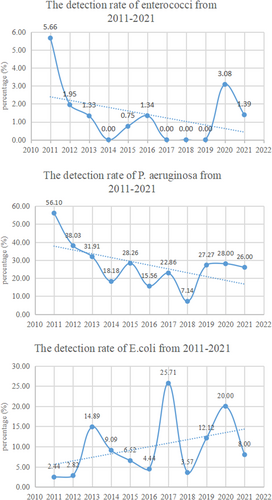
A total of 1709 bacterial strains including 1244 gram-positive bacterial strains and 465 gram-negative bacterial strains were detected among clinical specimens from patients treated in our hospital from January 2011 to December 2021. Staphylococcus aureus was the primary pathogen of gram-positive bacteria (783/1244), followed by CoNS (366/1244). Other gram-positive bacteria such as Streptococcus and Enterococci accounted for less than 10%, as shown in Table 1. Of the gram-negative bacteria, P. aeruginosa had the highest percentage (137/465) (Table 2). Overall, from 2011 to 2021, the detection rates of Enterococci and P. aeruginosa showed a downward trend (p < 0.05), while the detection rate of E.coli showed an upward trend (p < 0.05). There was no obvious variation trend in other strains (p > 0.05), as shown in Figure 1.
| Species | S. aureus | CoNS | Streptococcus | Enterococci | Else | Total |
|---|---|---|---|---|---|---|
| 2011 | 36 (67.92) | 13 (24.53) | 1 (1.89) | 3 (5.66) | 0 (0) | 53 (4.26) |
| 2012 | 99 (64.29) | 44 (28.57) | 6 (3.90) | 3 (1.95) | 2 (1.30) | 154 (12.38) |
| 2013 | 89 (59.33) | 47 (31.33) | 11 (7.33) | 2 (1.33) | 1 (0.67) | 150 (12.06) |
| 2014 | 75 (55.97) | 43 (32.09) | 12 (8.96) | 0 (0) | 4 (2.99) | 134 (10.77) |
| 2015 | 90 (67.16) | 36 (26.87) | 9 (6.72) | 1 (0.75) | 6 (4.23) | 142 (11.41) |
| 2016 | 102 (68.46) | 37 (24.83) | 2 (1.34) | 2 (1.34) | 6 (4.03) | 149 (11.98) |
| 2017 | 77 (62.60) | 41 (33.33) | 4 (3.25) | 0 (0) | 1 (0.81) | 123 (9.89) |
| 2018 | 72 (61.54) | 39 (33.33) | 5 (4.27) | 0 (0) | 1 (0.85) | 117 (9.41) |
| 2019 | 59 (69.41) | 24 (28.24) | 2 (2.35) | 0 (0) | 0 (0) | 85 (6.83) |
| 2020 | 38 (58.46) | 22 (33.85) | 3 (4.62) | 2 (3.08) | 0 (0) | 65 (5.23) |
| 2021 | 46 (63.89) | 20 (27.78) | 1 (1.39) | 1 (1.39) | 4 (5.56) | 72 (5.79) |
| Total | 783 (62.94) | 366 (29.41) | 56 (4.50) | 14 (1.13) | 25 (2.01) | 1244 (100) |
| Χ 2 | 0.014 | 0.504 | 2.083 | 4.220 | 1.242 | 1.601 |
| p | 0.905 | 0.478 | 0.149 | 0.040 | 0.265 | 0.206 |
- Abbreviations: CoNS, coagulase-negative staphylococcus; S. aureus: staphylococcus aureus.
| Species | P. aeruginosa | K. pneumonia | E. cloacae | E. coli | A. baumannii | Else | Total |
|---|---|---|---|---|---|---|---|
| 2011 | 23 (56.10) | 2 (4.88) | 8 (19.51) | 1 (2.44) | 3 (7.32) | 4 (9.76) | 41 (8.82) |
| 2012 | 27 (38.03) | 9 (12.68) | 2 (2.82) | 2 (2.82) | 3 (4.23) | 28 (39.44) | 71 (15.27) |
| 2013 | 15 (31.91) | 3 (6.38) | 1 (2.13) | 7 (14.89) | 1 (2.13) | 20 (42.55) | 47 (10.11) |
| 2014 | 8 (18.18) | 7 (15.91) | 8 (18.18) | 4 (9.09) | 1 (2.27) | 16 (36.36) | 44 (9.46) |
| 2015 | 13 (28.26) | 4 (8.70) | 4 (8.70) | 3 (6.52) | 1 (2.17) | 21 (45.65) | 46 (9.89) |
| 2016 | 7 (15.56) | 10 (22.22) | 4 (8.89) | 2 (4.44) | 2 (4.44) | 20 (44.44) | 45 (9.68) |
| 2017 | 8 (22.86) | 6 (17.14) | 4 (11.43) | 9 (25.71) | 1 (2.86) | 7 (20.00) | 35 (7.53) |
| 2018 | 2 (7.14) | 5 (17.86) | 7 (0.25) | 1 (3.57) | 6 (21.43) | 7 (25.00) | 28 (6.02) |
| 2019 | 9 (27.27) | 3 (9.09) | 4 (12.12) | 4 (12.12) | 3 (9.09) | 10 (30.30) | 33 (7.10) |
| 2020 | 7 (28.00) | 3 (12.00) | 2 (8.00) | 5 (20.00) | 0 (0.00) | 8 (32.00) | 25 (5.38) |
| 2021 | 13 (26.00) | 3 (6.00) | 8 (16.00) | 4 (8.00) | 1 (2.00) | 21 (42.00) | 50 (10.75) |
| Total | 137 (29.5%) | 56 (12.0%) | 50 (10.8%) | 43 (9.2%) | 17 (3.7%) | 162 (34.8%) | 465 (100%) |
| Χ 2 | 24.698 | 0.414 | 0.016 | 9.692 | 0.679 | 0.827 | 2.428 |
| p | 0.000 | 0.520 | 0.898 | 0.002 | 0.410 | 0.363 | 0.119 |
- Abbreviations: A. baumannii, Acinetobacter baumannii; E. cloacae, Enterobacter cloacae; E. coli, Escherichia coli; K. pneumonia, Klebsiella pneumonia; P. aeruginosa, Pseudomonas aeruginosa.
3.2 The change trends of MRSA and MRCNS
From 2011 to 2021, the detection rates of MRSA and MRCNS in our hospital showed an upward trend, as shown in Table 3.
| Year | S. aureus | CoNS | ||||
|---|---|---|---|---|---|---|
| Cases (n) | MRSA (n) | Proportion (%) | Cases (n) | MRCNS (n) | Proportion (%) | |
| 2011 | 36 | 5 | 13.89 | 13 | 2 | 15.38 |
| 2012 | 99 | 20 | 20.20 | 44 | 31 | 70.45 |
| 2013 | 89 | 18 | 20.22 | 47 | 24 | 51.06 |
| 2014 | 75 | 18 | 24.00 | 43 | 21 | 48.84 |
| 2015 | 90 | 17 | 18.89 | 36 | 19 | 52.78 |
| 2016 | 102 | 19 | 18.63 | 37 | 21 | 56.76 |
| 2017 | 77 | 18 | 23.38 | 41 | 24 | 58.54 |
| 2018 | 72 | 23 | 31.94 | 39 | 25 | 64.10 |
| 2019 | 59 | 16 | 27.12 | 24 | 17 | 70.83 |
| 2020 | 38 | 14 | 36.84 | 22 | 16 | 72.73 |
| 2021 | 46 | 11 | 23.91 | 20 | 10 | 50.00 |
| Total | 783 | 179 | 22.86 | 366 | 210 | 57.38 |
| Χ 2 | — | — | 12.755 | — | — | 29.471 |
| p | — | — | 0.000 | — | — | 0.000 |
- Abbreviations: CoNS: coagulase-negative staphylococcus; S. aureus, Staphylococcus aureus.
3.3 The pathogen distribution in different sites
As shown in Table 4, E. coli (18/76) and P. aeruginosa (9/76) were the most common pathogen of perineum. In oral cavity, S. aureus (13/61) was the most frequent pathogen, followed by P. aeruginosa (12/61). In other parts in our study, the predominant pathogen was S. aureus, and secondly CoNS.
| Part | Staphylococcus aureus | CoNS | Streptococcus | Enterococci | E. coli | E. cloacae | P. aeruginosa | A. baumannii | K. pneumonia | Else | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ear | 198 | 133 | 7 | 1 | 2 | 5 | 22 | 2 | 3 | 33 | 406 |
| Nose | 107 | 44 | 3 | 1 | 4 | 7 | 17 | 0 | 10 | 37 | 230 |
| Face | 104 | 53 | 15 | 2 | 1 | 3 | 19 | 2 | 11 | 20 | 230 |
| Trunk | 59 | 22 | 4 | 4 | 7 | 7 | 14 | 4 | 2 | 14 | 137 |
| Lower limbs | 52 | 16 | 1 | 0 | 3 | 2 | 3 | 1 | 0 | 4 | 82 |
| Upper limbs | 42 | 14 | 1 | 0 | 0 | 1 | 7 | 0 | 3 | 9 | 77 |
| Perineum | 6 | 5 | 3 | 4 | 18 | 5 | 9 | 2 | 2 | 22 | 76 |
| Neck | 35 | 13 | 2 | 0 | 0 | 2 | 3 | 1 | 2 | 5 | 63 |
| Scalp | 37 | 16 | 0 | 0 | 1 | 3 | 5 | 0 | 0 | 9 | 71 |
| Lower mandible | 25 | 9 | 8 | 0 | 0 | 2 | 6 | 1 | 7 | 8 | 66 |
| Oral cavity | 13 | 2 | 4 | 0 | 1 | 5 | 12 | 3 | 7 | 14 | 61 |
| Breast | 24 | 13 | 2 | 0 | 1 | 0 | 4 | 0 | 0 | 2 | 46 |
| Lip | 16 | 9 | 1 | 0 | 0 | 3 | 2 | 1 | 3 | 2 | 37 |
- Abbreviations: A. baumannii, Acinetobacter baumannii; CoNS, coagulase- negative staphylococcus; E. cloacae, Enterobacter cloacae; E.coli, Escherichia coli; K. pneumonia, Klebsiella pneumonia; P. aeruginosa, Pseudomonas aeruginosa; S. aureus, Staphylococcus aureus.
3.4 Antimicrobial resistance of the predominant bacterial pathogens in different parts
3.4.1 Antimicrobial Resistance of S. aureus in different parts
As mentioned,S. aureus was the most common types of gram-positive bacteria in all parts, all of them were completely sensitive to vancomycin. As shown in Table 5, no S. aureus strains of oral and perineal were resistant to cotrimoxazole and linezolid. The strains of ear, nose, lower mandible, face, and lower limb had more than 90% sensitiveness to linezolid, and the sensitive rate of neck, scalp, trunk, and breast was higher than 85% to linezolid. The sensitive rate of lip and scalp to chloramphenicol was 93.8% and 86.5%, respectively. The rate of upper limb sensitive to levofloxacin was 83.3%. The resistance rate of S. aureus of all parts to penicillin was the highest (over 80%), among them the resistance rate of the perineal was as high as 100%. The resistance rate of S. aureus strains to erythromycin and clindamycin was also high.
| Part | Cefoxitin | Cotrimoxazole | Penicillin | Clindamycin | Tetracycline | Levofloxacin | Erythromycin | Vancomycin | Chloramphenicol | Gentamicin | linezolid | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ear | S | 71.2 | 87.4 | 3.5 | 27.8 | 77.3 | 85.9 | 22.2 | 100 | 84.9 | 78.8 | 90.4 |
| R | 24.2 | 6.1 | 96.0 | 70.2 | 18.2 | 5.6 | 75.3 | 0 | 9.6 | 17.7 | 1.0 | |
| Nose | S | 79.4 | 78.5 | 9.4 | 37.9 | 79.4 | 82.2 | 29.9 | 100 | 80.4 | 60.8 | 97.2 |
| R | 18.7 | 12.2 | 90.7 | 60.8 | 15.9 | 12.2 | 66.4 | 0 | 13.1 | 36.5 | — | |
| Oral cavity | S | 81.8 | 100 | 18.2 | 27.3 | 72.7 | 72.7 | 27.3 | 100 | 81.8 | 63.6 | 100 |
| R | 18.2 | 0 | 81.8 | 72.7 | 18.2 | 18.2 | 72.7 | 0 | 9.1 | 18.2 | 0 | |
| Lip | S | 81.3 | 68.8 | 12.5 | 31.3 | 68.8 | 81.3 | 25.0 | 100 | 93.8 | 75.0 | 87.5 |
| R | 6.3 | 6.3 | 81.3 | 62.5 | 18.8 | — | 62.5 | 0 | — | 6.3 | — | |
| Lower mandible | S | 68.0 | 88.0 | 8.0 | 32.0 | 72.0 | 92.0 | 28.0 | 100 | 88.0 | 80.0 | 96.0 |
| R | 28.0 | 8.0 | 92.0 | 64.0 | 24.0 | 8.0 | 72.0 | 0 | 4.0 | 16.0 | — | |
| Face | S | 70.2 | 82.7 | 3.8 | 25.0 | 73.1 | 75.0 | 18.3 | 100 | 78.8 | 62.5 | 93.3 |
| R | 21.2 | 8.7 | 94.2 | 70.2 | 20.2 | 11.5 | 76.0 | 0 | 7.7 | 32.7 | — | |
| Scalp | S | 48.6 | 86.5 | 8.1 | 18.9 | 51.4 | 75.7 | 13.5 | 100 | 86.5 | 70.3 | 86.5 |
| R | 43.2 | 5.4 | 89.2 | 70.3 | 40.5 | 8.1 | 75.7 | 0 | 8.1 | 21.6 | — | |
| Neck | S | 60.0 | 74.3 | 8.6 | 25.7 | 65.7 | 77.1 | 25.7 | 100 | 82.9 | 57.1 | 85.7 |
| R | 25.7 | 17.1 | 91.4 | 65.7 | 25.7 | 14.3 | 74.3 | 0 | 8.6 | 40.0 | — | |
| Upper limbs | S | 73.8 | 78.6 | 9.5 | 26.2 | 71.4 | 83.3 | 21.4 | 100 | 76.2 | 52.4 | 81.0 |
| R | 19.0 | 4.8 | 85.7 | 66.7 | 21.4 | 9.5 | 73.8 | 0 | 7.1 | 42.9 | — | |
| Lower limbs | S | 76.9 | 82.7 | 3.8 | 21.2 | 78.8 | 75.0 | 19.2 | 100 | 69.2 | 61.5 | 98.1 |
| R | 23.1 | 13.5 | 94.2 | 76.9 | 15.4 | 21.2 | 69.2 | 0 | 25.0 | 32.7 | — | |
| Trunk | S | 62.7 | 88.1 | 1.7 | 22.0 | 66.1 | 76.3 | 15.3 | 100 | 74.6 | 57.6 | 86.4 |
| R | 30.5 | 8.5 | 94.9 | 69.5 | 28.8 | 16.9 | 79.7 | 0 | 16.9 | 39.0 | — | |
| Breast | S | 79.2 | 87.5 | 8.3 | 37.5 | 75.0 | 70.8 | 25.0 | 100 | 79.2 | 62.5 | 87.5 |
| R | 8.3 | 8.3 | 91.7 | 54.2 | 20.8 | 12.5 | 66.7 | 0 | 8.3 | 29.2 | — | |
| Perineum | S | 66.7 | 100 | 0 | 33.3 | 66.7 | 66.7 | 33.3 | 100 | 83.3 | 50.0 | 100 |
| R | 33.3 | 0 | 100 | 66.7 | 33.3 | 16.7 | 66.7 | 0 | — | 16.7 | 0 | |
- Abbreviations: R, resistance rate; S, sensitive rate.
3.4.2 Antimicrobial resistance of CoNS in different parts
As to analysis of the sensitivity of pathogens to common antibiotics in the drug susceptibility test, the sensitive rate of all parts to vancomycin was 100%. Except for mandibular, the rate of CoNS sensitive rate to linezolid in other parts was only lower than that of vancomycin, among them the sensitive rate of lip, breast, and perineum to linezolid was 100%. The susceptibility rate of mandible to gentamicin was 88.9%. The CoNS had the highest resistance to penicillin and erythromycin in all parts. The resistance rate of oral cavity, lip, mandible, neck, lower limb, breast, and perineum to penicillin was 100%. In addition, the erythromycin resistance rate of neck and perineum was also 100%, as shown in Table 6.
| Part | Cefoxitin | cotrimoxazole | Penicillin | Clindamycin | Tetracycline | Levofloxacin | Erythromycin | Vancomycin | Chloramphenicol | Gentamicin | linezolid | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ear | S | 38.3 | 48.9 | 9.0 | 37.6 | 69.9 | 58.6 | 16.5 | 100 | 63.9 | 65.4 | 93.2 |
| R | 57.1 | 38.3 | 91.0 | 55.6 | 24.8 | 17.3 | 78.2 | 0 | 24.8 | 28.6 | — | |
| Nose | S | 27.3 | 52.3 | 4.5 | 29.5 | 61.4 | 56.8 | 13.6 | 100 | 65.9 | 56.8 | 97.7 |
| R | 70.5 | 38.6 | 95.5 | 68.2 | 36.4 | 27.3 | 86.4 | 0 | 27.3 | 34.1 | 2.3 | |
| Oral cavity | S | 100 | 100 | 0 | 50.0 | 100 | 100 | 50.0 | 100 | 100 | 100 | 100 |
| R | 0 | 0 | 100 | 50.0 | 0 | 0 | 50.0 | 0 | 0 | 0 | 0 | |
| Lip | S | 33.3 | 11.1 | 0 | 33.3 | 55.6 | 66.7 | 11.1 | 100 | 55.6 | 66.7 | 100 |
| R | 66.7 | 77.8 | 100 | 44.4 | 33.3 | 22.2 | 88.9 | 0 | 33.3 | 33.3 | 0 | |
| Lower mandible | S | 44.4 | 66.7 | 0 | 44.4 | 55.6 | 66.7 | 22.2 | 100 | 33.3 | 88.9 | 77.8 |
| R | 33.3 | 33.3 | 100 | 55.6 | 22.2 | 22.2 | 77.8 | 0 | 33.3 | — | — | |
| Face | S | 35.8 | 45.3 | 9.4 | 41.5 | 58.5 | 58.5 | 24.5 | 100 | 62.3 | 60.4 | 90.6 |
| R | 50.9 | 49.1 | 84.9 | 54.7 | 32.1 | 18.9 | 71.7 | 0 | 28.3 | 34.0 | — | |
| Scalp | S | 43.8 | 56.3 | 6.3 | 50.0 | 81.3 | 68.8 | 25.0 | 100 | 75.0 | 37.5 | 93.8 |
| R | 50.0 | 37.5 | 93.8 | 37.5 | 18.8 | 25.0 | 75.0 | 0 | 18.8 | 43.8 | — | |
| Neck | S | 30.8 | 23.1 | 0 | 7.7 | 61.5 | 53.8 | 0 | 100 | 46.2 | 38.5 | 84.6 |
| R | 61.5 | 46.2 | 100 | 76.9 | 38.5 | 46.2 | 100 | 0 | 23.1 | 53.8 | — | |
| Upper limbs | S | 28.6 | 42.9 | 7.1 | 35.7 | 64.3 | 35.7 | 7.1 | 100 | 28.6 | 35.7 | 92.9 |
| R | 71.4 | 42.9 | 85.7 | 57.1 | 28.6 | 35.7 | 85.7 | 0 | 42.9 | 50.0 | — | |
| Lower limbs | S | 43.8 | 50.0 | 0 | 43.8 | 50.0 | 50.0 | 31.3 | 100 | 50.0 | 56.3 | 62.5 |
| R | 56.3 | 43.8 | 100 | 56.3 | 25.0 | 25.0 | 68.8 | 0 | 37.5 | 37.5 | — | |
| Trunk | S | 31.8 | 54.5 | 4.5 | 36.4 | 63.6 | 72.7 | 13.6 | 100 | 72.7 | 45.5 | 95.5 |
| R | 63.6 | 36.4 | 95.5 | 63.6 | 27.3 | 18.2 | 86.4 | 0 | 13.6 | 54.5 | — | |
| Breast | S | 46.2 | 61.5 | 0 | 53.8 | 61.5 | 69.2 | 30.8 | 100 | 76.9 | 69.2 | 100 |
| R | 38.5 | 15.4 | 100 | 46.2 | 23.1 | 23.1 | 61.5 | 0 | 7.7 | 23.1 | 0 | |
| Perineum | S | — | — | 0 | 20.0 | 40.0 | 20.0 | 0 | 100 | 40.0 | 20.0 | 100 |
| R | 80.0 | 80.0 | 100 | 80.0 | 60.0 | 80.0 | 100 | 0 | 20.0 | 60.0 | 0 | |
- Abbreviations: R, resistance rate; S, sensitive rate.
4 DISCUSSION
Infections have always been one of the common and feared complications after plastic surgery. Two main factors contribute to the postoperative infections. Frist, some treatment area of plastic surgery such as ear, nose, oral cavity, breast, and perineum are colonized by opportunistic pathogens. Once the colonization bacteria break through the barrier, they can invade the body and cause infection.5-7 Second, infection is a common complication after the application of foreign bodies such as expanders and prostheses.8, 9 In addition, some studies also revealed that the implants contaminated with bacteria were the main cause of infection.10, 11 Hence, the infection has become a great challenge for plastic surgery. In recent years, the non-standard use of antibiotics and the emergence of bacterial resistance have greatly increased the difficulty of clinical antibacterial treatment. It is very important to understand the distribution of drug-resistant strains and drug susceptibility characteristics to formulate reasonable antibacterial treatment plan.
In our study, gram-positive cocci were the primary pathogen over the last decade, accounting for 72.79% of the total positive isolates. Among them, S. aureus was the main bacteria, followed by CoNS, which may be related to the widespread colonization of staphylococcus in the skin and mucosa, and the majority of plastic surgery sites on the body surface. Pseudomonas aeruginosa and K. pneumonia were the most common gram-negative bacteria. Contrary to our study, in a study of pathogenic bacterial infections after head and face surgery in plastic surgery, gram-negative bacteria outnumbered gram-positive bacteria.12 In addition, the results reported by China Antimicrobial Resistance Surveillance System (CARSS) in 2020 also revealed that gram-negative bacteria accounted for 71.1%, while gram-positive bacteria only accounted for 28.9%.
As a result of the difference in the anatomical characteristics, treatment methods and common colonization bacteria in different parts, it is quite necessary to analyze the characteristics of bacterial distribution and drug resistance in different parts in order to improve the accuracy of antibacterial treatment and reduce the emergence of drug-resistant strains. The study found that the most common bacteria of perineal pathogens were E.coli and P. aeruginosa, which may be related to the perineum being adjacent to the lower gastrointestinal often colonized by E. coli.13 The predominant pathogenic bacteria in other parts were S. aureus, accounting for more than 40% of the total infection strains except oral cavity and lower mandible. But in another study performed by Supasid Jirawatnotai and other scholars, CoNS (82.4%) were the most frequently isolated pathogen after rhinoplasty, while S. aureus (17.6%) only ranks second.14 Staphylococcus aureus was the most frequent microbial agents of outbreak and significantly increased infection morbidity and mortality, which was easy to develop drug resistance, and difficult to control.15 The treatment of MRSA bacteria had always been a troublesome challenge. In recent years, owing to the implementation of control measures for the antibiotic, a significant downward trend was observed in the rate of MRSA detection in China year by year. In this study, the general detection rate of MRSA (28.1%) in the past 5 years was lower than the national level (29.4%) at the same period reported in 2020 CARSS. The study indicates that the occurrence rate of MRSA was constantly at a high level (43.2%) in scalp, while the MRSA prevalence of lips and breasts was lower (6.3% and 8.3%). Fortunately, all S. aureus strains were sensitive to vancomycin, and the sensitive rates of S. aureus to linezolid were all above 80%, among which the sensitive rates of oral and perineal strains to linezolid and cotrimoxazole reached 100%, indicating that vancomycin and linazolamine are still the most effective drugs in the current clinical treatment of S. aureus infection. However, it should be noted that two linezolid-resistant strains were discovered in the ear. The sensitive rate of S. aureus in some parts to cotrimoxazole, chloramphenicol, and levofloxacin was also high, which could provide reference for the instruction empirical clinical medication. However, S. aureus of all parts had high resistance to penicillin, erythromycin, and clindamycin, which should be avoided or used with caution.
Coagulase-negative staphylococcus were the second most common pathogen in this study, which were considered avirulent constituents of the human and warm-blooded animal microbiota in the past. However, at present, some strains of CoNS are recognized as opportunistic pathogens, which is the most common bacteria in clinical nursing and closely related to the application of medical equipment due to the emergence and rapid spread of methicillin resistance.16, 17 The second most common pathogen in the breast infection in our study was CoNS, accounting for more than 25%. However, Mesa et al.18 believed that P. aeruginosa was the second cause of breast implant infections after S. aureus. Furthermore, Lohmeyer JA et al. identified CoNS as the primary cause of clinically insignificant infections of breast implants.9 In our study, highest rates of MRCNS were predominately found in nose, upper limb, and perineum infection sites. The resistance rate of CoNS to methicillin in oral cavity, breast, and lower mandible infection sites was 0%, 38.5%, and 33.35%, respectively, much lower than the national level (74.7%) at the same period reported in 2020 CARSS. Studies have shown that the most common CoNS including Staphylococcus epidermidis and Staphylococcus haemolyticus, were resistant to methicillin at a rate of up to 80%.19 In this study, the infection sites of CoNS all had a highly sensitive rate to linezolid, and the sensitive rate of oral cavity, lip, breast, and perineum strains to linezolid reached 100%. However, it is worth to note that one linezolid-resistant strain had been found in the nose site. In line with our research, some study also found the linezolid-resistant strains as well as linezolid-dependent strains in other countries because of high dosage and long-term therapeutic course of medication.20 Although currently linezolid can be used for severe gram-positive infections with methicillin resistance and has been used in a variety of diseases with CoNS infections, we cannot ignore the phenomenon of linezolid resistance. Therefore, it is more necessary to refine medication principle according to infection site and formulate strict and reasonable medication indications.
The proper use of antibiotics is critical to reducing the emergence of resistant strains. The selection of antibiotics is often based solely on the type of pathogen in the past; however, we found that there were differences in pathogen species and drug sensitivity characteristics in different parts of infections. Therefore, summarizing the common types of pathogens, understanding the characteristics of drug sensitivity of pathogens in different parts, and taking the differences of anatomic site as one of the references for medication are helpful to improve the accuracy of empirical use of antibiotics and provide suggestions for formulating prevention and control measures.
AUTHOR CONTRIBUTIONS
Sien Zhan and Jintian Hu developed the idea and provided the methodology for the study. Fengli Jiang analyzed the data and wrote the paper. Xueshang Su collected the data and wrote the paper. The remaining authors contributed to refining the ideas, discussing the results, and revising the manuscript.
FINANCIAL DISCLOSURE STATEMENT
The authors have no financial interest to declare in relation to the content of this article.
ACKNOWLEDGEMENT
We are grateful that this work was supported by the National Natural Science Foundation of China (81671933) and the Key Medical Discipline Research Project of Beijing Shijingshan District.
DATA AVAILABILITY STATAEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
ETHICAL STATEMENT
The authors confirm that the ethical policies of the journal, as noted on the journal’s author guidelines page, have been adhered to and the medical ethics review board of Plastic Surgery Hospital, Chinese Academy of Medical Sciences approved the proposal (2022170).