Genome-wide DNA methylation profiling of gastric cardia cancer
Declaration of conflict of interest: The authors have no conflicts of interest to declare.
Author contributions: RL, YQ, GC and SL participated in the design and performance of the study, review of results, analysis and discussion. GC, JZ, DG, YH and MX enrolled subjects, collected samples for the study and contributed clinical data. DX, BQ and WL conducted the experiment. LH contributed to the diagnosis of pathological changes. The manuscript was drafted by RL, YQ, GC and SL, and reviewed by all authors. All authors read and approved the final manuscript. RL and YQ contributed equally to this paper.
Ethical approval: Written informed consent was obtained from the participants, and this study was approved by the Ethics Committee of Shantou University Medical College (SUMC-2017-06). All methods were performed in accordance with the Declaration of Helsinki.
Financial support: This study was supported by the National Natural Science Foundation of China under Grant No. 81702717 and Grant No. 81902634, Natural Science Foundation of Guangdong Province under Grant No. 2014A030310139, The Innovation Strong School Project of Department of Education of Guangdong Province under Grant No. 2019KTSCX037, the Medical Science Foundation of Guangdong Province under Grant No. A2022084, the Medicine and Health Science and Technology Project of Shantou, China (Grant No. [2022]88-6, Grant No. [2022]88-9 and Grant No. [2022]88-44), The Open Fund of Guangdong Provincial Key Laboratory of Infectious Diseases and Molecular Immunopathology (Grant No. GDKL202205), and the research grants from Shantou Science and Technology Bureau (Grant No. 210712186880511).
Abstract
Background and Aim
Aberrant DNA methylation has been found in various cancer types including gastric cancer, yet the genome-wide DNA methylation profile of gastric cardia cancer (GCC) remains unclear. Therefore, we aimed to profile the DNA methylation pattern of GCC and identify promising diagnostic epigenetic biomarkers.
Methods
We investigated the genome-wide DNA methylation pattern in eight pairs of GCC and adjacent normal tissues using Illumina 850K microarrays. Subsequently, bisulfite-pyrosequencing and quantitative real-time PCR were performed on eight pairs of GCC-adjacent normal tissues for validation. Finally, we performed immunohistochemistry to examine ADHFE1 expression on 126 pairs of GCC-adjacent normal samples.
Results
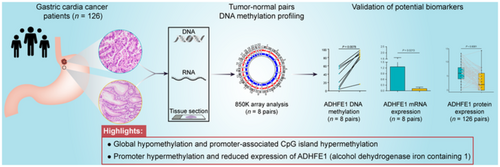
DNA methylome analysis showed global hypomethylation and local hypermethylation of promoter cytosine-phosphate-guanine (CpG) islands (CGIs) in GCC tissues compared with gastric cardia normal mucosa (P < 2.2 × 10−16). Differential methylation analysis identified a total of 91 723 differentially-methylated probes (DMPs), and the candidate gene with the largest average DNA methylation difference mapped to ADHFE1 (mean Δβ = 0.53). Subsequently, three DMPs in the ADHFE1 promoter were validated by pyrosequencing. Notably, the mean methylation level of the three candidate DMPs (ADHFE1_cg08090772, ADHFE1_cg19283840, and ADHFE1_cg20295442) was negatively associated with ADHFE1 mRNA expression level (Spearman rho = −0.64, P = 0.01). Moreover, both mRNA (P = 0.0213) and protein (P < 0.0001) expression of ADHFE1 were significantly decreased in GCCs compared with the adjacent normal tissues.
Conclusions
Our results reveal DNA methylation aberrations in GCC and that ADHFE1 gene DNA methylation contributes to the risk of GCC, thus providing novel mechanistic insights into gastric cardia cancer carcinogenesis.
Graphical Abstract
Lin et al. perform genome-wide DNA methylation analyses on 8 pairs of normal gastric cardia mucosa and gastric cardia cancer specimens, and identify global hypomethylation and aberrant hypermethylation in gene promoter regions in gastric cardia cancer. They find aberrant down-regulation of ADHFE1 gene in gastric cardia cancer, which is associated with promoter hypermethylation.
Open Research
Data Availability Statement
The raw data used and analyzed in this study are deposited in NCBI's Gene Expression Omnibus (GEO) and are accessible at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE200484, through GEO Series accession number GSE200484.