Microsatellite markers for the fungal banana pathogens Mycosphaerella fijiensis, Mycosphaerella musicola and Mycosphaerella eumusae
Abstract
We developed a total of 50 microsatellite markers for the three fungal pathogens causing the most important leaf spot diseases of banana: 32 loci for Mycosphaerella fijiensis are presented, and nine loci each for Mycosphaerella musicola and Mycosphaerella eumusae. All these loci were polymorphic within each species on a sample of isolates collected from various locations around the world. Within M. fijiensis and M. musicola, most of the loci tested (> 80%) in a sample of isolates from a single location in Cameroon were also polymorphic. Multiplex polymerase chain reaction systems were developed with 15 loci for M. fijiensis.
The most important leaf spot diseases of banana are caused by three related haploid Ascomycetes belonging to the genus Mycosphaerella (Jones, 2000): M. musicola (causal agent of Sigatoka disease), M. fijiensis (black leaf streak disease) and M. eumusae (eumusae leaf spot disease). In order to conduct evolutionary studies of these three fungal pathogens, it is important to use the same class of genetic markers. Reliable genetic markers are also needed to develop a genetic map of M. fijiensis, currently the most aggressive and predominant species. Some microsatellite markers have already been developed for M. fijiensis (Neu et al. 1999) and M. musicola (Molina et al. 2001). The present work describes new microsatellite markers for M. musicola and M. fijiensis, including a particularly large number for M. fijiensis, as well as the first reported markers for M. eumusae.
A genomic library enriched with GA and GT microsatellite sequences was produced for each of the three species according to the protocol described by Billotte et al. (1999). Libraries of isolates CIRAD009 (M. fijiensis), CIRAD090 (M. musicola) and CIRAD487 (M. eumusae) were constructed from total DNA extracted using the method described in Carlier et al. (1996). For each genomic library, inserts from several hundred clones were hybridized with γ32P-radiolabelled (GA)15 and (GT)15 probes. Satisfactory positive signals indicating the presence of a microsatellite region were obtained in 185/491 (37.7%), 133/629 (21.1%), and 162/519 (31.2%) clones from the M. fijiensis, M. musicola, and M. eumusae libraries, respectively. Positive clones for each species were sequenced by the Centre National de Séquençage (CNS, Génoscope, Evry, France); 67/115 (58.3%), 28/63 (44.4%), and 34/58 (58.6%) of these M. fijiensis, M. musicola, and M. eumusae sequences, respectively, were unique and contained a microsatellite region. Using the software primer 3 (Rozen & Skaletsky 2000), primer pairs were designed from 52 unique sequences for M. fijiensis, 18 sequences for M. musicola and 19 sequences for M. eumusae. Polymerase chain reaction (PCR) amplification using these primers was first tested for each species with the reference isolate used to produce the library, as described by Rivas et al. (2004). Of the primer pairs tested, 45/52 (86.5%; M. fijiensis), 11/18 (61.1%; M. musicola), and 11/19 (57.9%; M. eumusae) gave rise to a single amplification product of the expected size. The polymorphism of these sequences was then tested within each species on a global sample of 15 isolates [collected from different banana-producing regions (Asia, Oceania, America and Africa) to maximize diversity]. PCRs were performed as described by Roy et al. (1996) using automated infrared fluorescence technology (LI-COR IR2 sequencer). For each locus, one member of the primer pair had a 5′-end M13 extension (5′-CACGACGTTGTAAAACGAC-3′). PCRs were performed in a PTC-100 thermocycler (MJ Research) in a reaction mixture (15 µL) containing 15 ng of fungal genomic DNA, 1× reaction buffer (Eurobio), 0.2 mm of each dNTP (Sigma), 2 mm MgCl2, 0.08 µm of the M13-labelled primer, 0.1 µm of the other primer, 0.06 µm M13 primer-fluorescent dye IR700 or IR800 (Biolegio, the Netherlands) and 0.1 U of Taq-DNA polymerase (Eurobio). PCR was performed under the following conditions: 5 min at 94 °C; 34 cycles of 1 min at 94 °C, 1 min at 55 °C, 1 min 30 s at 72 °C; and a final elongation of 8 min at 72 °C. IR700- or IR800-labelled PCR products were diluted 10-fold or fourfold, respectively, subjected to electrophoresis on a 6.5% polyacrylamide gel and then sized using the IR fluorescence scanning system of the sequencer. For M. fijiensis and M. musicola, the polymorphism of 15 and seven loci, respectively, was further tested on samples of 43 and 66 isolates collected from a single location in Cameroon, Kumba and Bambui, respectively. Finally, we developed a multiplex PCR protocol for M. fijiensis using a capillary sequencer system. Two pools of six and nine primer pairs were used (Table 1) to amplify M. fijiensis genomic DNA as follows: 30 ng of genomic DNA was used as template in PCRs containing 0.2 µm of each primer [with one primer labelled with one of the following fluorescent dyes: NED, HEX or FAM (Applied Biosystems; see Table 1)], 5 µL 2× QIAGEN multiplex mastermix [containing HotStar Taq DNA polymerase, KCl (NH4)SO4, TrisHCl, 3 mm MgCl2, and 0.2 µm of each dNTP], 1 µL 10× primer mix, 0.5× Q-solution, and sterile distilled water to give a final volume of 10 µL. Cycling was performed in a thermocycler (PTC100) under the following conditions: 15 min at 95 °C; 45 cycles of 30 s at 94 °C, 90 s at 57 °C, 60 s at 72 °C; and a final extension of 10 min at 72 °C. Two microlitres of diluted amplified products (1/40) were mixed with 8 µL of MegaBACE ET 400-R size standard (diluted 1/40). PCR products were separated on a MegaBACE 1000 DNA sequencing apparatus (Amersham).
| Locus name | GenBank Accession no. | Repeat motif of cloned allele | Primer name | Sequences | Expected size | PCR | Diversity | Cross-amplification‡ | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Global scale* | Intrapopulation† | ||||||||||||||
| T a (°C) | Labelling dye (multiplex pool) | N nu (%) | N | H E | N nu(%) | N a (Range size) | H E | ||||||||
| Mycosphaerella fijiensis | M. musicola | M. eumusae | |||||||||||||
| MfSSR301 | EU200696 | (GT)7(GA)3 | FA02 | F: CAGCCCAGCCTCCTTTTACTR: TGTTCAGCCACCACCCTCTC | 113 | 55 | 0 | 3 | 0.5 | 6/10 | 0/15 | ||||
| MfSSR303 | EU200697 | (GT)3(...)(GA)3 | F3 | F: ATGTTTTTGCATGCGTGTGTR: TCCTCAAAGACAAGGACATGA | 268 | 55 | 0 | 2 | 0.32 | ||||||
| MfSSR305 | EU200698 | (CA)6 | F5 | F: AGTGGCATCTTTCTGCTCGTR: ATTTTGCAAGTCGGGCTAT | 302 | 55 | 1 (6.7) | 3 | 0.61 | ||||||
| MfSSR306 | EU200699 | (GT)15 | FB05 | F: GGCTTCGGCAAATAAATAR: GCTGATGCCGCTTCGTAG | 123 | 55 | HEX (2) | 0 | 8 | 0.83 | 2 (4.6) | 2 (124/126) | 0.21 | 0/10 | 0/15 |
| MfSSR313 | EU200700 | (TA)5 | F13 | F: CAACCTATAGCAACGCATCGR: GAGTGCGGATTAGGGAAACA | 209 | 55 | 2 (13.3) | 3 | 0.38 | ||||||
| MfSSR314 | EU200701 | (GGT)2GT(GTT)2(GT)4 T(GT)2(...)(AC)3 | F14B | F: CACTGTGCCAAAAGAAACCAR: AGTGACTTGATGCGGATGTG | 142 | 55 | 0 | 7 | 0.8 | ||||||
| MfSSR315 | EU200702 | (AT)4(...)(AC)8 | FC12b | F: GAGCGACCTTCCTCTTTCAAR: CGAGAGCTTGGGAAAAAGTG | 178 | 55 | HEX (1) | 0 | 6 | 0.71 | 0 | 2 (178/179) | |||
| MfSSR316 | EU200703 | (CAA)3(...)(TG)3C(GT)4 (TG)3(...)(GA)3 | FD01 | F: AACCACGCACAACAACAAGCR: ATCAACATCAGCCTCACACG | 143 | 55 | 0 | 3 | 0.24 | 9/10 | 2/15 | ||||
| MfSSR319 | EU200704 | (CT)17C(CT)1 | F19 | F: CGTCATCGGGCTTTATGTCTR: ATCATGGGTAGGGGGAGAAC | 270 | 55 | FAM (1) | 0 | 3 | 0.48 | 0 | 1 (271) | 0 | ||
| MfSSR322 | EU200705 | (AC)7 | FE05B | F: GCGCAACAGGTCACATAAGAR: AGTCCTCTGGGAGGCATTCT | 203 | 55 | HEX (1) | 0 | 4 | 0.72 | 0 | 3 (197/202) | 0.57 | ||
| MfSSR323 | EU200706 | (CTT)3(...)(GT)6(...) (GA)4 A (GA)1 | FE07B | F: GGCTGGGTTTGATGTTGAGTR: CGCGACGTCTTCTCTCTCTT | 220 | 55 | 1 (6.7) | 2 | 0.14 | ||||||
| MfSSR324 | EU200707 | (TG)14 | FE09 | F: ACAGACGAACGACCGACGACR: ATTACGCCCAGAACACCTTT | 115 | 55 | NED (1) | 0 | 6 | 0.53 | 1 (2.3) | 3 (100/120) | 0.38 | 0/10 | 0/15 |
| MfSSR328 | EU200708 | (GT)2G(GT)5 | F28 | F: ATCACTGACGGTCGGTAAGGR: CTGGTCAGAGCGTCTTTTCC | 150 | 55 | HEX (2) | 0 | 3 | 0.42 | 2 (4.6) | 2 (150/153) | 0.5 | ||
| MfSSR329 | EU200709 | (AG)9(TG)7 | F29 | F: GTCGATAGAGCCAGCGGTAGR: GGGAAGGGGAATGGAATTTA | 184 | 55 | 2 (13.3) | 4 | 0.63 | ||||||
| MfSSR333 | EU200710 | (AG)7(TG)9 | FG08 | F: GCATGAGCAGGCGAGATAR: GCAAGGACAAGGATTAGC | 140 | 55 | 3 (20) | 7 | 0.79 | 0/10 | 0/15 | ||||
| MfSSR340 | EU200711 | (CT)3(...)(CA)9 | F40 | F: CCCACTCTTACTCCCCACAAR: CACACGACAAAAACCCCTTT | 270 | 57 | HEX (2) | 0 | 4 | 0.59 | 0 | 2 (271/273) | 0.44 | ||
| MfSSR343 | EU200712 | (TG)6C(TG)1 | F43 | F: ATGTCCAAAGTATGTGTGR: CAACGAAATCCCTCTCCA | 78 | 55 | 0 | 2 | 0.32 | ||||||
| MfSSR347 | EU200713 | (AC)3(...)(CA)7 | FD10B | F: CCAATGCAAACCAGTCAATGR: AAACGACAGCAACAGCACAG | 153 | 55 | HEX (1) | 0 | 4 | 0.68 | 0 | 4 (152/155) | 0.46 | ||
| MfSSR350 | EU200714 | (GAA)3(...)(CA)9 | FF09 | F: AAGGCGTTGGTTGTGTAAAR: GCTTGCGGTTCTGTCGTCA | 222 | 55 | FAM (2) | 0 | 5 | 0.52 | 0 | 2 (224/226) | 0.04 | 0/10 | 0/15 |
| MfSSR353 | EU200715 | (GT)6 | F53 | F: AATGGCAACCGCTTATGAR: ACTCATAGATGTAAATACACAC | 130 | 55 | 0 | 3 | 0.52 | ||||||
| MfSSR354 | EU200716 | (GT)7 | Fb12b | F: AGACCCATTCTTCATGACACGR: GCAGAAACAACAGACCCACA | 221 | 55 | 3 (20) | 5 | 0.68 | ||||||
| MfSSR355 | EU200717 | (GT)7 | FD11 | F: CGACCATGCCAAGCGTTTCR: TGCCACTAGAGGAGACAGC | 149 | 55 | 0 | 4 | 0.59 | 0/10 | 0/15 | ||||
| F96 | F: TGTTTTCTTGCACAGCCATCR: CTCGTGTGTCTTCGGTGCTA | 195 | 55 | FAM (2) | 1 (6.7) | 6 | 0.73 | 0 | 2 (195/197) | 0.04 | |||||
| MfSSR356 | EU200718 | (CA)1C(CA)8AAA(CA)5 | fb10 | F: GCGCATGATGAGAAAAAR: GGATGTTGAGGAGTTGT | 224 | 50 | NED (1) | 0 | 4 | 0.58 | 1 (2.3) | 2 (210/224) | 0.46 | 0/10 | 0/15 |
| MfSSR357 | EU200719 | (TG)5(...)(GT)3GC(GT)3 GC(GT)11(...)(GT)4 | FC06B | F: GAATGGAAGCTTTGCAGGAGR: GCCTAAGACACACACGCACA | 223 | 55 | 0 | 9 | 0.82 | ||||||
| MfSSR358 | EU200720 | (AC)23 | FF01 | F: GGAAGTGCCAAGAAAAGGR: GACAGTGAGGAGATGAGC | 154 | 55 | NED (2) | 1 (6.7) | 6 | 0.69 | 1 (2.3) | 6 (131/155) | 0.66 | 0/15 | 0/15 |
| MfSSR359 | EU200721 | (AC)6 | F59 | F: ACGTAACCACATCGGCCAR: GTCCAGTGCCAAGGCTTG | 100 | 55 | 1 (6.7) | 3 | 0.5 | ||||||
| MfSSR362 | EU200722 | (GA)4(...)(GT)6 | F62 | F: TTCCCACTACATCCGGAAAGR: CCCTCCTTTTCCTCAACACA | 182 | 57 | FAM (1) | 1 (6.7) | 5 | 0.72 | 0 | 3 (183/187) | 0.34 | ||
| MfSSR364 | EU200723 | (GT)28 | FH12b | F: TGCAATTTCGACTGCTGTTCR: GCTAGCAACCAAAGAGAGACTG | 132 | 55 | FAM (1) | 5 (33.3) | 6 | 0.78 | 0 | 7 (117/161) | 0.75 | ||
| MfSSR375 | EU200724 | (TG)7(GT)2 | F75 | F: CTAAATTCCCCAATGCTGGAR: AGCCCAAAGATGAAGTGTGC | 120 | 55 | HEX (1) | 0 | 4 | 0.43 | 0 | 1 (122) | 0 | ||
| MfSSR381 | EU200725 | (TA)3(...)(CA)3C(CA)3 | F81 | F: ACTTTTACCTCGGCGATCCTR: CCGGAGATGATGAAGGTTGT | 126 | 55 | 2 (13.3) | 3 | 0.38 | ||||||
| MfSSR392 | EU200726 | (TG)34 | F92 | F: GCCATTGCTATTGATTTGTGR: GCCAGCTTTTCTCCCACA | 162 | 55 | 0 | 8 | 0.83 | ||||||
| MfSSR394 | EU200727 | (TA)4(ACT)3 | F94 | F: GACGACGGACTTATTACCAGCTAR: CGAACTCCTATCTAACGCTATTTC | 99 | 55 | 0 | 2 | 0.44 | ||||||
| Means | 4.6 | 0.59 | 2.8 | 0.32 | |||||||||||
| Mycosphaerella musicola | M. fijiensis | M. eumusae | |||||||||||||
| MmSSR101 | EU200730 | (GT)8 | MC07 | F: TGTCCGGCCATTTTGATAGR: ACTGCAGGTAGGTCTTTAG | 86 | 50 | 0 | 2 | 0.32 | ||||||
| MmSSR103 | EU200731 | (CAAG)4CAC(CA)8 CGCC(GCAC)5 | MD09 | F: CAAGCAAGCAAGCAAGCACR: TGGAAATGGGTCGGAAACG | 92 | 60 | 0 | 5 | 0.7 | 0 | 2 | 0.3 | 0/16 | 10/15 | |
| MmSSR104 | EU200732 | (GT)25 | MC03 | F: TGAGAAGATGAATGTGAAGTR: CTTGCGTAGGCGTGATA | 119 | 55 | 0 | 7 | 0.8 | 3(4.5) | 3 | 0.54 | 0/16 | 0/15 | |
| MmSSR105 | EU200733 | (CA)7CC(CA)2 | MF08 | F: GAAAGTGTTGGCGTGGTAGGR: AGCGAAGATCAGAGGTTATG | 122 | 55 | 1 (6.7) | 2 | 0.34 | 16/16 | 4/15 | ||||
| MmSSR106 | EU200734 | (GT)3T(GT)3GC(GT)7 | MD10 | F: AGCCTTGTAGATGTTTGTGTR: ATGGAAGTTGCGAGAATGTT | 141 | 55 | 0 | 2 | 0.48 | 4 (6.0) | 3 | 0.57 | 3/16 | 15/15 | |
| MmSSR108 | EU200735 | (GT)8 | MF12 | F: CCCGCTAAATGCCTATCTCGR: AGTTTGCGTAATCTAAGTCG | 159 | 50 | 1 (6.7) | 3 | 0.36 | 3 (4.5) | 3 | 0.51 | 1/16 | 1/15 | |
| MmSSR109 | EU200736 | (CAG)4(...)(TG)19 | MC12 | F: TCGAACAGCGACTTTATTCR: CAAAAGCGGAAAATGGTCA | 186 | 55 | 0 | 4 | 0.75 | 0 | 5 | 0.63 | 0/16 | 0/16 | |
| MmSSR110 | EU200737 | (AG)11(TG)9 | MA06 | F: TCCGCATTTTCCCCAGTCCTR: GCTATTCTTTCCACGCACAC | 199 | 60 | 0 | 4 | 0.66 | 2 (3.0) | 4 | 0.67 | 0/16 | 0/16 | |
| MmSSR111 | EU200738 | (GT)13CTCC(CT)4 | MG04 | F: TTTCCTTGTCACCTGTTTGCR: TTCCTGGGAATGCTCTACGG | 220 | 60 | 6 (40) | 7 | 0.84 | 5 (7.6) | 4 | 0.62 | 1/16 | 1/16 | |
| Means | 4.0 | 0.58 | 3.4 | 0.55 | |||||||||||
| Mycosphaerella eumusae | M. fijiensis | M. musicola | |||||||||||||
| MeSSR001 | EU200739 | (GT) 9 | EA03 | F: GTGACTGGGGTTTCGTGTR: TCCGTCCTTGTTGCTGTA | 134 | 58 | 0 | 5 | 0.78 | 0/16 | 0/10 | ||||
| MeSSR002 | EU200740 | (CA)7 | EA06 | F: TTCACTCATTCCCAACTCR: GGCGTGTCATTCCTCTCT | 110 | 55 | 0 | 2 | 0.4 | 0/16 | 0/10 | ||||
| MeSSR003 | EU200741 | (GT)4(...)(GT)5AT(GT)4 AT(GT)4 | EB08 | F: GGCGGCAGTAACAAGAAGR: AATAGAACTGGCACATCC | 124 | 55 | 0 | 3 | 0.54 | 0/16 | 0/10 | ||||
| MeSSR006 | EU200742 | (GT)8 | ED07 | F: CGAGGGCGGTATGACTTGAAR: CCCCTTTTAGCGAGCGACTT | 87 | 60 | 0 | 2 | 0.5 | 1/16 | 0/10 | ||||
| MeSSR010 | EU200743 | (GT)9(...)(AT)4 | ED09 | F: CATTGCGGCCAGGATAAGR: CCCCAGCAATACTCACATA | 92 | 55 | 0 | 2 | 0.5 | 0/16 | 0/10 | ||||
| MeSSR014 | EU200744 | (GT)8 | EG07 | F: CGTCGCAGCAAGGATGTGR: ACCTCCTCCCAAAGATGA | 238 | 58 | 0 | 3 | 0.61 | 2/16 | 0/10 | ||||
| MeSSR015 | EU200745 | (AC)8 | EG12 | F: CCATTGCATAACTACTACACCR: TGACTGATGATACCCGACTTG | 179 | 55 | 1 (6.7) | 4 | 0.69 | 2/16 | 0/10 | ||||
| MeSSR016 | EU200746 | (AC)5(TC)6 | EH01 | F: AAAAGAAACAAGACACCACAR: GGGATACGAGATACCACAAG | 88 | 55 | 0 | 2 | 0.33 | 0/16 | 0/10 | ||||
| MeSSR019 | EU200747 | (CA)9 | EF12 | F: CCTTCAAAGGTCTCGTCTACR: ATGTTGCTTGCTCACTAATG | 175 | 55 | 0 | 3 | 0.57 | 0/16 | 0/10 | ||||
| Means | 2.9 | 0.55 | |||||||||||||
- * sample of 15 isolates from different banana-producing regions (Asia, Oceania, America and Africa).
- † sample of 43 and 66 isolates of M. fijiensis and M. musicola, respectively, from a single location in Cameroon.
- ‡ for each combination of related species and locus, ratio between positive isolates and total number of isolates tested.
The characteristics of the selected microsatellite markers are listed in Table 1. To name the loci for M. fijiensis and M. musicola, we followed the nomenclature and continued the numeric series of Neu et al. (1999) and Molina et al. (2001), respectively. We adopted a similar nomenclature for M. eumusae. In total, 32 loci for M. fijiensis and nine each for M. musicola and M. eumusae are presented. All these loci were polymorphic on global samples with the mean number of alleles (Na) per locus ranging from 2.9 to 4.6 over all species. The gene diversity (HE = 1 –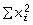 , where xi is the frequency of the ith allele) was calculated with the software genetix 4.05.2 (Belkhir et al. 2001). Over all species in global samples, HE ranged from 0.55 to 0.59. More than 80% of the 15 and seven loci tested on samples of M. fijiensis and M. musicola from a single location were polymorphic (2.8 < Na < 3.4 and 0.32 < HE < 0.55). Thus, for each of these three species, we inferred that most of the loci that proved polymorphic on global samples could be used to analyse any single population. Furthermore, because a high level of genetic differentiation has been detected in these fungi on both a global and a local scale (Carlier et al. 1996; Rivas et al. 2004; Hayden et al. 2005), some loci that were polymorphic in the global sample could be polymorphic in some populations but not in others. For M. fijiensis species, the 15 loci tested in the sample from a single location in Cameroon were chosen to develop two novel multiplex pools of markers for population structure studies. However, the remaining loci polymorphic on the global sample could be useful for population studies demanding a larger number of markers or for mapping purposes. Linkage disequilibrium (LD) between pairs of loci tested in population samples of M. fijiensis and M. musicola was determined with exact tests using genepop 3.4 (Raymond & Rousset 1995). No significant LD at the ‘table-wide’ 5% level (Rice 1989) was detected among all pairs of loci tested. Finally, cross-amplification of nine to 10 loci from each species was tested on samples of 10 to 16 isolates of the two other species from various locations (Table 1). Only 11.3% of the 848 species/primer pair combinations tested among the three species were successful in amplifying bands of the expected size (Table 1). However, it should be noted that, in contrast to other organisms, it is not unusual to observe such a low level of cross-species transferability in fungi (Dutech et al. 2007).
, where xi is the frequency of the ith allele) was calculated with the software genetix 4.05.2 (Belkhir et al. 2001). Over all species in global samples, HE ranged from 0.55 to 0.59. More than 80% of the 15 and seven loci tested on samples of M. fijiensis and M. musicola from a single location were polymorphic (2.8 < Na < 3.4 and 0.32 < HE < 0.55). Thus, for each of these three species, we inferred that most of the loci that proved polymorphic on global samples could be used to analyse any single population. Furthermore, because a high level of genetic differentiation has been detected in these fungi on both a global and a local scale (Carlier et al. 1996; Rivas et al. 2004; Hayden et al. 2005), some loci that were polymorphic in the global sample could be polymorphic in some populations but not in others. For M. fijiensis species, the 15 loci tested in the sample from a single location in Cameroon were chosen to develop two novel multiplex pools of markers for population structure studies. However, the remaining loci polymorphic on the global sample could be useful for population studies demanding a larger number of markers or for mapping purposes. Linkage disequilibrium (LD) between pairs of loci tested in population samples of M. fijiensis and M. musicola was determined with exact tests using genepop 3.4 (Raymond & Rousset 1995). No significant LD at the ‘table-wide’ 5% level (Rice 1989) was detected among all pairs of loci tested. Finally, cross-amplification of nine to 10 loci from each species was tested on samples of 10 to 16 isolates of the two other species from various locations (Table 1). Only 11.3% of the 848 species/primer pair combinations tested among the three species were successful in amplifying bands of the expected size (Table 1). However, it should be noted that, in contrast to other organisms, it is not unusual to observe such a low level of cross-species transferability in fungi (Dutech et al. 2007).
Acknowledgements
We thank Ange-Marie Risterucci, Claire Billot, Fabien Halkett and Andy James for their advice and help. DNA sequencing of genomic clones was carried out under a grant from Génoscope (Evry, France).




