Oxynoemacheilus marmaraensis, a new species from the Susurluk River, Türkiye (Teleostei: Nemacheilidae)
Abstract
Oxynoemacheilus marmaraensis, new species, is restricted to the Susurluk River. It is distinguished from all the named species of Oxynoemacheilus in the northwestern Anatolian by the flank with a vermiculate pattern and the presence of a suborbital groove in males, and no axillary lobe at the base of the pelvic fin. It also differs from the closest species, Oxynoemacheilus kentritensis, by having 58 nucleotide substitution sites. The genetic distance is 10.49% between O. marmaraensis and O. kentritensis. Phylogenetic analyses and species delimitation tests (Poisson tree processes and assemble species by automatic partitioning) support the validity of O. marmaraensis as a distinct species.
1 INTRODUCTION
Nemacheilid loaches of the genus Oxynoemacheilus are very common fish all over the eastern Mediterranean, the southern Caucasus, Anatolia, Mesopotamia, and central Iran (Freyhof et al., 2011, 2022; Kottelat, 2012). This genus is represented by a total of 64 valid species (Freyhof et al., 2021; Turan et al., 2023). Forty-one of these species are found in Türkiye, and 32 of them are endemic (Bektaş et al., 2022; Çiçek et al., 2018; Erk'akan, 2012; Freyhof et al., 2011, 2017, 2019, 2021; Freyhof & Geiger, 2021; Freyhof & Özuluğ, 2017; Kaya et al., 2020; Saygun et al., 2021; Sungur et al., 2017; Turan et al., 2019, 2023; Yoğurtçuoğlu et al., 2021a,b, 2022).
Cytochrome oxidase subunit I (COI), described as DNA barcodes, has been used for the genetic identification of many Oxynoemacheilus species in Türkiye. Recently, the COI barcoding gene has been largely used for the taxonomy and identification of loaches (Bektaş et al., 2022; Kaya et al., 2020; Rakıcı et al., 2020; Sayyadzadeh et al., 2016; Turan et al., 2019).
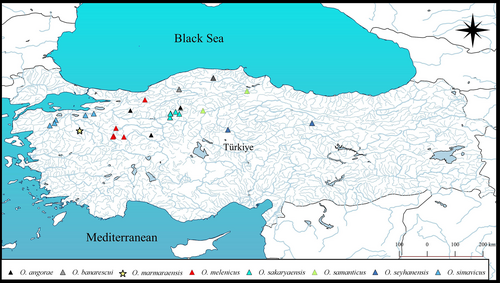
In our study on Oxynoemacheilus species distributed in the northwestern Anatolia, a total of seven species were found to be distributed in this region: Oxynoemacheilus angorae (the Sakarya, Sususurluk, and Kızılrmak rivers, and Eber and Ilgın lakes), Oxynoemacheilus banarescui (Devrekhani and Filyos streams), Oxynoemacheilus seyhanensis and Oxynoemacheilus samanticus (the Kızılırmak River), Oxynoemacheilus simavicus (the Susurluk River), Oxynoemacheilus sakaryaensis (the Sakarya River), and Oxynoemacheilus melenicus (the Sakarya and Melen rivers) (Erk'akan et al., 2007, 2008; Freyhof et al., 2011, 2017, 2022; Freyhof & Özuluğ, 2017; Kuru, 2004; Nalbant & Bianco, 1998; Turan et al., 2023; Yoğurtçuoğlu et al., 2022).
In addition to these species, an additional species, which occurs sympatrically with O. simavicus, was discovered in the Susurluk River.
2 MATERIALS AND METHODS
The care and use of experimental animals complied with the Republic of Türkiye animal welfare guidelines, laws, and policies as approved by the Republic of Türkiye Recep Tayyip Erdoğan University Local Ethics Committee for Animal Experiments (permit reference number 2020/4). Following anesthesia, the samples were fixed in 5% formaldehyde and then in 70% ethanol where possible. Alternatively, samples were fixed directly in absolute ethanol. A dial caliper, precisely set to 0.1 mm, was used for measurements. Stringent measurement procedures were applied point-to-point by following the guidelines of Kottelat and Freyhof (2007). The holotype was involved in the process of calculation of means and SD. The majority of male Oxynoemacheilus specimens exhibit an exposed lachrymal bone, which is known as a suborbital flap or groove. Particular attention was paid to distinguish between a suborbital flap and a suborbital groove. Thus, all Oxynoemacheilus specimens investigated within the scope of the current study possess an exposed lachrymal bone as well as a suborbital groove.
2.1 DNA extraction, PCR, sequencing, and molecular analysis
Total DNA isolation was made using the DNA/RNA isolation system (Qiacube) using Qiagen DNeasy Blood & Tissue Kit. The DNA quality was controlled by running the sample on 1% agarose gel electrophoresis at 90 V for 30 min. The DNA barcode region was amplified using the COI barcoding gene primer set (FishF1 and FishR1) designed by Ward et al. (2005). PCRs were applied in a 50 μL reaction volume, including 100 ng of DNA, 10× PCR buffer, 3 mM MgCl2, 5 μL of 0.5 mM dNTPs mix, 1 u Taq DNA polymerase (Thermo Scientific Inc.) and 0.5 mM of each primer. The PCR conditions were as follows: 95°C for 30 s, 95°C for 30 s, 58°C for 45 s, 72°C for 1 min through 35 cycles, and 72°C for 7 min using a Biorad T100 thermal cycler. The PCR products were displayed on a 1% agarose gel and monitored using a Vilber Lourmat, UV Quantum–Capt ST4 system. PCR products used for purification and gene sequencing were manufactured by Macrogen Europa Inc.
For molecular taxonomy analysis, six newly produced COI barcodes and 122 sequences from previously published studies (Bektaş et al., 2022; Freyhof et al., 2022; Geiger et al., 2014; Turan et al., 2019) were used. Outgroup taxa, Seminemacheilus lendlii (GenBank number: MT077008) and Turcinoemacheilus kosswigi (GenBank number: KJ179258), were chosen for rooting the phylogenetic tree. Bioedit v7.2.5 software (Hall, 1999) was used for the alignment of sequences with the option of Clustal W multiple alignments (Thompson et al., 1994). The sequences were submitted to NCBI GenBank via an online tool submission portal within the OQ657337-OQ657342 accession numbers. Maximum likelihood (ML), maximum parsimony (MP) analysis, and the estimate of pair-wise genetic distances among species were conducted using MEGA 11 (Tamura et al., 2021). The TN93 + G + I model (Tamura & Nei, 1993) was chosen as the substitution model of nucleotide according to the BIC in MEGA 11 (Tamura et al., 2021). In addition, the Bayesian inference (BI) analysis was run using a Metropolis-coupled Markov chain Monte Carlo (MCMC) algorithm for one million generations in the MrBayes 3.1.2 software (Ronquist & Huelsenbeck, 2003), and the initial 25% of the saved trees sampled in each MCMC run were discarded as burn-in.
Two methods were implemented for species delimitation: Poisson tree processes (PTP) (Zhang et al., 2013) and assemble species by automatic partitioning (ASAP) (Puillandre et al., 2021). The ASAP analysis was conducted via a web server (https://bioinfo.mnhn.fr/abi/public/asap/asapweb.html, accessed on March 12, 2023) with default parameters. PTP with maximum likelihood solution was conducted via a web server (http://mptp.h-its.org/#/tree) (accessed on March 12, 2023) to test candidate species.
3 RESULTS
3.1 Key to species in the northwestern Anatolia
1a-No suborbital groove in males………………………………O. seyhanensis.
1b-There is a suborbital groove in males…………………….2.
2a-Caudal peduncle depth more than 10%SL……………7.
2b-Caudal peduncle depth than less 10%SL.…………3.
3a-The snout length longer than the postorbital length…………O. samanticus.
3b-The snout length shorter than the postorbital length…………4.
4a-Maxillary barbels equal to or smaller than outer rostral barbels……………………O. banarescui.
4b-Maxillary barbels greater than outer rostral barbels………….5.
5a-The flank with plain yellowish or with numerous irregularly shaped pale brown bars in most individuals………………O. sakaryaensis.
5b-The flank dark brownish with 2–13 irregular shaped dark brownish bars or blotches……………….6.
6a-The flank with 10–13 irregularly shaped brown bars or blotches; (6) 7–8 irregularly shaped dark brown saddles on back………………O. melenicus.
6b-The flank with 2–8 irregularly shaped brown bars or blotches; 4–5 (6) irregularly shaped dark brown saddles on back……O. simavicus.
7a-Distance between pelvic-fin origin and pectoral-fin origin 25%–28% SL……………………O. angorae.
7b-Distance between pelvic-fin origin and pectoral-fin origin 29%–34% SL………………………O. marmaraensis.
3.2 Oxynoemacheilus marmaraensis, sp. nov.
urn:lsid:zoobank.org:act:923576E9-9DA4-4C90-B959-47636D3366D8.
3.3 Holotype
FFR 15631, 56 mm SL; Türkiye: Balıkesir Province: stream Dursunbey 10 km east of Dursunbey, 39° 36′ 32.4″ N, 28° 45′ 01.9″ E.
3.4 Paratypes
FFR 1511, 12, 46–59 mm SL; same data as holotype.
3.5 Material used in molecular genetic analysis
FFR DNA 15631, 6, Türkiye: Balıkesir Province: stream Dursunbey 10 km east of Dursunbey, 39° 36′ 32.4″ N, 28° 45′ 01.9″ E (GenBank accession numbers: OQ657337-OQ657342).
3.6 Diagnosis
Oxynoemacheilus marmaraensis differs from the species found in the Susurluk River and adjacent basins as follows. It differs from O. simavicus, O. banarescui, O. sakaryaensis, and O. melenicus by having the flank with a vermiculate pattern (vs. 2–8 dark brownish blotches on the flank in O. simavicus, 7–9 brownish blotches on the flank in O. banarescui, the flank plain or with numerous irregularly shaped pale brownish bars in O. sakaryaensis, and the flank with 10–13 irregular shaped brownish bars or blotches in O. melenicus), no axillary lobe at base of pelvic fin (vs. presence) and caudal peduncle depth 1.2–1.6 times in its length (vs. 2.2–3.1 in O. simavicus, 2.2–2.7 in O. banarescui, 2.8–3.2 in O. sakaryaensis, 1.9–2.8 in O. melenicus). It further differs from O. banarescui by having a greater distance between pectoral-fin origin and pelvic-fin origin (29%–34% SL, vs. 26–28) and a shorter caudal peduncle (13%–19% SL, vs. 19–23). It is distinguished from O. angorae by having the flank with a vermiculate pattern (vs. showing a dark-brown mid lateral stripe or a series of fused, dark-brown blotches interrupted by a whitish or pale-brown lateral line), having a greater distance between pectoral-fin origin and pelvic-fin origin 29%–34% SL, vs. 25–28) and a smaller distance between pelvic-fin origin and anal-fin origin (20%–23% SL, vs. 23–28).
3.7 Description
For general appearance see Figures 1 and 2; morphometric data are given in Table 1. Small-sized species. Body slender and body depth at dorsal-fin origin 14.7%–18.6% SL. Head slightly pointed and flattened on the ventral and dorsal surfaces. Caudal peduncle length 1.2–1.6 greater than its depth. No axillary lobe at the base of the pelvic fin. Distance between the anus and anal fin origin about 0.5–0.9 times the eye diameter. Body covered small scales. The lateral line complete, almost reaching to caudal-fin base. A suborbital groove in males. Mouth small and arched. Lips thin, with poorly marked furrows. A deep median interruption in the lower lip. No median incision in the upper lip. Barbels short, the outer one rarely reaching to vertical of the anterior margin of the eye in most individuals; the inner rostral barbel not reaching or rarely reaching to base of the maxillary barbell, and the maxillary barbel not reaching the vertical of the posterior margin of the eye. Largest known specimen 56 mm SL.


| O. marmaraensis | O. simavicus | ||||
|---|---|---|---|---|---|
| H | Range (mean) | SD | Range (mean) | SD | |
| Standard length (mm) | 46–53 | 46–71 | |||
| In percentage of standard length | |||||
| Head length | 23.3 | 21.1–24.5 (23.0) | 0.9 | 19.2–22.4 (21.0) | 0.9 |
| Body depth at dorsal-fin origin | 16.6 | 14.7–18.6 (16.6) | 1.0 | 12.1–16.5 (14.0) | 1.5 |
| Body width at dorsal-fin origin | 12.9 | 10.5–12.9 (12.0) | 0.9 | 8.9–14.1 (11.2) | 1.3 |
| Predorsal length | 52.6 | 47.5–52.6 (50.6) | 1.4 | 46.3–51.2 (49.7) | 1.2 |
| Postdorsal length | 35.5 | 32.8–40.8 (35.6) | 2.4 | 31.8–37.4 (34.9) | 1.7 |
| Pre-anal length | 78.4 | 73.2–78.4 (75.6) | 1.3 | 69.4–75.9 (72.4) | 1.9 |
| Prepelvic length | 54.7 | 50.3–55.7 (53.1) | 1.5 | 48.0–53.5 (50.3) | 1.7 |
| Distance between pectoral and pelvic-fin origins | 33.9 | 29.3–34.3 (32.1) | 1.5 | 23.3–32.4 (28.7) | 2.5 |
| Distance between pelvic and anal-fin origins | 23.6 | 20.4–23.6 (21.9) | 1.1 | 18.7–27.9 (21.1) | 2.8 |
| Distance between ventral and anal-fin origins | 4.2 | 2.3–4.2 (3.2) | 0.6 | 1.8–5.6 (3.2) | 1.1 |
| Depth of caudal peduncle | 11.6 | 10.0–13.1 (11.2) | 0.9 | 0.6–10.0 (7.2) | 1.0 |
| Length of caudal peduncle | 14.7 | 13.2–19.1 (16.2) | 1.7 | 16.0–19.9 (17.9) | 1.2 |
| Dorsal-fin depth | 21.3 | 15.8–21.6 (18.5) | 1.8 | 15.9–19.0 (17.7) | 0.9 |
| Anal-fin base length | 14.7 | 13.2–19.1 (16.3) | 1.8 | 11.3–19.4 (14.7) | 1.9 |
| Pectoral-fin length | 19.1 | 18.0–23.5 (20.5) | 1.6 | 17.9–23.3 (20.5) | 1.7 |
| Pelvic-fin length | 15.5 | 13.8–18.4 (15.8) | 1.6 | 13.1–17.4 (15.7) | 1.1 |
| In percentage of head length | |||||
| Head depth at eye | 42.5 | 38.7–56.6 (46.4) | 5.1 | 37.9–62.2 (44.6) | 6.5 |
| Snout length | 42.7 | 35.3–47.6 (42.0) | 2.9 | 32.3–51.1 (41.0) | 5.2 |
| Eye diameter | 19.3 | 19.3–26.1 (23.1) | 2.1 | 14.4–25.9 (19.8) | 3.7 |
| Postorbital distance | 51.0 | 45.2–63.2 (53.4) | 5.8 | 34.7–56.7 (47.3) | 5.7 |
| Maximum head width | 59.0 | 51.0–63.6 (57.6) | 4.1 | 40.4–68.1 (58.0) | 6.8 |
| Interorbital width | 24.7 | 24.0–34.3 (28.5) | 3.3 | 14.6–33.9 (21.1) | 5.1 |
| Length of inner rostral barbel | 23.8 | 20.2–39.3 (28.9) | 6.0 | 14.9–29.0 (21.2) | 4.1 |
| Length of outer rostral barbel | 34.0 | 28.7–47.1 (34.3) | 5.4 | 17.7–39.1 (29.2) | 6.4 |
| Length of maxillary barbel | 30.7 | 23.4–37.3 (30.8) | 4.0 | 13.2–34.4 (25.3) | 5.8 |
- Note: The calculations include the holotype.
Anal fin with 3 simple and 5½ branched rays, distal margin straight or slightly convex, and not reaching caudal-fin base. Pectoral fin with 1 simple and 9–10 branched rays, outer margin straight in most individuals. Dorsal fin with 3 or 4 simple and (7½) 8½ branched rays, outer margin straight. Pelvic fin with 6–7 rays, outer margin straight, and almost reaching to the anus. Caudal fin forked and lopes slightly rounded.
3.8 Sexual dimorphism
The pectoral fin length in males is longer than that in females. Pectoral fin length 23.0%–26.1% SL in males and 19.6–22.9 in females. No suborbital groove in females but present in females.
3.9 Colouration
General body color brownish in life and preserved individuals. Head and cheeks with small, plain brown mottling on top and cheeks, without color pattern ventrally. Dense pigmentation below a line from pectoral-fin base to the anus. A small, irregularly shaped, dark-brown blotch at the dorsal fin origin. The flank with a vermiculate pattern. Four or five faded blotches on the back in front of the dorsal fin. Three or four irregularly shaped blotches, not fused with mid-lateral blotches on the upper part of the caudal peduncle. One or two irregularly shaped small black spots on the caudal-fin base. Dorsal-fin with 2 and caudal fin with 2–3 fine, irregularly shaped black bands on rays. Anal and pelvic fins plain yellowish, and pectoral fin yellowish, with numerous small black spots on the rays.
3.10 Distribution
O. marmaraensis was found in a tributary of the Susurluk River drainage in the Marmara Sea basin (Figure 3).
3.11 Etymology
The name of “marmara” represents both the name of the “Marmara Sea Basin” and “Marmara Region” where the new species was found. A noun in apposition.
3.12 Remarks
Morphological and molecular data presented here suggest that O. marmaraensis is unrelated to the Oxynoemacheilus bergianus species group.
3.13 Molecular identification of O. marmaraensis
COI barcode region sequences (614 bp) of 65 Oxynoemacheilus species distributed in Asia and Europe were analysed. In the phylogenetic analysis, Oxynoemacheilus genus was divided into several clades promoted by high bootstrap values. One of the clades with a high bootstrap value (ML: 100) is constituted by O. marmaraensis (Figure 4). Kimura-2 Parameter distances between species ranged from 0.12% (O. mediterraneus, O. nasreddini) to 18.72% (O. pindus and O. isauricus). The Kimura-2 Parameter distance is 10.49% between O. marmaraensis and its closest relative, O. kentritensis, and between O. marmaraensis and O. çiçeki is 10.57% (Data S1). O. marmaraensis differs from its most closely related congeners, O. kentritensis, and O. çiçeki, by having 58 and 57 nucleotide substitution sites, respectively.
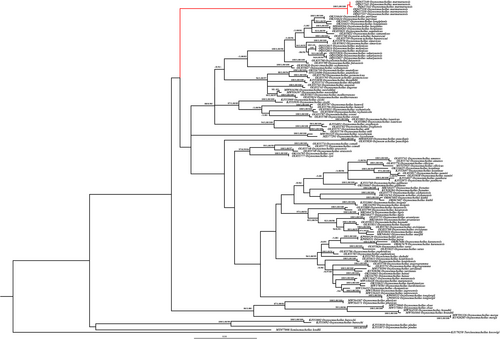
PTP was indicated in 68 species of studied members of the genus Oxynoemacheilus. The maximum likelihood partition support value of O. marmaraensis is 0.820. The best partition of ASAP (score = 3.5) results from a p-distance threshold of 1.01% and estimates 65 subsets. Both of the delimitation analyses predict O. marmaraensis to be a candidate species.
4 DISCUSSION
In this study, besides morphological differences, the results from the species delimitation tests (PTP and ASAP) and phylogenetic analyses support the validity of O. marmaraensis as a distinct species. Also, morphological data presented here suggest that the Oxynoemacheilus bergianus species group have a slender caudal peduncle (less than 10% SL, vs. more than 10). So far, seven species (O. banarescui, O. simavicus, O. angorae, O. seyhanensis, O. samanticus, O. sakaryaensis, and O. melenicus) have been reported from the northwestern Anatolia (Erk'akan et al., 2007, 2008; Freyhof et al., 2011, 2017; Freyhof et al., 2021; Turan et al., 2023; Yoğurtçuoğlu et al., 2022). Of these O. banarescui, O. simavicus, O. samanticus, O. sakaryaensis and O. melenicus belong to the O. bergianus species group, whereas O. angorae, O. seyhanensis, and O. marmaraensis do not belong to this group. O. marmaraensis is distinguished from other species in the northwestern Anatolia as follows. It differs from O. seyhanensis by having the flank with a vermiculate pattern (vs. the body with marmorate pattern or numerous small irregularly shaped and spaced dark-brown bars on flank), a suborbital groove in males (vs. absent), a forked caudal fin (vs. slightly emarginated), and lacking dorsal and ventral adipose crests on the caudal peduncle (vs. present). O. marmaraensis is distinguished from O. samanticus by having the flank with a vermiculate pattern (vs. the flank with 5–8 irregular shaped brownish blotches along the lateral line), the snout length smaller than the postorbital length (vs. the snout length longer than the postorbital length), and deeper caudal peduncle (caudal peduncle depth 1.2–1.6 times in its length, vs. 2.2–2.7).
This study reflected that the phylogenetic relationships of Oxynoemacheilus based on the COI barcoding gene region are in agreement with the previous research (Bektaş et al., 2022; Geiger et al., 2014; Sayyadzadeh et al., 2016; Turan et al., 2019). Similar to the previous study by Bektaş et al. (2022) Oxynoemacheilus genus was determined to be polyphyletic. Bektaş et al. (2022) determined the COI-based threshold value of 1.46% based on the genetic distance range. Also, Sayyadzadeh et al. (2016) suggested an appropriate species-level threshold for the Oxynoemacheilus COI genetic divergences of 1.4%. In this study, the K2P distance is 10.49% between O. marmaraensis and its closest relative, O. kentritensis. Considering the COI gene-based threshold, the genetic differences between O. marmaraensis and its closest relative are considerably higher than the threshold value given in previous studies.
4.1 Comparative materials
AUTHOR CONTRIBUTIONS
DT designed the study plan, conducted fieldwork, and wrote the manuscript draft. EB conducted fieldwork and performed morphological analysis. GK performed the molecular genetic study and wrote part of the manuscript. All authors read, edited, and approved the final version of the manuscript.
ACKNOWLEDGMENTS
We are pleased to thank Dr. Cüneyt Kaya for sampling and morphological laboratory studies. Also, we thank Dr. Münevver Oral for proofreading the manuscript. This study was supported by the Scientific Research Project Coordination Unit of Recep Tayyip Erdogan University (project no: FBA-2022-1419).




