Rapid and Simultaneous Detection of Avian Influenza and Newcastle Disease Viruses by Duplex Polymerase Chain Reaction Assay
Summary
A duplex reverse transcription-polymerase chain reaction (dRT-PCR) assay has been developed for the simultaneous, rapid and specific detection/discrimination of avian influenza virus (AIV) and Newcastle disease virus (NDV). Primers targeting the matrix protein gene (M) of AIV and the fusion protein gene (F) of NDV were evaluated experimentally with 13 AIV and 19 NDV strains. PCR products of the expected size of 144 bp and 316 bp were amplified from AIV/NDV samples, respectively, while no cross-reaction was observed with negative controls or with 16 other avian pathogens. The endpoint of detection was defined as approximately 10+0.5 50% egg infectious dose (EID50)/0.2 ml for AIV and 10+2.2 EID50/0.2 ml for NDV. The assay was able to detect AIV/NDV with similar sensitivity in spiked stool samples and in specimens from vaccinated birds. The developed dRT-PCR assay is a rapid, cost-effective tool, which provides powerful novel means for the early diagnosis of avian influenza and Newcastle disease.
Introduction
Avian influenza (AI) and Newcastle disease (ND) are viral diseases of birds with considerable economic impact on domestic poultry production. Type A influenza viruses, the causative agents of AI, belong to the family Orthomyxoviridae and they are classified into low-pathogenic (LPAIV) and highly pathogenic (HPAIV) strains on the basis of their distinct virulence properties. Similarly, Newcastle disease virus (NDV), which is a member of the Avulavirus genus within the Paramyxoviridae family, also has strains of different virulence (lentogenic/mesogenic/velogenic) causing symptoms from unapparent infection to acute disease and death up to 100% (Alexander, 2004). Infection by either HPAIV or NDV may result in similar clinical signs and epidemiology in poultry. Therefore, measures to control highly pathogenic avian influenza (HPAI) and ND are included in the same national contingency plans as legislated according to Council Directives of the European Community (CEC, 1992a,b).
In order to diagnose and confirm primary HPAI and ND cases, the isolation and characterization of the causative virus is necessary, which needs considerable time (at least 4–7 days) to accomplish (Alexander, 2004). Since it has economical and epidemiological significance to identify avian influenza virus (AIV)/NDV as soon as possible, several PCR-based assays have been developed for the detection of these viruses (Spackman et al., 2003; Dybkaer et al., 2004).
In this paper we describe the construction of a duplex RT-PCR test capable of detecting and distinguishing AIV and NDV simultaneously in clinical samples within a reasonably short period of time, with the purpose of eliminating diagnostic uncertainties and avoiding delays. The specificity/sensitivity of the method was tested with archive isolates and clinical samples and it was found to be appropriate for the rapid, accurate and cost-effective detection of AIV and NDV.
Materials and Methods
Virus isolates
Virus strains and bacteria used in the specificity assay of duplex reverse transcription-polymerase chain reaction (dRT-PCR) are listed in Table 1. NDV strains B1, La Sota (PhylavacTM, VitapestTM), V4 Queensland and AIV strain A/tk/Turkey/8/11/05 were kindly provided by Ceva-Phylaxia (Budapest, Hungary) and the Central Veterinary Institute (CVI, Budapest, Hungary), respectively. Other AIV/NDV field isolates were collected from clinical samples in the Virology Department of the CVI, Debrecen, Hungary, in the period of 1969–1990. Virus samples were identified by propagation in embryonated chicken eggs and haemagglutination activity (HA) test according to the OIE manuals (Alexander, 2004).
| NDV vaccine strains | Virulence | Test result by | |
|---|---|---|---|
| AIV PCR | NDV PCR | ||
| LaSota | L | − | + |
| B1 | L | − | + |
| V4 Queensland NDV field isolates | L | − | + |
| NDV/ch/Hu/Milota/69 | V | − | + |
| NDV/ch/Hu/H.hadház/107/69 | V | − | + |
| NDV/ch/Hu/H.hadház/108/69 | V | − | + |
| NDV/ch/Hu/Debrecen/168/69 | V | − | + |
| NDV/ch/Hu/Túrkeve/48/70 | V | − | + |
| NDV/ch/Hu/H.böszörmény/114/70 | L | − | + |
| NDV/ch/Hu/Debrecen/332/70 | V | − | + |
| NDV/ch/Hu/Püspökladány/368/71 | L | − | + |
| NDV/ch/Hu/Tiszavárkony/25/72 | L | − | + |
| NDV/ch/Hu/Ebes/14/89 | L | − | + |
| NDV/tk/Hu/H.nánás/348/89 | L | − | + |
| NDV/ch/Hu/Körösszakáll/393/89 | L | − | + |
| NDV/tk/Hu/Nádudvar/542/89 | L | − | + |
| NDV/ch/Hu/Rákóczifalva/308/90 | L | − | + |
| NDV/tk/Hu/Debrecen/573/90 | L | − | + |
| NDV/ch/Iraq/D514/2005 | V | − | + |
| AIV field isolates | Subtype | ||
| A/dk/Hu/1/70 | H6 | + | − |
| A/dk/Hu/3/70 | H4 | + | − |
| A/md/Hu/1/75 | H5 | + | − |
| A/gf/Hu/2/75 | H7 | + | − |
| A/dk/Hu/2/75 | H6 | + | − |
| A/dk/Hu/3/75 | H5 | + | − |
| A/gf/Hu/3/75 | H11 | + | − |
| A/dk/Hu/11/75 | H5 | + | − |
| A/dk/Hu/2/77 | H5 | + | − |
| A/gf/Hu/1/84 | H7 | + | − |
| A/tk/Hu/1/85 | H7 | + | − |
| A/tk/Hu/2/87 | H10 | + | − |
| A/tk/Turkey/8/11/05 | H5 | + | − |
| Heterologous virus/bacterial strains | |||
| Avian metapneumovirus | − | − | |
| Avian polyoma virus | − | − | |
| Avian reovirus | − | − | |
| Chicken anaemia virus | − | − | |
| Derzsy's disease virus | − | − | |
| Infectious bronchitis virus | − | − | |
| Infectious laryngotracheitis virus | − | − | |
| Mycoplasma gallisepticum | − | − | |
| Mycoplasma iowae | − | − | |
| Mycoplasma synoviae | − | − | |
| Ornithobacterium rhinotracheale | − | − | |
| Pasteurella multocida | − | − | |
| Psittacid herpesvirus 1 | − | − | |
| Riemerella anatipestifer | − | − | |
| Turkey adenovirus | − | − | |
| Turkey astrovirus | − | − | |
- Hu, Hungary; ck, chicken; dk, duck; gf, guinea fowl; md, muscovy duck; tk, turkey; L, lentogen; V, velogen.
Determination of virus titre
Virus stock solution was diluted 1 : 10 in sterile PBS supplemented with 100 U/ml of penicillin-G and 0.1 mg/ml of streptomycin (Sigma, St Louis, MO, USA). Embryonated chicken eggs were inoculated with 0.2 ml diluted virus preparation on the 10th day of incubation by the allantoic sac route (five eggs/dilution). Between days 3 and 6 post-inoculation allantoic fluid was collected and tested for haemagglutination with 0.5% chicken red blood cells (CRBCs) in PBS. The virus titre was determined by the Reed-Muench method (Reed and Muench, 1938).
RNA extraction and cDNA synthesis
RNA was extracted from 140 μl volume of sample (allantoic fluid/lyophilized virus stock reconstituted in distilled water/suspended stool sample) by using the QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) as recommended by the manufacturer. The reverse transcription (RT) of RNA to cDNA was performed using Moloney murine leukemia virus reverse transcriptase (M-MLV RT; Invitrogen, Carlsbad, NM, USA), pd(N)6 random hexamer 5′-phosphate and RNAguard (Amersham, Piscataway, NJ, USA) according to the manufacturer's instructions.
Primer design and polymerase chain reaction
Avian influenza virus and NDV specific oligonucleotide primers were designed by using nucleotide sequences available in GenBank, the software package DNASTAR and the program Primer Designer Version 2.0 (Scientific and Educational Software).
Primer pair AIV-MP37-For (5′-GATGAGTCTTCTAACCGAGG-3′) and AIV-MP180-Rev (5′-GTCTTTAGCCAYTCCATGAG-3′) amplifies a 144 bp long fragment from the AIV matrix protein gene (M), while primers NDVF-for2 (5′-TGGGAAGATGCAGCARTTTG-3′) and NDVF-rev1 (5′-RCCCAAGAGTTGAGTCTGTGA-3′) are specific for the fusion protein gene (F) of NDV and the size of their amplification product is 316 bp (Y: pyrimidine; R: purine).
The optimization of PCR conditions was performed using the cDNA from AIV strain A/gf/Hu/2/75 (H7) and NDV vaccine strain La Sota as template in the MyCycler (Bio-Rad, Budapest, Hungary) and the RapidCycler (Idaho Technology, Idaho Falls, ID, USA) devices. Optimal PCR conditions are as follows:
Amplification in the MyCycler instrument was performed in a final volume of 20 μl, the reaction mix consisted of 2 μl AmpliTaq Gold 10x PCR buffer, 3.2 μl 25 mm MgCl2, 0.1 μl AmpliTaq Gold polymerase (5 U/μl; Applied Biosystems, Foster City, CA, USA), 0.4 μl 10 mm dNTP blend (0.2 mm final concentration of each), 1 μl 5 mg/ml BSA (Promega, Madison, WI, USA), 0.2–0.2 μl of primers AIV-MP37-For; AIV-MP180-Rev; NDVF-for2 and NDVF-rev1 (25 pmol/μl; Biomi, Gödöllő, Hungary), 1 μl template cDNA and 11.9 μl PCR grade H2O (Life Technologies, Gaithersburg, MD, USA). The PCR cycling program was: 95°C 7 min; 40 cycles of 95°C 30 s, 58°C 30 s, 72°C 60 s; followed by 72°C 7 min.
Amplification in the RapidCycler was carried out as described for MyCycler. The total reaction volume was 10 μl and the quantity of PCR components was reduced accordingly, except that 1 μl 5 mg/ml BSA; 0.1 μl AmpliTaq Gold polymerase and 1 μl template cDNA was added to the reaction. The thermal profile was: 95°C 7 min; 40 cycles of 94°C 20 s, 58°C 20 s, 72°C 20 s; followed by 72°C 7 min. PCR grade water was used in the reactions as negative control.
DNA sequencing and nucleotide sequence accession numbers
The amplified DNA fragments were purified with the QIAquick PCR purification kit (Qiagen) according to the manufacturer's instructions. Double-stranded sequencing reaction was performed by using the BigDye Terminator Cycle Sequencing v3.1 kit (Applied Biosystems) and the same primers as in the dRT-PCR. Samples were analysed in an automated sequencer ABI3100 (Biomi). The nucleotide sequences of four AIV and four NDV amplicons have been deposited in GenBank (Acc. No.: DQ533836-39 and DQ455008-455011).
PCR specificity and sensitivity assay
To determine the specificity of the dRT-PCR assay, RNA/DNA was extracted from AIV/NDV strains and 16 heterologous avian pathogens (Table 1) by using QIAamp Viral RNA Mini Kit and DNeasy Tissue Kit (Qiagen). Nucleic acid samples were processed and tested as described above.
The sensitivity of the dRT-PCR assay was determined by using AIV/NDV stock solutions with known egg infectious dose (EID)50 values (AIV strain A/gf/Hu/2/75: 10+7.5 EID50/0.2 ml and NDV strain La Sota: 10+9.2 EID50/0.2 ml). Serial 10-fold dilutions were prepared of the AIV/NDV stock solutions with sterile H2O, then RNA was extracted and cDNA was synthesized from each dilution as described above. First, the detection limit was measured separately for AIV and NDV by adding only AIV or NDV cDNA to the reaction (101–108× dilutions). To determine the ability of the dRT-PCR to detect and differentiate AIV/NDV in the same reaction, different dilutions of AIV and NDV cDNAs were mixed in various combinations (in 1 : 1 ratio) and the mixture was used as template. In the first set of experiment, AIV and NDV stock solutions were diluted in parallel (101–108× dilutions, Fig. 1a), while in the second set the titre of one virus was constant (103–104× dilutions) and the quantity of the other virus was changing (101–108× dilutions, Fig. 1b).
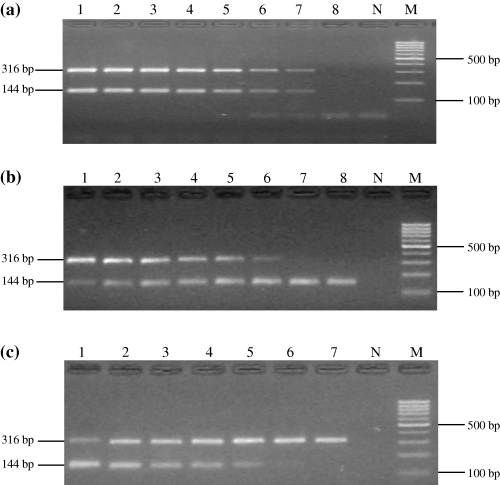
Sensitivity test of AIV-NDV dRT-PCR (MyCycler; 30-30–60 s cycle). (a) Template: cDNA purified from serial 10-fold dilutions of AIV strain A/gf/Hu/2/75 and NDV strain La Sota stock solutions. Lane 1–8, template dilution series (lane 1: ×101- lane 8: ×108); N: negative control, M: molecular size marker (Fermentas 100 bp ladder). (b) Template: cDNA purified from ×104 dilution of AIV strain A/gf/Hu/2/75 and serial 10-fold dilution of NDV stock solution. Lane 1–8, template dilution series (lane 1: ×101- lane 8: ×108). c-Template: cDNA purified from NDV positive stool samples (from vaccinated chickens) that were spiked with a serial 10-fold dilution of AIV stock solution (lane 1: ×102- lane 7: ×108).
Detection of AIV/NDV from experimentally infected stool specimens
Chicken stool samples previously giving negative test results in the dRT-PCR were collected and pooled. To 0.2 g pooled faeces 50 μl of the 101–108× dilutions of AIV (A/gf/Hu/2/75) or NDV (La Sota) stock solutions was added, then samples were suspended in 450 μl sterile H2O, thoroughly vortexed and centrifuged (2 min; 4700 g; RT). Viral RNA was extracted from the supernatant and processed as described above. The 10-fold dilution applied during the preparation of samples was calculated in the results.
Detection of AIV/NDV in stool samples from vaccinated chickens
A breeder flock was vaccinated with Avinew (Merial, Lyon, France) at day 1 and with TAD ND vac HB1 (Lohmann, Cuxhaven, Germany) at 21 days of age. Vaccines contained VG/GA and Hitchner B1 live NDV strains, respectively. Control chickens were left untreated and kept separately. After 2 weeks of the second vaccination, stool specimens were collected and pooled according to the EC Regulation No. 1003/2005 (CEC, 2005). To 0.2 g pooled faeces 0.5 ml sterile H2O was added, then samples were thoroughly vortexed and centrifuged (2 min; 4700 g; RT). Viral RNA was extracted from the supernatant and processed as described above. Results were confirmed by a PCR assay using universal and pathotype specific primer pairs able to differentiate lentogenic and velogenic NDV strains (Kant et al., 1997). In order to examine the effect of double infection on the sensitivity of dRT-PCR serial, 10-fold dilutions of AIV stock solution were made and aliquotes were added to the pooled stool samples collected from vaccinated birds. Viral RNA was extracted, processed and tested in dRT-PCR as described above (see details of spiking in previous section). The 10-fold dilution applied during the preparation of spiked stool samples was calculated in the results.
Results
Duplex RT-PCR identification of AI and ND viruses in different thermocyclers
The nucleotide sequences of known AIV and NDV isolates were compared in multiple alignments and primers targeting highly conserved regions, the matrix protein gene (M) of AIV and the fusion protein gene (F) of NDV, were designed. The optimal conditions for the duplex PCR assay were determined in two thermal cyclers using the cDNA from AIV and NDV as template. A conventional heat block thermal cycler, MyCycler, was tested to assure that the dRT-PCR can be applied on the currently most prevalent system. In the RapidCycler, cycling system is based on the heat transfer by ventilating hot/cool air to the samples. Rapid heat exchange, the use of 1.5 mm glass reaction cuvettes (capillaries), small reaction mix volume (10 μl) allows the use of relatively short cycling times. Optimal reaction conditions were almost identical in the two instruments, there were notable exceptions only in the total volume of reaction mix (MyCycler: 20 μl; RapidCycler: 10 μl), cycling times (MyCycler: 30–30–60 s; RapidCycler: 20–20–20 s) and BSA concentration (MyCycler: 0.25 mg/ml; RapidCycler: 0.5 mg/ml).
Specificity of duplex RT-PCR assay
In the first step, to examine the specificity of the dRT-PCR assay, 13 AIV isolates (subtypes: H4; H5; H6; H7; H10; H11), 16 NDV isolates (nine lentogenic and seven velogenic strains) and three NDV vaccine strains were subjected to dRT-PCR, as shown in Table 1. The expected PCR products of 144 bp and 316 bp length, respectively, were amplified from all AIV/NDV samples. In the second step, the specificity was further investigated by testing other potential avian pathogens, including bacteria. No cross-reaction was observed with the heterologous disease-producing agents. The identity of the 144 bp (AIV) and 316 bp (NDV) PCR products was confirmed by means of DNA sequence analysis and comparative search against GenBank.
Sensitivity of dRT-PCR assay
The sensitivity of the dRT-PCR assay was determined using serial 10-fold dilutions of AIV and NDV stock solutions. The detection limit of the duplex PCR was approximately 10+0.5 EID50/0.2 ml for AIV and 10+2.2 EID50/0.2 ml for NDV when cDNA templates were added separately to the reaction (data not shown). No alteration was observed in the sensitivity when both target cDNAs (AIV and NDV) were present in the PCR mix and they were diluted in parallel (Fig. 1a). However, when one viral cDNA was added to the reaction in excess a slight decrease (maximum one order of magnitude) was observed in the detection limit (Fig. 1b).
The type of the thermal cycler did not influence the endpoint of detection, as we had identical results in the MyCycler and in the RapidCycler.
Detection of AIV/NDV from experimentally infected stool specimens
We evaluated the ability of dRT-PCR to amplify the corresponding genomic regions from viral RNA extracted from faecal samples. Chicken stool samples were experimentally infected with a dilution series of AIV or NDV stock solutions. The detection limit of duplex RT-PCR was approximately 10+0.5 EID50/0.2 ml for AIV and 10+2.2 EID50/0.2 ml for NDV in this reaction. The assay was able to detect the corresponding virus(es) from each specimen with the same sensitivity as from allantoic fluid, in spite of the fact that stool samples are reported to contain several PCR inhibitory compounds (bile salts, haemoglobin degradation products, etc.) that seriously decrease the efficiency of nucleic acid detection (Higuchi, 1989; Radstrom et al., 2004).
Detection of AIV/NDV in stool samples from vaccinated chickens
Stool samples collected from a chicken flock previously inoculated with NDV vaccine strains were all positive by dRT-PCR for the presence of NDV, while no amplification product was observed in the negative controls. Positive samples were subjected to a pathotype specific PCR (Kant et al., 1997) and the results proved that specimens contained lentogenic NDV RNA (data not shown).
When the effect of double infection was investigated on the sensitivity of dRT-PCR by the addition of serial 10-fold dilutions of AIV stock solutions to the stool samples collected from vaccinated birds, NDV remained detectable in all samples, though high concentration of AIV was sometimes present in the reaction (Fig. 1c, lane 1–2: 10+5.5–10+4.5 EID50/0.2 ml). The endpoint of detection for AIV was calculated as approximately 10+1.5 EID50/0.2 ml.
Discussion
Highly pathogenic avian influenza and Newcastle disease are of major concern from veterinary and economic aspects. In the case of HPAIV, public health factors also became very important in the last years, considering that these viruses are capable of causing avian-to-human transmissions. It is widely accepted that HPAIV genetic variants gaining human-to-human transmission capacity would pose the potential threat of a pandemic among people with devastating consequences (Li et al., 2003; Reid et al., 2004). Therefore, the accurate detection and distinction of the two viruses is crucial for timely initiation of control when it becomes necessary.
The conventional methods of diagnosing AI and ND (Alexander, 2004) are time consuming. Therefore, more rapid procedures are certainly required in the laboratories involved in their investigation. The various amplification methods, like RT-PCR and real-time RT-PCR based detecting assays strongly facilitate the improved detection of AIV and NDV (Fouchier et al., 2000; Spackman et al., 2002, 2003; Belák and Thorén, 2004; Wise et al., 2004; Pham et al., 2005).
The duplex RT-PCR described herein is targeting highly conserved regions of the viral genomes (AIV M gene, NDV F gene) and it is capable of detecting a broad range of AIV and NDV variants in the same reaction. Compared with other multiplex PCR protocols designed to detect AIV/NDV (Pang et al., 2002; Malik et al., 2004; Soares et al., 2005), a wider range of causative agents (AIV subtypes, NDV strains, heterologous avian pathogens) and sample types (cDNA extracted from virus suspension, spiked stool samples, faecal specimens from vaccinated chickens) was involved in the specificity/sensitivity testing of the assay. Being a gel-based PCR, the simplicity of the method facilitates the immediate and front-line diagnosis of AI and ND close to outbreak cases. The assay was shown to be capable of detecting either virus in faecal samples that makes it an appealing tool among the diagnostic procedures used in monitoring programmes. Therefore, the described dRT-PCR provides a useful and practical supportive method for the effective diagnosis of avian influenza and Newcastle disease.
Acknowledgements
This work was supported by a grant from the 6th Framework Programme of the European Union (Contract No.: SSPE-CT-2004–513645, http://www.labonsite.com). Archive AIV isolates were collected by Dr J. Tanyi (CVI, Debrecen, Hungary). We wish to thank É. Székely, A. Incze for excellent technical assistance and S. Egedi (Biomi, Gödöllő, Hungary) for the support in oligonucleotide synthesis and DNA sequencing.




