Effects of input data sources on species distribution model predictions across species with different distributional ranges
Handling Editor: François Munoz
Abstract
Aim
A major source of uncertainty in the application of species distribution models (SDMs) is related to input data quality. Citizen-collected species occurrence data are often used for fitting SDMs when data from standardized and expert-supported surveys are unavailable. Macroclimate variables are much more commonly used as predictors in SDMs than other sources coming from remote sensing data. Here, we assess the effects of using different data sources (in both response and predictor variables) on SDM performance across a wide range of species with contrasting distributional ranges.
Location
Iberian Peninsula.
Taxon
Birds.
Methods
A SDM ensemble-forecasting approach was implemented using bird data from two different data sources: the eBird project and Atlases. We fitted SDMs with three predictor types: macroclimate, remotely sensed ecosystem functional attributes (EFAs) and their combination. Species were grouped in four range size classes. We assessed the uncertainty of model predictions by different evaluation metrics. Generalized linear mixed-effects models tested the effect on model performance of input data source across distributional range sizes while accounting for different accuracy metrics. Pairwise comparisons between range projections were used to assess their spatial similarity.
Results
Data source, size class, predictor and accuracy metric showed significant effects on SDM performance. eBird-based models outperformed those built with Atlas data for less widespread species. Climate predictors yielded models with the best performance, especially when combined with EFAs. However, the predictor contribution was consistent across bird datasets, being mostly driven by the species range.
Main Conclusions
Our models demonstrated the usefulness and complementarity of different input data sources when modelling species distribution across different distributional ranges. These findings highlight the need to integrate different data sources to improve the model predictions at regional scale. Our framework also underlines that model uncertainty should be examined more exhaustively at early stages of the modelling process.
1 INTRODUCTION
Species distribution models (SDMs) are a popular tool in theoretical and quantitative ecology (Guisan et al., 2017; Peterson et al., 2011) and constitute the most widely used modelling framework in global change science and biodiversity conservation (Peterson et al., 2011). This popularity is mainly because SDMs are readily accessible due to the availability of different software packages, guidelines and application contexts, as well as having comparatively low data requirements for calibration (Franklin et al., 2013; Guisan et al., 2017; and references within). As main data sources, SDMs require georeferenced biodiversity observations as a response or dependent variable (e.g. species occurrence, species richness, etc.) and geographical layers of environmental information as predictors or independent variables (e.g. climate, land cover, vegetation indices derived from remote sensing, etc). As an incentive in the application of SDMs, such information is freely available in digital format (e.g. GBIF for biodiversity data and WorldClim for climate data). However, although SDMs have become one of the most important quantitative tools for addressing regular and timely biodiversity assessments worldwide, these techniques are still subject to different sources of uncertainty that have been unequally assessed (Beale & Lennon, 2012; Thuiller et al., 2019). While simplicity or complexity in modelling algorithms and model evaluation metrics for describing species–environment relationships or complex processes have garnered more attention, quality and biases in biodata and environmental variables still are less explored (Čengić et al., 2020; Mod et al., 2016). Thus, despite uncertainty related to niche-based or distribution-based models has been addressed at different stages in the modelling process (Gould et al., 2014; Keil et al., 2014), an analysis of the effect of uncertainty coming from alternative data sources on the predictive ability of SDMs is still limited.
The increasing availability of spatially referenced species occurrence records (Bayraktarov et al., 2019), as well as spatially comprehensive environmental data, has enabled researchers to generate quickly and easily SDMs (Zimmermann et al., 2010). The main current data types available range from systematic/standardized and well-structured scientific data (e.g. Atlases) (Jetz et al., 2012), to the fast-growing, more opportunistic, semi-structured citizen science projects (e.g. eBird) (Johnston et al., 2021; Kelling et al., 2019). Although both are valuable for addressing a wide range of socio-ecological research questions and biological modelling and monitoring approaches (Chandler et al., 2017; Neate-Clegg et al., 2020), doubts still exist about data quality from citizen science surveys (Aubry et al., 2017; Jiménez et al., 2019). The data combination from different sources also proved to be a useful tool in ecological modelling (Miller et al., 2019; Suhaimi et al., 2021), specifically to improve estimates of species distributions (Isaac et al., 2020; Robinson et al., 2020), abundance and population trends (Boersch-Supan et al., 2019; Hertzog et al., 2021). However, there is still much to explore on the data quality (Johnston et al., 2019; Van Eupen et al., 2021) and usefulness of each input data source through the modelling process (Kobori et al., 2016; Kosmala et al., 2016), as well as the uncertainty hosted in each modelling step should also be controlled more exhaustively.
In addition to observed distribution data of species, the predictor variables or covariate data is another critical aspect of modelling that could affect the quality of model outputs (Manzoor et al., 2018; Synes & Osborne, 2011). Considering that predictor variables used in SDMs may be collected from different sources (e.g. field-sampled data, or interpolations from meteorological stations, derived from remote sensing, among others), and at different spatial and temporal scales (from broad extent/coarse grain to more local extent/fine grain) (Austin & Van Niel, 2011; Franklin et al., 2013), the uncertainty associated with variable selection should be carefully addressed (Varela et al., 2015; Waltari et al., 2014). For instance, while climate data have been traditionally used as predictors in SDMs to forecast changes in biodiversity, recent integration of satellite-derived remote sensing data as predictors in SDMs offers some novel ecological insights (Arenas-Castro & Sillero, 2021; Regos et al., 2022; and references within), but also generates additional uncertainty (Barsi et al., 2019; Borg et al., 2011).
The other main uncertainty source in SDMs relates to model building and model evaluation/validation. The former includes testing the relationship between species occurrence and environmental data by mathematical or statistical analysis. Although many techniques have already been tested (Elith et al., 2006; Feng et al., 2019), this rapidly developing field still lacks consensus regarding which algorithms are the best for which purposes. Among others, the high uncertainty source regarding model calibration lies in quantifying the degree to which variability in the final predictions is introduced by the modelling methods themselves, and depends on data sources, both species occurrences and environmental data (Buisson et al., 2010). Another major criticism and source of uncertainty in SDM calibration is the lack of true absences for accurate species distribution predictions (Hirzel et al., 2002; Wisz & Guisan, 2009). Since the absence information is not often available, it is possible to calculate pseudo-absences (PAs) through different methods (Hertzog et al., 2014; Senay et al., 2013). However, PAs must be created with caution, as their placement may strongly affect the results of models (Sillero et al., 2021). Both source and number of PAs depend on the modelling technique (Barbet-Massin et al., 2012). Thus, model results are more affected by sources of bias and by the number of PAs than by the distribution of PAs (Lobo & Tognelli, 2011; Sillero et al., 2021).
Model evaluation is an integral part of the model development process that helps to find the best model, it is an additional source of uncertainty when appropriate metrics or different validation tools are not used (Konowalik & Nosol, 2021). In SDMs, the cross-validation is a widely used tool for model evaluation. As in the case of model building, many different metrics for modelling evaluation have been tested considering their intrinsic characteristics and specifications, as they are sensitive to the nature of the input data and the type of algorithm used (Jiménez & Soberón, 2020). On the other hand, model validation is the task of confirming that the outputs of a statistical model have enough fidelity to the outputs of the data-generating process. For instance, SDM validation can be approached through an independent occurrence dataset. While there are available some robust evaluation indices developed for presence-only data such as the Boyce Index (Boyce et al., 2002) or the minimal predicted area index (Engler et al., 2004), it should be noted that the best way to validate the performance of a model is through an independent dataset, although that may not always be available. The validation of SDM models is probably the least developed task, particularly in the case of presence-only and presence-background modelling algorithms (Watling et al., 2015).
We introduce here a framework to elucidate uncertainty derived from the most early-state sources in the model process: the input data, including both the available biodiversity data and the environmental predictors, while accounting for well-known sources of variability in SDM predictions related to modelling techniques and evaluation metrics. We aimed to assess the effects of using different input data on model performance, and if these effects differ between species with different distributional ranges. In particular, we compared SDMs fitted with semi-structured citizen science data (eBird) against those calibrated with standardized and well-structured scientific data (Bird Atlas), considering only macroclimate data, remotely sensed ecosystem functional descriptors and their combination. The results were then analysed by grouping species according to their distributional range (that varied from narrow-ranged to widespread species). We also applied a multi-technique approach (ensemble forecasting) to account for the uncertainty arising from the modelling technique and considered several metrics for model evaluation.
2 MATERIALS AND METHODS
2.1 Study area and bird data
We tested our approach in the Iberian Peninsula (IP; southern Europe) since it covers a wide range of environmental gradients. The IP (581,200 km2) is administratively divided between Portugal (PT; 89,015 km2) and Spain (SP; 492,175 km2) and is characterized by a combination of natural and human history, geologic and topographic heterogeneity, and strong climatic gradients, offering a wide range of environmental conditions for hosting a broad variety of endemic and rare species (Underwood et al., 2009).
To fit SDMs at the first stage, we used bird occurrence data from two different data sources of biodiversity (Table 1): (i) a standardized dataset based on national Bird Atlases (Atlas) and (ii) a citizen science (i.e. non-standardized) dataset based on the EOD—eBird Observation Dataset from the Global Biodiversity Information Facility—GBIF (eBird). To reduce the potential geographical errors in species records that can strongly influence the results of models (Hijmans, 2012), we filtered the original dataset removing duplicates and positional/spatial errors such as outliers using R and qgis programs, and nomenclature errors and taxa misidentification supported by expert knowledge. In addition, we harmonized the species records in grid cells with a resolution higher than 10 km before the modelling procedures. We adjusted eBird data to the spatial resolution of both Atlases (10-km UTM square) to standardize input data and make both datasets comparable. Predictive variables were aggregated at 10-km UTM square (see below). We also matched the eBird data to the years and months within the Atlas data (1999–2012), from late February to mid-August, the breeding season in the IP (SEO/BirdLife, 2020).
| Dataset | Name | Number of species | Time period | Source |
|---|---|---|---|---|
| Atlas | Atlas of Breeding Birds in Portugal | 251 | 1999–2005 | Sociedade Portuguesa para o Estudo das Aves (SPEA; Portugal) |
| Databases of the Spanish Inventory of Terrestrial Species | 1999–2012 | Ministerio de Medio Ambiente, y Medio Rural y Marino (MAGRAMA; Spain) | ||
| eBird | EOD—eBird Observation Dataset | 236 | 1999–2012 | Auer et al. (2022). EOD—eBird Observation Dataset. Cornell Lab of Ornithology. Occurrence dataset https://doi.org/10.15468/aomfnb accessed via GBIF.org on 2021-06-16 |
Additionally, to perform subsequent comparisons, we built a full dataset from the combination of both Atlas and eBird datasets. This full dataset represents the best knowledge of bird distribution in Iberia as it includes all occurrences recorded in the two datasets. To assess the effect of species distributional range (from narrow-ranged to widespread species) on modelling performance, we grouped the species records from the full dataset in four sets of size classes based on number of occurrences at 10-km UTM squares: (i) Class I (10–100); Class II (101–500); (ii) Class III (501–1000); Class IV (>1000). We also considered the conservation categories of species in the IUCN Red List to assess the potential effects of model uncertainty on decision-making for bird conservation. Therefore, each species always belongs to the same group (n = 236) because the full dataset represents the actual distributional range of each species, and it is not affected by potential sampling biases of each dataset (e.g. in the eBird dataset).
2.2 Environmental predictors
We selected environmental factors that are known to influence bird physiology, distribution and local habitat preferences, as close as possible to the time period covered by the species occurrence datasets (1999–2012) (Table S1 in Appendix S1).
We derived 19 (bio-) climate predictors from monthly temperature and rainfall data. This climatic dataset was obtained for historical conditions (1979–2013) from the CHELSA 1.2 database at a spatial resolution of 30 arc-sec (~1-km pixel size).
Satellite remote sensing data from the Moderate Resolution Imaging Spectrometer (MODIS) on-board the Terra satellite platform were used to derive remotely sensed ecosystem functional attributes (EFAs) (Regos et al., 2022, and references within), to characterize species habitat dynamics as a counterpoint/complement of climate data. To compute EFAs, we used the MODIS Enhanced Vegetation Index (EVI) (MOD13Q1.v006; 232 m pixel every 16 days) as a proxy of vegetation greenness, biomass and leaf area index—with values ranging from −1 to 1, with healthy vegetation generally holding values between 0.20 and 0.80, and for the 2000–2012 time period. For that, we used Google Earth Engine (GEE) cloud-based platform (Gorelick et al., 2017) to derive originally 11 metrics of the EVI seasonal dynamics (Table S1 in Appendix S1). These statistical measures were calculated for each complete year. To capture the multi-year normal conditions of each EFA variable, thus reducing the effect of stochastic interannual climatic fluctuations, we computed the overall mean. EFAs were exported from GEE at 1-km squares of final spatial resolution.
All environmental, climate and remotely sensed variables were aggregated from its original spatial resolution (1 × 1 km) by computing the mean values within each 10-km UTM square, to match to the spatial resolution of bird datasets, 10 × 10 km. To avoid including highly correlated variables in model fitting, we conducted a multicollinearity analysis by testing Pearson pairwise correlations and variance inflation factors (VIF). Based on these multicollinearity analyses, we retained those predictors with Pearson's correlation coefficients of <0.7 and VIF of <5 to calibrate models in following steps (Figures S1.1–S1.4 and Table S1 in Appendix S1). Based on the pairwise correlations and the collinearity assessment from initial 19 (bio-) climate variables, three temperature-related (bio4—Temperature Seasonality; bio8—Mean Temperature of Wettest Quarter; bio9—Mean Temperature of Driest Quarter) and two precipitation (bio16—Precipitation of Wettest Quarter; bio17—Precipitation of Driest Quarter) variables were selected (Table S1 in Appendix S1) to build the climate dataset. Similarly, we selected five remotely sensed EFAs as descriptors of species habitat dynamics: EVI annual mean (EVImean as surrogate of annual total amount of primary production), EVI annual minimum (EVImin as an indicator of the annual extremes), EVI seasonal standard deviation (EVIsd as descriptor of variations between seasons), and dates of maximum (EVIdmax) and minimum (EVIdmin) EVI (indicators of phenology—growing season) (Table S1 in Appendix S1).
2.3 Model fitting
We used the species occurrences as response variables in modelling processes within each 10 × 10 km UTM square (Figure 1).
We designed three groups of models: (1) climate-based models (CLIM_only)—using variables extracted from the monthly (bio-) climate dataset; (2) models based on habitat/ecosystem functioning-related predictors (EFAs_only); and (3) the combined model, combining the most significant five (and uncorrelated) predictors selected from the previous partial climate and EFA-based models (CLIM_EFAs).
We calibrated SDMs using an ensemble forecasting approach based on the nine modelling techniques implemented in the ‘biomod2’ R package (Thuiller, 2014): (1) artificial neural networks (ANN); (2) classification tree analysis (CTA); (3) flexible discriminant analysis (FDA); (4) generalized additive model (GAM); (5) generalized boosted models (GBM); (6) generalized linear model (GLM); (7) multivariate adaptive regression splines (MARS); (8) maximum entropy using Phillip's Maxent software (MAXENT) and (9) random forests (RF). Default parameters were used for all modelling techniques, with the exception of the smoothing degree term in GAM algorithm which was set to k = 4, and the number of boosting trees in GBM (n.trees = 2000) to prevent over-fitting issues (Guisan et al., 2002). We also generated a total of 10 sets of randomly distributed PAs in the model calibration. PAs were generated by assigning unoccupied grid cells with the following constraints: (1) generating the same number of PAs as of presences to avoid potential bias caused by different levels of prevalence in the presence/absence datasets (as recommended by Barbet-Massin et al., 2012; Manel et al., 2001) and (2) defining a minimum distance between PAs, corresponding with the grain size (10 km), and without overlapping with presences (Wisz & Guisan, 2009), to avoid spatial autocorrelation and to cover the different ecological conditions in the study area. Each model was fitted using 70% of the data and tested using the remaining 30%. On the other hand, considering that there is not consensus about the minimum number of species records to fit models (Breiner et al., 2015; Sillero et al., 2021), we followed the criterion provided by Franklin (2010) and the species with less than 10 occurrences at 10 km grid cells were excluded to avoid model overfitting and ensure the most threatened species can be included in the analysis.
2.4 Model evaluation and performance
We employed hold-out cross-validation to evaluate the models with 10 evaluation rounds for each PAs set. The predictive performance and discrimination ability of individual models were evaluated using two metrics (Baasch et al., 2010): (i) the area under receiver operating characteristic curve (AUC) and (ii) and the true skill statistic (TSS). The AUC ranges between 0 and 1 (models with AUC ≥0.7 were considered good), while TSS ranges from 0 or less to 1 (models with TSS ≥0.4 were considered good). Finally, to deal with uncertainty in our models coming from the single-algorithm techniques, we built ensemble (consensus) models among those satisfying the conditions AUC ≥0.7, and TSS ≥0.4. We used the weighted mean of all the partial projections (Marmion et al., 2009), a consensus method that considers the weights proportional to the selected evaluation scores (i.e. the higher the AUC of the model, the greater the importance in the ensemble modelling; Konowalik & Nosol, 2021).
Once the predictions of the ensemble models by each species were obtained for each dataset, all model comparisons (namely data source, range class, predictor type and evaluation metric) were carried out on the full dataset resulting from the combination of both Atlas and eBird datasets (n = 236). Considering that Atlas could be treated as a presence–absence dataset and the eBird as only-presence dataset, we tested the performance of the resulting ensemble models for the species hosted in the subset of 236 species using two complementary measures of accuracy: the AUC and Boyce's Index (BoyceI). Unlike the TSS, a prevalence-independent and threshold-dependent binary measure of model accuracy but usually highly correlated to AUC (Shabani et al., 2016), the Boyce's Index is an appropriate metric in the case of presence-only models, measuring how much model predictions differ from a random distribution of the observed presences across the prediction gradients (Hirzel et al., 2006; Pearce & Boyce, 2006). Both accuracy measures (AUC and Boyce's Index) were calculated from the model predictions using the R packages ‘PresenceAbsence’ (Freeman & Moisen, 2008) and ‘ecospat’ (Di Cola et al., 2017), respectively.
In addition, to test the effect of data source, type of predictor (quantitative), size class and accuracy metric (namely AUC and Boyce's Index) on models performance, we developed generalized linear mixed-effects models (GLMMs; Bolker et al., 2009) with R package ‘lmer4’ (Bates et al., 2015). We fitted GLMMs with gamma probability distribution and inverse link (after checking for overdispersion effects in Poisson models and robust estimation via weighted likelihood), as the dependent variable (the values of AUC and the Boyce's Index) showed continuous probability distributions. We fitted ‘data source’, ‘predictor’, ‘size class’ and ‘accuracy metric’ as fixed effects, while ‘species’ was considered a random effect. We only fitted interactions between ‘data source’ and ‘size class’. Fixed effects were considered significant at p-values <0.01.
2.5 The most contributing predictors and spatial projections across set of models
To examine the contribution of satellite-derived EFAs as integrative predictors of habitat dynamics in SDMs for narrow-ranged and widely ranged bird species, we compared the relative importance of three sets of predictors for a total of six final ensemble models (CLIM_only, EFAs_only and CLIM_EFAs), ranging between 0 (no importance) and 1 (high importance) (Araújo & New, 2007). Only predictors with importance >0.1 were considered.
To assess the spatial confidence associated with model results, we compared the spatial projections of the six sets of models based on two data sources (Atlas vs. eBird). For that, we performed pairwise comparisons between spatial projections of each species distribution and obtained at each combination of data source, set of predictors and finally grouped by size class. To do so, we used the ‘raster. overlap’ function in the ‘ENMTools’ R 1.0.2 package (Warren et al., 2008, 2021). This function measures similarity in the geographical distribution of suitability scores from pairwise SDMs. Among other metrics (Warren et al., 2008), the niche overlap is calculated using Schoener's D Index which varies from 0 (complete divergence/no overlap) to 1 (high similarity/complete overlap). In addition, to map uncertainty from the different data sources, we examined the agreement of spatial projections between models obtained by different sets of predictors. For that, we first performed binary transformations of the habitat suitability predicted by the ensemble models into presence–absence maps based on the AUC optimized thresholds available on the ‘biomod2’ R package (Thuiller, 2014), since AUC it is a robust threshold-independent measure of a models ability to discriminate presence from absence (Lawson et al., 2014). Then, we combined the predictions by simply overlapping the binary maps, resulting in similarity maps that ranged from 0 (no prediction) to 6 (prediction based on the six models).
The entire process of obtaining, editing and analysing data, as well as model calibration, mapping and evaluations, was carried out in R software 4.0.1 available at CRAN (http://cran.r-project.org/), the GEE platform and in qgis 3.14 software.
3 RESULTS
3.1 Data structure
The number of species records available in the eBird dataset was, approximately, 30.7% higher than in the Atlas dataset (Figure S2.1 in Appendix S2). However, the number of 10-km UTM squares with data in the eBird dataset was 71.75% lower than in the Atlas dataset (Figure S2.1 in Appendix S2), which did not prevent to cover a similar environmental gradient than the Atlas (see maps of the multivariate environmental similarity surface—MESS analysis in Figure S2.2 in Appendix S2). The number of species recorded in the Atlas was 15 species higher than in the eBird (Figure S2.3 in Appendix S2). Considering the size classes based on species ranges, while the Atlas dataset hosted the highest number of narrow- and wide-ranged species (I: 10–100 and IV: >1000 size classes), eBird hosted the highest scores for the II (101–500) and III (501–1000) classes in the IP (Figures S2.3 and S2.4 [datasets combined] in Appendix S2). Overall, the most represented size classes were those that include more widely distributed or common species (IV, II and III, in this order). In terms of IUCN conservation categories, the Least Concern level was overwhelmingly more frequent in both datasets followed by Vulnerable level and considering the selected period (1999–2012) (Figures S2.5 and S2.6 [datasets combined] in Appendix S2).
3.2 Model evaluation and performance
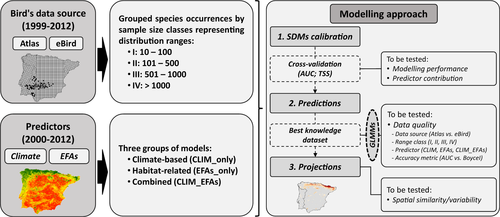
Overall, the ensemble SDMs yielded higher predictive ability for both bird datasets and all groups of predictors when compared to single-algorithm models as measured by median, and interquartile range to be compared across models (AUCmedian-Atlas: 0.89 ± 0.09 and BoyceImedian-Atlas: 0.83 ± 0.33; AUCmedian-eBird: 0.72 ± 0.19 and BoyceImedian-eBird: 0.82 ± 0.36) (Figure 2; Text S3.1 and Figures S3.1 and S3.2 in Appendix S3). Models showed contrasting results depending on the evaluation metric. For equal species number (n = 236), and for ensemble models, while AUC values were higher for models calibrated with Atlas data, the presence-only Boyce's Index indicates better performance of models fitted with eBird data (Table S3.1 in Appendix S3). Differences in model accuracy were found between both data sources and size classes (teBird-IV: 7.869; p-value <0.05) and evaluation metrics (tBoyceI: 4.838; p-value <0.05) (Table S3.1 in Appendix S3 and see accuracy metrics per each species in Table S3.2).
Based on GLMM results, and considering the species range (size classes) and type of predictors, climate-based models (individually or in combination with EFAs) fitted with Atlas data showed the highest AUC values for narrow-ranged species (AUCCLIM_only-I: 0.99 ± 0.02; AUCCLIM_EFAs-I: 0.99 ± 0.03) (Figure 2a; Table S3.1 in Appendix S3), while climate- and EFA-based models alone fitted with eBird data showed the highest performances for the same size class when the BoyceI was used (BoyceICLIM_only-I: 0.93 ± 0.22; BoyceIEFAs_only-I: 0.93 ± 0.10) (Figure 2b; Table S3.1 in Appendix S3). In addition, although the BoyceI showed higher performance than AUC for the less widespread species (size class I, II, III), Atlas-fitted models showed higher values than eBird-fitted models. Predictor types also affected modelling performance (tEFAs_only: 3.434; p-value <0.05) (Table 2).
| Fixed effects | |||||
|---|---|---|---|---|---|
| Variable | Estimate | SE | t-value | p-value | |
| Intercept | 1.164 | 0.051 | 22.793 | <2e-16*** | |
| Data source | eBird | −0.002 | 0.039 | −0.069 | 0.944 |
| Class | II | 0.028 | 0.058 | 0.482 | 0.629 |
| III | 0.043 | 0.065 | 0.671 | 0.502 | |
| IV | −0.017 | 0.052 | −0.323 | 0.746 | |
| Predictor | CLIM_EFAs | 0.006 | 0.013 | 0.455 | 0.649 |
| EFAs_only | 0.046 | 0.013 | 3.434 | 5.9e-04*** | |
| Metric | BoyceI | 0.053 | 0.011 | 4.838 | 1.3e-06*** |
| Data source:Class | eBird:II | −0.068 | 0.045 | −1.497 | 0.134 |
| eBird:III | −0.039 | 0.051 | −0.766 | 0.443 | |
| eBird:IV | 0.332 | 0.042 | 7.869 | 3.6e-15*** | |
| Random effects | |||||
|---|---|---|---|---|---|
| σ 2 | τ00Sps_code | ICC | NSps_code | ||
| 0.04 | 0.01 | 0.15 | 236 | ||
| Observations | 2770 | ||||
| Marginal R2 | 0.338 | ||||
| Conditional R2 | 0.436 | ||||
- *** 0.001 (Significance codes).
Regarding the IUCN conservation categories, while the Atlas showed the highest values of the AUC for all classes (Figure S3.3a in Appendix S3), the highest Boyce's Index values were found for the most threatened species when modelled with the eBird dataset (Figure S3.3b in Appendix S3).
3.3 The variable importance ranking
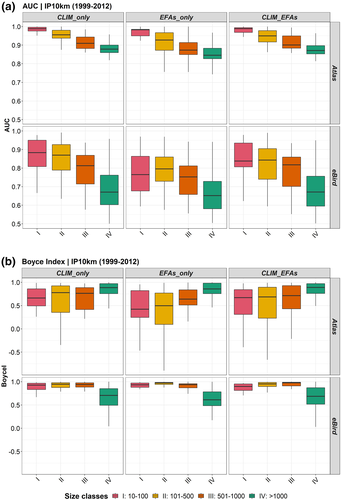
The contribution of each independent variable to models by type of predictors was also affected by the combination of data source, type of predictor and size class. Overall, bio17 (Precipitation of Driest Quarter), bio4 (Temperature Seasonality) and bio9 (Mean Temperature of Driest Quarter) were the most contributing predictors of climate group to model performance across size classes, while EVImean, EVImin and EVIsd, descriptors related to productivity and seasonality in primary productivity, were the most important ones within the group of the habitat attributes (EFAs_only) (Figure 3). However, when we combined both types of predictors (CLIM_EFAs) in the models, bioclimate predictors (bio17 and bio4) held the highest importance across size classes, followed by EVImean (Figure 3).
3.4 Overlap and uncertainty in spatial projections
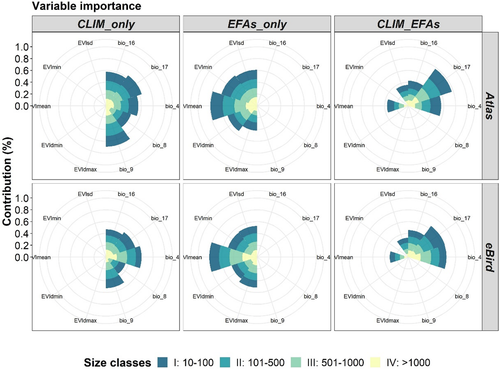
Overall, the similarity test based on Schoener's D Index indicated that pairwise niche overlaps between models based on eBird and Atlas datasets for each species were high for the tested models (Doverall-mean = 0.82 in all cases). However, the similarity between spatial predictions increased with the distributional range, from narrowly distributed species (IDmean = 0.78 and IIDmean = 0.81) to more widespread species (IIIDmean = 0.82, IVDmean = 0.83) (Figure 4). The similarity test also showed that, in general, the overlap between species niches, from both Atlas and eBird datasets, predicted by the EFAs was higher than the predicted by the climate and the combination of climate and EFAs, being highly significant in the case of widely distributed species (based on Kruskal–Wallis test [K–W]: class II: K–W, p = 3.3e-5; class III: K–W, p = 0.0057; and class IV: K–W, p = 0.00012; Figure 4).
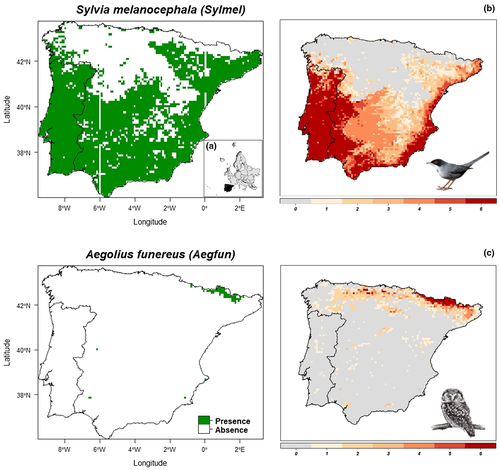
The species-specific spatial projections of the similarity maps derived from the Atlas and eBird datasets and the three groups of predictors (CLIM_only, EFAs_only and CLIM_EFAs) allowed the identification of UTM squares where the predictions of the six models overlapped (Class 6), and therefore were the most consistent across models, for widespread (Figure 5b) and narrow-ranged (Figure 5c) species. Maps for all species in Figure S3.4 are found in Appendix S3.
4 DISCUSSION
We assessed the effects of using different data sources (both in the response and predictor variables) in SDMs while accounting for well-known sources of uncertainty. Overall, for the same environmental predictors (interpolated macroclimate vs. remote sensing ecosystem attributes) and evaluation metrics (AUC vs. BoyceI), we have demonstrated that SDMs fitted with citizen science data (eBird) performed as well as standardized data (Atlas) at a regional scale (Iberian Peninsula). eBird-based models provided more accurate predictions for less common species (the narrow range) than Atlas data that recorded much more widespread species, with strong implications for species conservation. Our results also confirmed that model predictions benefit from the combination of macroclimate data and remote-sensing-derived EFAs, which has also implications for both conservation (Arenas-Castro et al., 2019; Regos et al., 2020) and management (Carvalho-Santos et al., 2018). Among other drawbacks, we also showed that there are important biases in model estimation as well as overconfidence about results accuracy or quality (e.g. through evaluation metrics) (Hernandez et al., 2006; Johnston et al., 2019).
4.1 The added value of citizen science occurrence data for more accurate predictions
Albeit eBird has evolved from a citizen-science project into cooperative partnerships among experts in a wide range of fields, it still hosts several challenges that can inhibit robust ecological inferences (e.g. species and spatial biases, variation in effort and observer skills, among other). A priori, observers (citizens) may be more likely to misidentify or miss, especially rare or not very abundant species, but overall identify target species as accurately as trained researchers (Aceves-Bueno et al., 2017). This, therefore, suggests that citizen science (such as eBird) data may complement more traditional standardized occurrence datasets (such as Atlases), or even substitute for other presence-only datasets (Cox et al., 2012).
Despite differences in sampling designs and protocols between data sources (e.g. a lower number of 10-km UTM squares with data in eBird than Atlas), our results confirmed the usefulness of both citizen science project eBird and standardized and well-structured Atlas datasets to predict bird species distributions with different distributional ranges in the Iberian Peninsula. Overall, Atlas-based ensemble models showed better performance through the AUC, while eBird-based ensemble models showed better performance when the specific presence-only metric BoyceI was used (Figure 2; Table 2; Table S3.1 in Appendix S3). These results can be related to the type of evaluation metric and the nature of the response variable. The AUC is a metric for models fitted with presence/absence data, which are better represented by standardized surveys such as those performed in Atlas projects than opportunistic records from citizen-science programs. Considering species grouped by distributional range through sample size classes, models fitted with Atlas data showed higher BoyceI values for widespread species for the same group of predictors, while the eBird-based ensemble models showed higher BoyceI values across all range sizes except the most widespread species (Figure 2; Table S3.1 in Appendix S3). Our results suggest that SDMs fitted with eBird data can predict species distributions as accurately as those based on Atlas data.
4.2 Contribution of predictors to the performance and predictions of SDMs
Unlike the quality of species observations and their effect on the performance of SDMs, which have been extensively documented (Fei & Yu, 2016), the uncertainty associated with environmental variables as predictors should have been more deeply addressed considering its key role in the process of calibrating and evaluating SDMs (Petitpierre et al., 2017; Scherrer & Guisan, 2019). Overall, predictors showed different effects depending on data source and size class. In terms of performance, our results showed that models fitted with Atlas-based data showed higher AUC values than models fitted by eBird-based data for the same group of predictors (Figure 2a). However, results were different when the BoyceI metric was used. Climate-based models showed the highest performance for the Atlas dataset based on the AUC, while EFAs-based models, and the combined model (CLIM_EFAs), showed the highest performances for the presence-only eBird dataset through the BoyceI (Figure 2b). Considering the contribution of predictors to model predictions, the variable importance overall seems to be consistent across bird datasets used to fit the models (Figure 3), being mostly driven by the species range (size classes) than the data source (eBird and Atlas) (Table 2).
Although SDMs have traditionally relied on climatic data as abiotic factors, among other, more integrative descriptors of high ecological relevance for species in terms of habitat dynamics and ecosystem functions have been neglected (Gonzalez et al., 2020). Satellite-derived EFAs stand up as integrative predictors of SDMs, thank its quicker spatiotemporal response of ecosystem dynamics to environmental drivers and changes than interpolated climate data and structural/compositional attributes. EFAs also timely capture shifts in habitat conditions that may trigger changes in populations and communities (Arenas-Castro et al., 2018; Regos et al., 2021; Vila-Viçosa et al., 2020; and references within). Our results showed that EFA-based models alone or in combination with climate data, performed similarly, or even better, than models only based on climate. Thus, our findings support the need for mainstreaming other key environmental factors for improving model performance, and hence, improving ecological inferences for decision-making or conservation planning.
4.3 Similarity and stability across spatial projections in SDMs
Equally important to the environmental and statistical modelling space is the confidence or scepticism related to the spatial projections or maps derived from SDMs, as they are usually inferred into geographical space from the previously fitted environmental space (Goberville et al., 2015; Thuiller et al., 2019). Overall, the similarity between our spatial predictions of species from the Atlas and eBird datasets was high and consistent across species range and compared models (Figure 4). As expected, the similarity between overlapping species niches was progressively increased as the species ranges were larger, but it was significantly higher between widely distributed species in EFAs-based models. The low similarity between overlapped spatial predictions for species with narrow distributions (Class I) are in line with the BoyceI-based predictive capacity (Figure 2) that was also higher for eBird-based data than for Atlas-based data for this same size class. The high similarity measurements for niche overlaps (D statistic) between eBird-based and Atlas-based models suggest that occurrence data derived from the eBird dataset can be an effective presence-only sample data source in ensemble SDMs for the Iberian Peninsula. In addition, the species compatibility/uncertainty maps built on the base of both Atlas and eBird datasets and calibrated with the three group of predictors (CLIM_only, EFAs_only and CLIM_EFAs) clearly represent a valuable resource to visualize the level of uncertainty and confidence hosted when modelling species distributions with different data sources (Figure 5; Figure S3.4 in Appendix S3).
5 CONCLUSIONS
This study provided further evidence on uncertainty-related gaps associated with SDMs from the most early-state sources of the model process (such as input data) to the final stages and outputs (such as model performance and spatial projections assessments). Our results showed that both the predictive capacity and performance of fitted regional (Iberian Peninsula) SDMs are influenced by the type of biodiversity data used for model calibration (presence/absence vs. only-presence data), the nature of the predictor variables used (interpolated macroclimate vs. remote sensing data), as well as model evaluation metrics (AUC vs. BoyceI). Our models confirmed the overall usefulness of presence-only citizen-collected data (eBird) for spatial range modelling, but also specific robustness for rare (or less widespread) species since eBird outperformed standardized Atlas datasets in predicting narrowly distributed species. This means that taxa that enjoy less attention (in terms of sampling effort, resulting in poor data availability) benefit from structured monitoring programs, while less abundant (or rare) species benefit from more opportunistic data. These findings might have implications for species conservation as models of less widespread or rare species with clear conservation concerns benefited from the inclusion of citizen science data. Despite ongoing concerns, citizen science data are becoming increasingly valuable research tools in biodiversity modelling and monitoring due to their increasing prevalence and broad spatiotemporal scope. However, and because neither data source is complete, data integration joining both structured sampling (Atlas) and opportunistic data (eBird) will leverage the strengths of each source of data and provide better predictions of species distributions and their drivers. Our models also showed that variable importance overall was consistent across bird datasets used to fit the models, being mostly driven by the species range. These results highlight the need for mainstreaming other key environmental factors into SDMs for increasing their predictive accuracy, and hence, improving ecological inferences for decision-making or conservation planning. In the light of our results, we suggest that the integration of different data sources (for both the response and predictor variables) in modelling frameworks will strongly contribute to a better knowledge and update of the distribution of biodiversity at regional scales. In this sense, we encourage careful attention must be paid at early stages of the modelling process, to all aspects of the ecological inference based on model-assisted approaches that use semi-structured or unstructured biodiversity data.
ACKNOWLEDGEMENTS
This research work was funded by PORBIOTA-Portuguese e-Infrastructure for Information and Research on Biodiversity (POCI-01-0145-FEDER-022127). S.A.-C. was financially supported by PORBIOTA grants and is currently supported by the María Zambrano fellowship program funded by the Spanish Ministry of Universities and the EU-NextGenerationEU fund. A.R. was funded by the Xunta de Galicia (Spain) grants (postdoctoral fellowship ED481B2016/084-0) and is currently supported by Juan de la Cierva fellowship program funded by the Spanish Ministry of Science and Innovation (IJC2019-041033-I). I.M. was financially supported by the Portuguese Foundation for Science and Technology-FCT (SFRH/BD/145676/2019), funded by the Portuguese Ministry of Science, Technology and Higher Education, and the European Social Fund (ESF-European Union) through NORTE2020. No permits were needed to carry out this work. Funding for open access charge: University of Córdoba/CBUA
CONFLICT OF INTEREST
The authors state that they have no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available in Dryad Digital Repository at 10.5061/dryad.qfttdz0jm.
REFERENCES
BIOSKETCH
Salvador Arenas-Castro is a broad-spectrum ecologist with interesting in different integrative perspective of the fundamental ecology, macroecology and biogeography with their both application and relationship to climate and land management, exploring other research sources in agroecology, spatial biology and earth observation techniques applied to natural resources. His ongoing work focuses on the detection of changes in biodiversity patterns, from the species to the ecosystem and landscape levels, according to forecasted climate and land uses at different multi-scale approaches. Please check his webpage for further information: https://salvadorarenascastro.wordpress.com/
Author contributions: All authors conceived and designed the idea; Salvador Arenas-Castro and Adrián Regos collected the bird datasets; Salvador Arenas-Castro performed the RS-EFAs computation and Adrián Regos derived the climate dataset; Salvador Arenas-Castro performed the modelling process and analysed the results with the advice of Adrián Regos, Ivone Martins, João Honrado and Joaquim Alonso; Adrián Regos and João Honrado provided advice in interpreting habitat models and the variables contribution; Ivone Martins and Joaquim Alonso provided help in analysing and discussing data quality assessment and model accuracy; Salvador Arenas-Castro led the writing and format editing, and all co-authors reviewed, edited and provided critical comments and feedback on drafts and approved the manuscript for publication.




