Surveys reveal the occurrence of phytoplasmas in plants at different geographical locations in Peru
Abstract
Two independent surveys were performed in Peru during February and November 2007 to detect the presence of phytoplasmas within any crops showing symptoms resembling those caused by phytoplasmas. Molecular identifications and characterisations were based on phytoplasma 16S and 23S rRNA genes using nested PCR and terminal restriction fragment length polymorphism (T-RFLP). The surveys indicated that phytoplasmas were present in most of the locations sampled in Peru in both cultivated crops, including carrots, maize, native potatoes, improved potato, tomato, oats, papaya and coconut, and in other plants such as dandelion and the ornamental Madagascar periwinkle (Catharanthus roseus). Phylogenetic analysis of the sequences confirmed that while most of the isolates belong to the 16SrI aster yellows group, which is ubiquitous throughout other parts of South America, one isolate from potato belongs to the 16SrII peanut witches’ broom group, and one isolate from tomato and one from dandelion belong to the 16SrIII X-disease group. The use of T-RFLP was validated for the evaluation of phytoplasma-affected field samples and provided no evidence for mixed infection of individual plants with more than one phytoplasma isolate. These data represent the first molecular confirmation of the presence of phytoplasmas in a broad range of crops in Peru.
Introduction
Phytoplasmas are members of the class Mollicutes that are known to cause disease in hundreds of plant species worldwide (Liefting et al., 2004). New disease reports are being published frequently, and hosts include economically important food, fibre, forage, fruit and ornamental plants (Hogenhout et al., 2008). Transmission of phytoplasmas between plants is by phloem-feeding insects of the order Hemiptera, primarily leafhoppers (Cicadelloidea), planthoppers (Fulgoroidea) and psyllids (Psylloidea) (Lee et al., 2000). However, there are still many phytoplasmas for which the vector(s) have not been confirmed, while others can be vectored by many species. There have also been reports in which phytoplasma DNA has been detected in coconut embryos (Cordova et al., 2003; Nipah et al., 2007), although as yet there is no clear evidence that they are transmitted through the seed to cause disease in the progeny plants. Seed transmission would however have major implications for global trade because most phytoplasma quarantine checks currently focus on movement of whole plants and not the seed.
Peru is a country with a wide range of habitats, which can be divided into three main regions, the coastal plain, the highland region and the jungle area. The coastal plain is an arid stretch of land extending the entire length of the country and is the economic centre of Peru where many of the nation’s crops for export grow. Parallel to and lying east of the coastal plain are the Andes, and to the northeast is the vast, flat tropical jungle, extending to the Brazilian border and forming part of the Amazon Basin. This presents a wide range of cropping environments and the country grows a range of crops, including carrots, potatoes, onions, maize and papaya, for both subsistence and export. Farming is very important to the economy, and farming practices are typically of very high intensity and use a mixed cropping system to use all the available land to its greatest potential.
Many of these crops are potentially prone to infection by phytoplasmas, but the only previous records of phytoplasmas in Peru have been the observation of maize bushy stunt-like symptoms in maize from one location north of Lima (Nault et al., 1979) and an association with virescence in the ornamental Madagascar periwinkle, Catharanthus roseus (Schneider et al., 1993; Lee et al., 2004). However, in neighbouring countries, 16SrI aster yellows phytoplasmas have been recorded in potato in Bolivia (Jones et al., 2005a) and in maize in Brazil (Bedendo et al., 1997, 2000; De Oliveira et al., 2002), where the vector has been identified as the leafhopper, Dalbulus maidis, and this vector species has also been found in Peru (Nault et al., 1979). Aster yellows phytoplasmas have been found in other crops such as alfalfa in Bolivia (Jones et al., 2005b), and 16SrIII group phytoplasmas have been reported in chinaberry trees (Melia azedarach) in Bolivia (Harrison et al., 2003); coffee (Coffea arabica L.) (Galvis et al., 2007) and cassava (Manihot esculenta) (Álvarez et al., 2007) in Colombia; and chayote (Sechium edule) (Montano et al., 2000) and tomato (Mello et al., 2006) in Brazil.
It has been suggested that one possible reason for the limited reports of phytoplasma diseases in crops in Peru is that the Andes may act as a physical barrier, preventing spread of the specific vectors for phytoplasma diseases to the coastal region. Hopper species known to be phytoplasma vectors in other regions have been collected as high as 4000 m in the Andes, but there have been no studies to confirm whether these harbour phytoplasmas, and the habitat/host-specific nature of most species could mean that the lack of a suitable habitat at these altitudes is a barrier to vector movement (M. Wilson, National Museum of Wales, personal communication). In addition, it has been suggested that the low temperatures in many inland areas of Peru during winter would make overwintering difficult (Nault et al., 1979). However, phytoplasmas could still be introduced to different regions of Peru by vegetative means, or overwinter in previous season’s crops that have not been removed by farming practices such as mixed cropping, and it has also been postulated that vectors could be blown inland by strong winds that blow from the coast early in the growing season (Nault et al., 1979).
As it is not possible to isolate and study phytoplasmas in pure cultures, PCR has become the method of choice for detection and diagnosis in plants and insect vectors, and phytoplasma-specific primers have been developed that amplify various regions of the rRNA operon (Lee et al., 1998, 2000). Owing to the low titres of phytoplasmas and their uneven distribution within infected plants, it is often appropriate to use nested PCR. Recently, a new diagnostic tool, terminal restriction fragment length polymorphism (T-RFLP) based on the sequence of the 23S rRNA gene has been developed for the identification of phytoplasmas present in plant tissue (Hodgetts et al., 2007, 2008), and this method has proved to be useful for simultaneous detection, taxonomic grouping and identification of mixed infections in plants.
To try and ascertain whether the lack of reported phytoplasma diseases in Peru is because of poor use of diagnostic techniques and reporting, or because phytoplasmas have been unable to spread into the main agricultural areas because of physical barriers such as the Andes, two surveys were undertaken to assess a range of crops in several regions of Peru for the presence of phytoplasmas and to identify the taxonomic groups to which they belong. These studies have been based on the use of the 16S and 23S rRNA genes using both conventional PCR and T-RFLP.
Materials and methods
Source of isolates and extraction of DNA
Details of the origins of material used in the two surveys and the locations of these sites within Peru are given in Table 1 and Fig. 1[link] respectively. In survey 1, plant material from both suspected phytoplasma-infected plants and healthy looking plants was sampled during February 2007, stored at 4°C after sampling [apart from time in transport at ambient temperature (25–30°C) for a duration of 1 day for Mantaro Valley and 3 days for San Martin] and transferred to the laboratories at CIP, Lima for extraction of DNA. Plants from on-site were processed on the day of sampling, those from papaya (Carica papaya) and coconut (Cocos nucifera) within 18 days of sampling and all remaining samples were processed within 12 days of sampling. In survey 2, plant material from suspected phytoplasma-infected plants was collected during November 2007 from two areas of the Mantaro Valley and stored at 4°C after sampling (apart from time in transport at ambient temperature) at the SENASA laboratories, before being transported to Rothamsted Research, UK. DNA was extracted from 300 to 500 mg of leaf tissue using the cetyl trimethyl ammonium bromide method of Doyle & Doyle (1990).
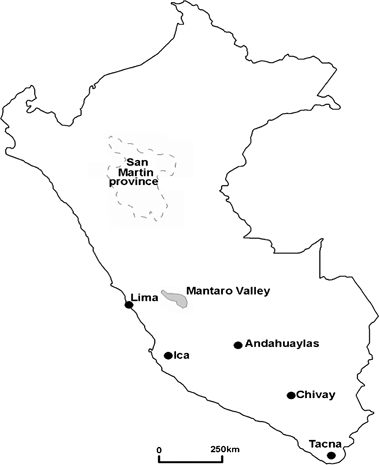
Map of Peru illustrating the sampling sites.
| Sample ID | Plant | Symptoms | DNA from | Sampling field/source | Area of Fig. 1 | Altitude | R16 resulta | 16Sr group |
|---|---|---|---|---|---|---|---|---|
| 1 | Alfalfa | No symptoms | Midribs | Pampa Sicaya | Mantaro Valley | 3350 | − | |
| 2 | Alfalfa | Yellowing, internode elongation, little leaf | Whole leaves | Pampa Sicaya | Mantaro Valley | 3350 | − | |
| 3 | Alfalfa | Stunting, little leaf | Whole leaves | Pampa Sicaya | Mantaro Valley | 3350 | Weak | I |
| 4 | Alfalfa | Stunting, little leaf | Whole leaves | Pampa Sicaya | Mantaro Valley | 3350 | − | |
| 5a | Carrot | Yellowing of leaves, stunted root and leaves | Leaves | Pampa Sicaya | Mantaro Valley | 3350 | Weak | I |
| 5b | Carrot | Yellowing of leaves, stunted root and leaves | Root | Pampa Sicaya | Mantaro Valley | 3350 | + | I |
| 6a | Carrot | Yellowing of leaves, stunted root and leaves | Leaves | Pampa Sicaya | Mantaro Valley | 3350 | + | I |
| 6b | Carrot | Yellowing of leaves, stunted root and leaves | Root | Pampa Sicaya | Mantaro Valley | 3350 | + | I |
| 7a | Carrot | No symptoms | Whole leaves | Huayao | Mantaro Valley | 3300 | − | |
| 7b | Carrot | No symptoms | Root | Huayao | Mantaro Valley | 3300 | + | I |
| 8a | Carrot | Yellowing of foliage, stunted root and foliage | Leaves | Huayao | Mantaro Valley | 3300 | + | I |
| 8b | Carrot | Yellowing of foliage, stunted root and foliage | Root | Huayao | Mantaro Valley | 3300 | − | |
| 9a | Carrot | Bronzing of foliage, stunted root and foliage | Leaves | Huayao | Mantaro Valley | 3300 | − | |
| 9b | Carrot | Bronzing of foliage, stunted root and foliage | Root | Huayao | Mantaro Valley | 3300 | − | |
| 10a | Carrot | Yellowing of foliage, stunted root and foliage | Leaves | Huayao | Mantaro Valley | 3300 | − | |
| 10b | Carrot | Yellowing of foliage, stunted root and foliage | Root | Huayao | Mantaro Valley | 3300 | + | I |
| 11a | Carrot | Bronzing of foliage, stunted root and foliage | Leaves | Huayao | Mantaro Valley | 3300 | − | |
| 11b | Carrot | Bronzing of foliage, stunted root and foliage | Root | Huayao | Mantaro Valley | 3300 | − | |
| 12 | Carrot | Fanged, stunted root. Bronzing of foliage tips | Root | Huayao | Mantaro Valley | 3300 | − | |
| 13a | Carrot | Fanged, stunted root. Yellowing of foliage | Root | Huayao | Mantaro Valley | 3300 | − | |
| 13b | Carrot | Fanged, stunted root. Yellowing of foliage | Leaves | Huayao | Mantaro Valley | 3300 | − | |
| 14a | Carrot | Fanged and cracked root. Yellowing of foliage | Root | Huayao | Mantaro Valley | 3300 | + | I |
| 14b | Carrot | Fanged and cracked root. Yellowing of foliage | Leaves | Huayao | Mantaro Valley | 3300 | + | I |
| 15 | Clover | No symptoms | Midribs | Pampa Sicaya | Mantaro Valley | 3350 | − | |
| 16a | Clover | Stunting, little leaf, proliferation | Midribs | Pampa Sicaya | Mantaro Valley | 3350 | + | I |
| 16b | Clover | Stunting, little leaf, proliferation | Roots | Pampa Sicaya | Mantaro Valley | 3350 | + | I |
| 17 | Coconut | Yellowing/mottling of leaves. Stunting? | Leaf | Tocache viejo | San Martin | Unknown | + | I |
| 18 | Coconut | Yellowing/mottling of leaves. Stunting? | Leaf | Tocache viejo | San Martin | Unknown | + | I |
| 19 | Coconut | Yellowing/mottling of leaves. Stunting? | Leaf | Tocache viejo | San Martin | Unknown | + | I |
| 20a | Maize | Stunting, yellowing deformed inflorescence | Leaves adjacent to cob | Pampa Sicaya | Mantaro Valley | 3350 | + | I |
| 20b | Maize | Stunting, yellowing deformed inflorescence | Maize grains | Pampa Sicaya | Mantaro Valley | 3350 | + | I |
| 21a | Maize | No symptoms | Leaves adjacent to cob | CIP Huancayo | Mantaro Valley | 3300 | + | I |
| 21b | Maize | No symptoms | Maize grains | CIP Huancayo | Mantaro Valley | 3300 | − | |
| 21c | Maize | No symptoms | Leaves adjacent to cob | CIP Huancayo | Mantaro Valley | 3300 | + | I |
| 21d | Maize | No symptoms | Maize grains | CIP Huancayo | Mantaro Valley | 3300 | − | |
| 22a | Maize | Yellowing, stunting, deformed inflorescence | Leaves adjacent to cob | Huayao | Mantaro Valley | 3300 | + | I |
| 22b | Maize | Yellowing, stunting, deformed inflorescence | Maize grains | Huayao | Mantaro Valley | 3300 | + | I |
| 22c | Maize | Yellowing, stunting, deformed inflorescence | Leaves adjacent to cob | Huayao | Mantaro Valley | 3300 | + | I |
| 22d | Maize | Yellowing, stunting, deformed inflorescence | Maize grains | Huayao | Mantaro Valley | 3300 | + | I |
| 23a | Maize | Bronzing, stunting, deformed inflorescence | Leaves adjacent to cob | Huayao | Mantaro Valley | 3300 | + | I |
| 23b | Maize | Bronzing, stunting, deformed inflorescence | Maize grains | Huayao | Mantaro Valley | 3300 | + | I |
| 23c | Maize | Bronzing, stunting, deformed inflorescence | Leaves adjacent to cob | Huayao | Mantaro Valley | 3300 | + | I |
| 23d | Maize | Bronzing, stunting, deformed inflorescence | Maize grains | Huayao | Mantaro Valley | 3300 | + | I |
| 24 | Onion | Stunting, yellowing | Leaf, stem white | Huayao | Mantaro Valley | 3300 | − | |
| 25 | Papaya | Yellowing, stunting, proliferation | Midribs | Armallari | San Martin | 286 | − | |
| 26 | Papaya | Yellowing, stunting, proliferation | Whole leaves | Chambira | San Martin | 376 | + | I |
| 27 | Papaya | Yellowing, stunting, proliferation | Leaves + immature fruit | Sauce | San Martin | 277 | Weak | I |
| 28 | Papaya | Yellowing, stunting, proliferation | Whole leaves | San Juan Salada | San Martin | 371 | + | I |
| 29 | Papaya | Yellowing, stunting, proliferation | Whole leaves | Leticia | San Martin | 286 | + | I |
| 30 | Papaya | Yellowing, stunting, proliferation | Midribs | Vainilla | San Martin | 251 | Weak | I |
| 31 | Papaya | Yellowing, stunting, proliferation | Whole leaves | San José | San Martin | 239 | − | |
| 32 | Papaya | Yellowing, stunting, proliferation | Whole leaves | San José | San Martin | 249 | + | I |
| 33 | Papaya | Yellowing, stunting, proliferation | Whole leaves | Pajarillo | San Martin | 247 | − | |
| 34 | Papaya | Yellowing, stunting, proliferation | Whole leaves | Viveres | San Martin | 266 | Weak | I |
| 35 | Papaya | Yellowing, stunting, proliferation | Whole leaves | Viveres | San Martin | 267 | Weak | I |
| 36 | Papaya | Yellowing, stunting, proliferation | Whole leaves | Camino Nuevo Egipto | San Martin | Unknown | − | |
| 37 | Papaya | Yellowing, stunting, proliferation | Whole leaves | Piscoyacu | San Martin | 297 | + | I |
| 38 | Papaya | Yellowing, stunting, proliferation | Leaves + flower bud | Armallari | San Martin | 276 | + | I |
| 39 | Papaya | Yellowing, stunting, proliferation | Leaves + flower bud | Sauce | San Martin | 286 | − | |
| 40 | Papaya | Yellowing, stunting, proliferation | Whole leaves | Santa Catalina | San Martin | 238 | − | |
| 41 | Papaya | Yellowing, stunting, proliferation | Whole leaves | Santa Rosa de Upakihua | San Martin | 224 | + | I |
| 42 | Papaya | Yellowing, stunting, proliferation | Whole leaves | San Juan Salada | San Martin | 371 | − | |
| 43 | Papaya | Yellowing, stunting, proliferation | Whole leaves | El Dorado | San Martin | 348 | + | I |
| 44 | Papaya | Yellowing, stunting, proliferation | Whole leaves | San José | San Martin | 229 | + | I |
| 45 | Papaya | Yellowing, stunting, proliferation | Whole leaves | El Codo | San Martin | 283 | + | I |
| 46a | Potato | No symptoms | Whole leaves | CIP Lima | Lima | 300 | + | I |
| 46b | Potato | No symptoms | Leaves | CIP Lima | Lima | 300 | + | I |
| 47 | Potato | Witches’ broom | Whole leaves | Tacna | Tacna | 400 | − | |
| 48 | Potato | Witches’ broom | Whole leaves | Tacna | Tacna | 400 | − | |
| 49 | Potato | Purple-top | Whole leaves | CIP Lima | Lima | 300 | − | |
| 50 | Potato | Purple-top | Whole leaves | CIP Lima | Lima | 300 | − | |
| 51 | Potato | Purple-top | Whole leaves | CIP Lima | Lima | 300 | − | |
| 52 | Potato | Purple-top | Whole leaves | CIP Lima | Lima | 300 | + | I |
| 53a | Potato (native) | Stunting, chlorotic leaves, no tubers | Leaves + midribs | Quillacocha | Mantaro Valley | 3500 | + | I |
| 53b | Potato (native) | Stunting, chlorotic leaves, no tubers | Whole leaves | Quillacocha | Mantaro Valley | 3500 | − | |
| 54 | Potato (native) | Stunting, chlorotic leaves, no tubers | Leaves + midribs | La Libertad | Mantaro Valley | 3600 | − | |
| 55 | Potato (native) | Witches’ broom (originally) | Whole leaves | Andahuaylas | Andahuaylas | 3700 | + | I |
| 56 | Potato (native)b | Small leaves, proliferation | Whole leaves | Chivay | Chivay | 300 | − | |
| 57a | Shiri | No symptoms | Whole leaves | Concepciòn | Mantaro Valley | 3700 | + | I |
| 57b | Shiri | No symptoms | Whole leaves | Concepciòn | Mantaro Valley | 3700 | − | |
| 58a | Shiri | Squat habit, yellowing, small leaves | Whole leaves | Concepciòn | Mantaro Valley | 3700 | + | I |
| 58b | Shiri | Squat habit, yellowing, small leaves | Whole leaves | Concepciòn | Mantaro Valley | 3700 | − | |
| 58c | Shiri | Squat habit, yellowing, small leaves | Whole leaves | Concepciòn | Mantaro Valley | 3700 | + | I |
| 59 | Tomato | No symptoms | Whole leaves | CIP Lima | Lima | 300 | − | |
| 60 | Tomatoc | Internode elongation, small leaves | Whole leaves | Ica | Ica | 300 | + | III |
| 61 | Catharanthus roseus | No symptoms | Midribs | CIP Lima | Lima | 300 | + | I |
| 62 | C. roseus | Yellowing, little leaf, deformed inflorescence | Whole leaves | CIP Lima | Lima | 300 | + | I |
| 63 | C. roseus | Yellowing, little leaf, deformed inflorescence | Whole leaves | CIP Lima | Lima | 300 | − | |
| 64 | C. roseus | Yellowing, little leaf, deformed inflorescence | Whole leaves | CIP Lima | Lima | 300 | − | |
| A | Carrot | Yellowing of leaves, stunted root and leaves | Whole leaves | Huayao | Mantaro Valley | 3350 | I | |
| B | Carrot | Yellowing of leaves, stunted root and leaves | Whole leaves | Huayao | Mantaro Valley | 3350 | I | |
| C | Carrot | Yellowing of leaves, stunted root and leaves | Whole leaves | Marcatuna | Mantaro Valley | 3350 | ||
| D | Carrot | Yellowing of leaves, stunted root and leaves | Whole leaves | Marcatuna | Mantaro Valley | 3350 | ||
| E | Oat | Witches’ broom and little leaf | Whole leaves + stem | Huayao | Mantaro Valley | 3350 | I | |
| F | Dandelion | Witches’ broom | Whole leaves + stem | Marcatuna | Mantaro Valley | 3350 | III | |
| G | Potato | Yellowing of leaves, stunting and little leaf | Whole leaves | Huayao | Mantaro Valley | 3350 | II |
- The unshaded region of the table represents samples from survey 1 and the shaded region, samples from survey 2.
- a Results are scored as + (positive), weak (weakly positive) or − (negative).
- b Potato (native) maintained in Datura metei.
- c Tomato maintained in Datura stramonium.
16S ribosomal DNA analysis
Amplifications of the 16S rRNA gene were performed in 25 μL reactions for all samples using ‘illustra PuRe Taq Ready-To-Go™ PCR beads’ (GE Healthcare Life Sciences, Amersham, UK). Survey 1 used 15-ng template DNA and 100 ng of each primer in an MJ Research PTC200 thermocycler (Waltham, MA, USA), and the phytoplasma universal primer pairs P1/P7 and R16F2/R16R2 were used for first and second PCR rounds, respectively, under conditions previously described (Hodgetts et al., 2007). Survey 2 used 20-ng template DNA and 0.4 μM of each primer in a TC-412 TECHNE thermocycler (Burlington, NJ, USA) and the phytoplasma universal primer pairs R16mF2/R1 (Gundersen & Lee, 1996) for first round PCR, and fU5/rU3 (Lorenz et al., 1995) for the second round PCR with conditions as stated in the respective papers. In addition, restriction endonuclease digestion with Sau3AI and HpaII, on the nested-PCR product was performed via the manufacturer’s procedure in a 10 μL final volume. Reference controls of 16SrI [aster yellows dwarf, Germany], 16SrII [Tomato big bud, Australia] and 16SrIII [Vaccinium witches’ broom, Germany] from the Rothamsted Research phytoplasma collection were used for RFLP comparisons. Following PCR (or RFLP), 5 μL (10 μL) of resultant products were separated on 1% (1.5%) agarose gels in 1× TBE (Tris Borate EDTA) buffer containing ethidium bromide and visualised under UV light.
Cloning and sequence analysis
PCR products (16S rRNA) obtained from surveyed material were cloned using the Promega pGEM®–T easy Vector System (Southampton, UK) via the manufacturer’s protocol. Colonies corresponding to samples from survey 1 were amplified by PCR with the universal primers M13For and M13Rev described in the kit, and these PCR products were processed on both strands using a Beckman Quickstart kit technology that uses WellRed Dye chemistry (infrared dyes), with a CEQ 8000 Genetic Analysis System, manufactured by Beckman Coulter (High Wycombe, UK). Colonies corresponding to samples from survey 2 were confirmed by PCR with fU5/rU3 primer pair. Plasmid DNA of recombinant colonies was purified (QIAprep® Spin Miniprep; Qiagen, Crawley, UK) and sequenced on both strands with M13 forward and reverse primers.
Terminal-restriction fragment length polymorphism
Samples from survey 1 were subjected to T-RFLP analyses using primers for the 23S rRNA gene as previously described (Hodgetts et al., 2007). After amplification, PCR products were transported to the University of Nottingham, digested with either MseI (New England Biolabs, Hitchin, UK) or Bsh12361 (Fermentas Life Sciences, York, UK) in 10 μL reaction volumes containing 1 U of restriction enzyme for 2 h at 37°C, and analysed on a CEQ 8000 DNA analysis system as previously described (Hodgetts et al., 2007).
Phylogenetic analysis
Complete sequences from primers R16F2 to R16R2 were obtained for all the phytoplasmas from survey 1, but samples from survey 2 were only sequenced between primers fU5 and rU3. BLAST searches (Altschul et al., 1990) were performed at the National Center for Biotechnology Information web site at http://www.ncbi.nlm.nih.gov/. Phylogenetic trees were constructed with partial 16Sr sequences obtained by trimming all sequences to the same region flanked by primers fU5 and rU3. Sequence alignments were performed using ClustalW (Thompson et al., 1994), and phylogenetic and molecular evolutionary analyses were performed using the MEGA version 3.1 neighbour-joining method and all default values, with 1000 replications for bootstrap analysis (Kumar et al., 2004).
Results
Surveys of field crops in Peru
In the first survey, sampling was performed in two areas of Peru, the Mantaro Valley and San Martin (Fig. 1). The Mantaro Valley is a high altitude region on the side of the Andes, and San Martin is a low altitude jungle area. These areas therefore represent varied habitats, growth conditions and types of crops in cultivation. In the Mantaro Valley, carrots (Daucus carota) were sampled from two adjacent fields where the crops were 2 and 4 months old, with the latter being harvested for sale at the time of collection. The symptoms displayed by these plants are listed in Table 1, with an example of cracking and fanging of roots shown in Fig. 2a. Fanging of roots could have been attributed partially to the stony nature of the soil and there was also suspicion of nematode damage although this could not be confirmed because of lack of appropriate facilities. Maize (Zea mays) plants at the early flowering stage also showed two distinct sets of symptoms (Table 1; Fig. 2b to Fig. 2d[link]). Some of these symptoms were possibly because of the presence of viruses and the corn stunt spiroplasma that are known to be widespread in Peru (Nault et al., 1979), but these were not tested for in this study. The symptom sets were distributed throughout the fields with no obvious clustering. Symptoms on native potato (Solanum tuberosum) and on Shiri (likely Solanum tuberosum cultivar group Curtilobum or Juzepczukii; Huamán & Spooner, 2002), a native Andean root and tuber crop, are given in Table 1 and shown in Fig. 2e and Fig. 2f[link]. In certain sampling fields, evidence of the previous season’s crop was visible and there were a small number of plants regrowing within the main crop. In the carrot fields, these crops were onions and in the maize fields they were carrots, and these crops were also sampled. In San Martin, a range of papaya plantations and coconut palms were sampled (Table 1; Fig. 2g and Fig. 2h[link]).
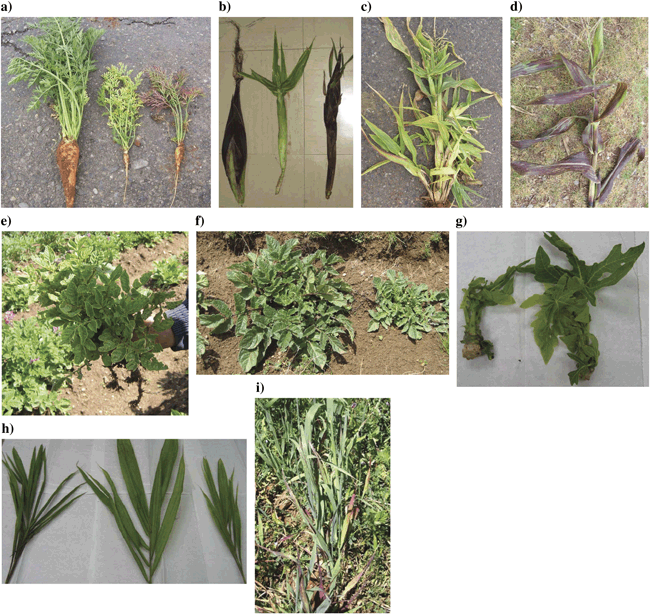
Photographs of plant material tested for the presence of phytoplasma. (a) Carrots aged 4 months, from Huayao; left, asymptomatic; centre, yellowing; right, bronzing. (b) Maize cobs; left, asymptomatic from Huancayo, centre, yellowing from Pampa Sicaya and right, bronzing from Huayao. (c) Maize from Pampa Sicaya showing yellowing, proliferation and deformation of cobs. (d) Maize from Huayao with bronzing. (e) Native potato from Quillacocha, with yellowing, stunting and lack of tuber production. (f) Shiri from Concepciòn, left asymptomatic and right with stunting. (g) Papaya from San Martin with yellowing, stunting and proliferation. (h) Coconut from San Martin with yellowing. (i) Oat from Huayao with a bushy growing habit and little leaf.
Additional samples were obtained from CIP Lima, this being an irrigated desert habitat in which potatoes are grown as part of the organisation’s routine work. CIP also maintain some potato and tomato material in culture suspected of harbouring phytoplasmas, and these were included in the study and are listed in Table 1 under the locations from which the samples were originally obtained, that is, Tacna (irrigated desert), Andahuaylas (Andes), Chivay (Andes), Ica (irrigated desert). Those from Chivay and Ica had been transferred by grafting and then maintained in Datura spp., and those from Tacna and Andahuaylas were maintained in the original plant species. CIP’s routine screening of potatoes for export also produced some samples of improved potato variety with leaf reddening and vein necrosis that were included in the survey. The C. roseus samples were from ornamental flower beds at CIP Lima where some plants were showing phytoplasma symptoms and were growing alongside healthy looking plants.
Control plants of tomato and improved potato were provided at CIP Lima where they had been grown from disease-free stock/seed in insect proof glasshouses. Asymptomatic plant samples were also collected at each sampling site for each crop with the exception of maize, where asymptomatic plants were sampled from a region close to the sampling area (CIP, Huancayo) because of all fields at the sites showing widespread symptoms with no convincing asymptomatic plants present. In the second survey, sampling was performed within two areas of the Mantaro Valley (Fig. 1), and the symptoms displayed are listed in Table 1, with symptoms on oats shown in Fig. 2i.
16S ribosomal RNA analysis
Approximately 20% of samples produced phytoplasma PCR products following first round PCR, and the results for nested PCR are shown in Table 1. The absence of any previous molecular analysis of phytoplasmas in Peru precluded the use of positive controls in survey 1, but negative controls were used at both stages of PCR and were clear from contamination. The 89 samples from survey 1 listed in Table 1 represent extractions from 64 different individual plants, of which 52 were positive and the remainder were negative with no ambiguity. Ten carrots were sampled, and all except one had extractions performed from both the foliage and root material. No phytoplasma DNA was detected in any of the samples with bronzing of foliage, while five of the six samples that had yellowing of foliage were positive for phytoplasma DNA in either roots, leaves or both roots and leaves. Roots that were cracked and fanged were found to be both positive and negative, and thus these symptoms may not be attributed to phytoplasma infection (fanging is not characteristic of phytoplasma infection in carrots). The asymptomatic carrot was also tested and was found to be positive for phytoplasma.
Four maize plants were tested, and all symptomatic plants were positive in both sample types (grain and leaves). The asymptomatic control was positive in the leaves but not grains, perhaps indicating that the plant was at an earlier stage of infection. The papaya and coconut samples represent individual plants from 16 plantations, and testing revealed that all but three plantations had phytoplasma infection (Table 1). All three coconut samples and 14 of the 21 papaya samples tested positive for phytoplasma DNA. Both Shiri plants (one with and one without symptoms) were positive for phytoplasma DNA, and two of the four C. roseus samples from within CIP Lima were also positive for phytoplasmas (one symptomatic and one asymptomatic). From the samples maintained at CIP, the tomato from Ica and potato from Andahuaylas were found to be infected with phytoplasmas. Those from Tacna and Chivay had no detectable phytoplasma DNA, although this does not rule out the possibility that the original samples were positive but the phytoplasma was lost during the grafting procedures that had been used to maintain the cultures. Other potatoes at CIP that gave a positive result in testing were a ‘healthy control’ plant and a symptomatic sample taken from routine quarantine testing.
Overall, in the Mantaro Valley, the crops found to be positive for phytoplasma DNA were carrot, maize, native potato and Shiri. The symptomatic samples of alfalfa (Medicago sativa) and clover (Trifolium sp.) were negative for phytoplasma DNA, and subsequent analysis in which a region of the 23S rRNA was PCR amplified and sequenced indicated the presence of other bacterial infections (an unculturable bacterium and an Actinobacterium spp.) (results not shown). In addition, a symptomatic onion sample (found in a carrot field) was negative for all PCR tests and was therefore suspected to be a viral infection.
Samples from survey 2 represent extractions from seven different individual plants, of which five were positive for phytoplasma DNA and the remainder were negative (Table 1). Four carrots were sampled, two each from two locations, and those from Huayao were found to be positive, while those from Marcatuna were free of phytoplasma infection. Both the oat and potato samples from Huayao along with the dandelion from Marcatuna were found to be positive for phytoplasma DNA. RFLP analysis using Sau3AI (Fig. 3) and HpaII (data not shown) indicated that the carrot samples and the oat sample showed RFLP profiles identical to those of the group 16SrI reference strain. However, the restriction profiles for the phytoplasma in the potato sample were identical to those of the 16SrII reference strain, while those from the dandelion were identical to the patterns of the 16SrIII X-disease group phytoplasma.
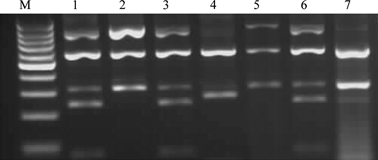
RFLP patterns for samples from survey 2 following digestion with Sau3AI. Lanes (M) 100-bp molecular weight marker (Fermentas Life Sciences); Lanes 1, carrot A; 2, potato G; 3, oat E; 4, dandelion F; 5, reference control tomato big bud; 6, reference control aster yellows dwarf; 7, reference control Vaccinium witches’ broom.
T-RFLP results
Terminal RFLP was performed on all samples from survey 1, which were positive with the 16S rRNA primers (R16F2/R16R2) as well as a negative sample for each plant species. All the control plant samples showed a single TRF (Terminal Restriction Fragment) at 461 bp, which is the size of the product expected from plant chloroplast DNA, indicating that the PCR had been successful and that there was no phytoplasma present (Fig. 4a). With the exception of one sample, all the infected isolates were identified by the technique to be 16SrI aster yellows group phytoplasmas by the presence of a TRF of 129 bp with MseI (Fig. 4d) and 305 bp with Bsh12361 (Fig. 4e) (Hodgetts et al., 2007). The isolate Tomato 60 from Ica, was identified as a 16SrIII X-disease group phytoplasma by the presence of a TRF of 129 bp with MseI (Fig. 4b) and 450 bp with Bsh12361 (Fig. 4c). For samples where 16S rRNA PCR had failed to identify phytoplasma infection, no phytoplasmas were identified by T-RFLP.
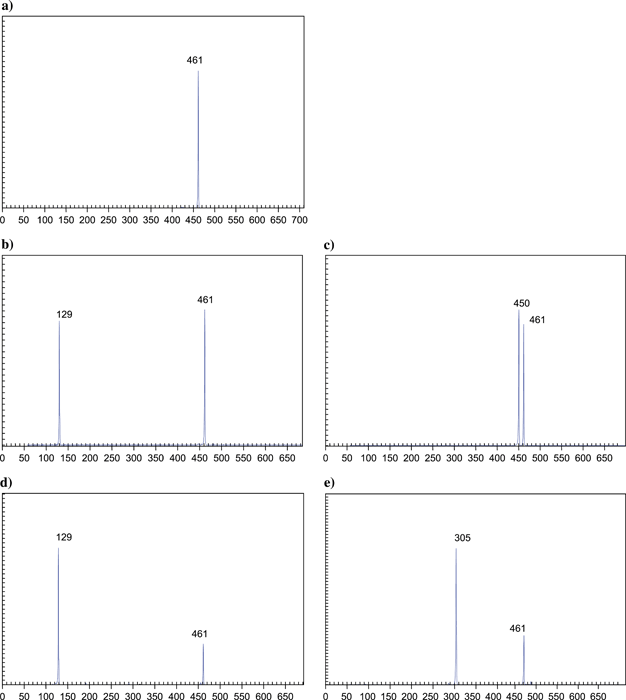
Terminal restriction fragment length polymorphism profiles for healthy control and infected plants amplified with MJD5 and labelled 23Srev, following digestion with either MseI or Bsh12361. (a) Sample 59 – healthy tomato (control plant) digested with MseI; (b) sample 60 – tomato digested with MseI; (c) sample 60 – tomato digested with Bsh12361; (d) sample 22b – maize digested with MseI; (e) sample 22b – maize digested with Bsh12361. The values on the x-axis are the sizes in base pairs (as are the numbers on the peaks). The y-axis gives the signal strength in arbitrary units.
Phylogenetic analysis
For those samples from survey 1, for which PCR and T-RFLP had confirmed the presence of phytoplasma DNA, four representative samples (papaya 34 from Viveres, native potato 55 from Andahuaylas, C. roseus 61 from CIP, Lima and Tomato 60 from Ica) were selected from which to clone and sequence the 16S rDNA. From the five positive samples from survey 2, where PCR and RFLP had confirmed the presence of phytoplasma DNA, one sample from each crop at each location (carrot A from Huayao, oat E from Huayao, potato G from Huayao and dandelion F from Marcatuna) were selected and the 16S rDNA was cloned and sequenced. Complete sequences from between the nested-PCR fU5/rU3 primers were obtained for all the samples. Sequencing and phylogenetic analysis (Fig. 5) indicated that the majority of the samples belonged to the 16SrI aster yellows group, with those from Andahuaylas, Viveres and CIP most closely aligned to the 16SrI-B subgroup, and those from Huayao to the 16SrI-A subgroup. The potato sample from Huayao had 99% sequence identity to members of the 16SrII peanut witches’ broom group, while the tomato sample from Ica and the dandelion sample from Marcatuna had 99% sequence identity with the Solanum quitoense machorreo phytoplasma from Colombia (accession number AY731819) that belongs to the 16SrIII X-disease group.

Dendrograms, constructed by the neighbour-joining method, showing the phylogenetic relationships for partial 16S rRNA sequences (primers fU5/rU3) for all samples surveyed, compared with representatives from other 16S rRNA groups. GenBank accession numbers for previously published sequences are shown in parenthesis alongside the names of the isolates. Bootstrap values greater than 50% (expressed as percentages of 1000 replications) are shown, and branch lengths are proportional to the number of inferred character state transformations. Bar, substitutions per base.
Discussion
These two independent surveys have demonstrated the presence of phytoplasmas in both wild and cultivated crops, including carrots, maize, native potatoes, improved potato, tomato, oats, papaya and coconut in Peru over a wide geographical area. Furthermore, phytoplasmas were identified within ornamental plants (C. roseus) and weeds (dandelion), perhaps indicative of a natural reservoir for transmission to crops. Three different groups of phytoplasmas, 16SrI, 16SrII and 16SrIII were identified, and of the eight regions visited, six were demonstrated to harbour phytoplasmas. No phytoplasmas were identified in samples from Tacna and Chivay, although this may have been because only three plants were tested at these locations. However, at three of the other regions (Marcatuna, Andahuaylas and Ica) only a single plant was tested and phytoplasmas were detected. The 16SrI phytoplasmas were found to be most widespread, and were present at four of the six positive sites where phytoplasma infections were found. A 16SrII phytoplasma was located in a single plant in an area (Huayao, Mantaro Valley) where infection with 16SrI seemed to be endemic, although no evidence of mixed infection was found using T-RFLP, a technique that was validated as being suitable for the analysis of field collected samples during this survey. This contrasts with the findings of Leyva-López et al. (2002) in Mexico, where 16SrI and 16SrII isolates were found to coexist not only in the same potato fields but also in the same plant. 16SrIII phytoplasmas were found in two areas, Mantaro Valley, where 16SrI and 16SrII phytoplasmas were also detected, and Ica, where neither of the other groups were found. A small number of samples that showed symptoms were negative for phytoplasma detection by PCR, which is consistent with results from other studies and is believed to result from the uneven distribution of phytoplasma cells within the plant such that they are sometimes missed by the sampling technique. Similarly, the presence of phytoplasma in asymptomatic plants is also consistent with previous studies and is generally attributed to the latent period between infection and symptom development (Wei et al., 2004).
All the plant species in this study have been well documented to harbour phytoplasmas in other countries throughout the world, including the American continent, and the symptoms found in Peru were consistent with those caused by phytoplasma infection in these crops worldwide. In particular, the molecular detection of a 16SrI phytoplasma in maize confirms the presence of a maize bushy stunt-type phytoplasma in Peru, which had previously been suggested at a different location through observation of symptoms (Nault et al., 1979). This is also the first report of a 16SrIII phytoplasma associated with dandelion in the American continent, although there are reports of 16SrI group phytoplasmas associated with dandelion diseases in Canada (Wang & Hiruki, 2001, 2005), group 16SrII in Italy (Marcone et al., 2001), and group 16SrIII in Lithuania (Jomantiene et al., 2002). Detection of a phytoplasma within the native Andean root and tuber crop Shiri is significant because of the altitude at which this sample was found (3700 m), which was thought to be too high for the presence and survival of most phytoplasma vector species, although D. maidis, the vector for the maize bushy stunt phytoplasma, has been found previously at 3200 m in Peru (Nault et al., 1979). It is also important because of the cultural value of Shiri, which is only grown in small areas of the country. These plants represent an important genetic resource and the presence of phytoplasma within one of these crops raises concern that the remaining crops need to be monitored closely to prevent phytoplasmas from destroying them. It is also not possible at this stage to identify whether the vectors for phytoplasmas have been able to transmit the diseases across the Andes, or whether the phytoplasmas have been transmitted into the country through whole plant imports and have then been spread within the country by the indigenous insect populations. Surveys of potential vector species collected at high altitude for the presence of phytoplasmas could help to resolve this.
Of particular interest during survey 1 was that one of the improved potatoes held at CIP for quarantine testing (sample 52) and one of the healthy looking control plants (sample 46) tested positive for phytoplasma. There was no evidence of any breach in the containment against insects, and these plants were raised from different sources, which suggests that there may be some phytoplasma contamination of the potato germplasm collections being used within Peru. This is obviously an issue that requires further evaluation to confirm whether there is contamination and the extent to which this is present.
The identification of phytoplasmas within the grains of maize plants raises an interesting issue of whether phytoplasmas can be transmitted through seed, and there have been previous reports of the detection of phytoplasma DNA in embryos of coconut (Cordova et al., 2003; Nipah et al., 2007), although there have been no confirmed reports that this leads to phytoplasma infection of the progeny plants. Although the phytoplasma DNA was detected in grains of the maize cobs, there would need to be further evaluation of matured seeds to ascertain if viable phytoplasmas were present, if the seeds themselves were viable and if the phytoplasmas could be transmitted to a plant germinated from such seed. Nevertheless, this indicates the need for further study into this area because of the potential issues for movement of quarantine pathogens especially where current legislation covers only plant material and not seeds. In addition, further work is required to identify the insect vectors responsible for phytoplasma transmission in Peru and to determine how widespread phytoplasmas are in Peruvian crops.
Acknowledgements
This work was performed as part of a DEFRA Plant Health Division-funded Fellowship (for J. H.). This work was funded by a British Society for Plant Pathology Junior Fellowship with additional funding from CIP, CSL and the University of Nottingham. The authors thank Prof. Phil Jones (Rothamsted Research, UK), Dr Jaraslava Přibylova (Institute of Plant Molecular Biology, Czech Republic) and Prof. Assunta Bertaccini (University of Bologna, Italy) for providing samples. The authors also thank Dominic Eyre, CSL, for producing the map used in Fig. 1, and Dr Mike Wilson, National Museum and Galleries of Wales, for useful discussions on phytoplasma vectors. Phytoplasmas in the UK were held under DEFRA Plant Health Licence numbers PHL 173B/5244 (12/2005) (Nottingham University) and PHL 174B/4612 (09/2003) (Rothamsted Research).




