Lethal yellowing-type diseases of palms associated with phytoplasmas newly identified in Florida, USA
Abstract
A nested polymerase chain reaction (PCR) assay was used to detect phytoplasmas in stem tissues from declining silver date (Phoenix sylvestris), Canary Island date (Phoenix canariensis), edible date (Phoenix dactylifera), Queen (Syagrus romanozoffiana) and Mexican fan (Washingtonia robusta) palms, all displaying symptoms similar to lethal yellowing (LY) disease, in a tri-county region of west central Florida. Restriction fragment length polymorphism analysis or sequencing of PCR-amplified rDNA products (1.6 kb) identified three distinct group 16SrIV phytoplasma strains among 38 infected palms. Most palms (86.8%) contained Texas Phoenix decline (TPD) phytoplasma, a subgroup 16SrIV-D strain, while two P. canariensis harboured palm LY phytoplasma, a subgroup 16SrIV-A strain. A novel phytoplasma, initially detected in W. robusta and tentatively classified as a subgroup 16SrIV-F strain, also occurred with LY phytoplasma as mixed infections in two P. dactylifera palms. Collectively, these findings extend the known geographic range of TPD in the USA to include Florida and the listing of TPD-susceptible palm species to include P. dactylifera, P. sylvestris and S. romanozoffiana. Moreover, discovery of a novel subgroup 16SrIV-F strain in W. robusta adds to the list of phytoplasma host palm species and complexity of phytoplasma-vector-palm pathosystems newly identified in the west central region of the state. The emergence of new lethal disease of palms beyond southern subtropical region of the state is unprecedented but follows similar developments in other regions where LY is known to occur.
Introduction
Lethal yellowing (LY) is a fast-moving, fatal phytoplasma disease of coconut and numerous other palm species (McCoy et al., 1983; Harrison & Jones, 2004). For almost two decades after its first appearance on mainland southern Florida during the early 1970s, LY was confined to just four contiguous southeastern counties bounded by Monroe county in the south and Palm Beach county in the north. Long distance dispersal to and establishment of the disease in Lee county on Florida’s southwest coast occurred during the late 1980s. Although further southward advance of LY into neighbouring Collier county, which contains the largest remaining population of about 80 000 ‘Jamaica tall’ coconut palms in the state, has occurred, palm mortality has been minimised by implementation of a rigorous disease management programme (Fedelem, 2000). More northerly spread of LY beyond the southern subtropical tier has not occurred. This well-established pattern of LY distribution also coincides with the native range of its neotropical cixiid vector Myndus crudus (Van Duzee) (Howard et al., 1982, 1984), which is not cold hardy.
Early detection of LY has involved the use of molecular diagnostics to confirm presence of the pathogen in diseased palms. For this purpose, polymerase chain reaction (PCR) assays capable of detecting LY phytoplasma strains in either a group-specific (Harrison et al., 2002a,c) or pathogen-specific manner (Harrison et al., 1994) have been deployed. Group-specific PCRs that are based on selective amplification of rRNA gene products from palm stem tissues have required restriction fragment length polymorphism (RFLP) analysis or sequencing of rDNA amplification products to ascertain phytoplasma identity. The uniformity of RFLP patterns resolved by this method has indicated that phytoplasmas associated with LY disease in southern Florida are a homogeneous population of subgroup 16SrIV-A strains (Harrison et al., 2002a).
In October 2006, an unusual foliar discolouration affecting 8- to 10-year-old, seed-grown Phoenix sylvestris, P. sylvestris ×Phoenix dactylifera L. and P. sylvestris ×Phoenix canariensis Chab. hybrids in production groves was first discovered at a palm field nursery in Hillsborough county, west central Florida. Discolouration began on the lowermost (oldest) leaves, which turned reddish-brown, starting at the tips and intensified to involve successively younger leaves in the mid-crown and upper crown. The advent of this symptom was accompanied by a premature shedding of most or all fruits as inflorescences on affected palms withered and died prematurely. Collapse and death of the newest (spear) leaf developed early in the foliar discolouration phase usually before it had progressed to leaves of the mid-crown. By this stage, mature roots of palms at or near the soil surface were found to be unusually soft in texture and easily severed as compared to roots of nearby symptomless palms that remained firm and resistant to breakage. Palms that had undergone these adverse changes could be easily pushed back and forth indicating a deterioration in the structural integrity of their root systems. Furthermore, foliar and root symptoms resembling those observed on P. sylvestris and hybrids were also found affecting immature Queen palm [Syagrus romanozoffiana (Cham.) Glassman] interplanted with P. sylvestris. Collectively, symptoms observed on both palm species most closely resembled those previously described for LY and the affected palms all died within a matter of months once foliar discolouration became apparent.
The present study was initiated to determine whether decline and mortality of P. sylvestris and S. romanozoffiana in Hillsborough county was because of LY. Furthermore, in response to concerns about disease spread, a survey of palms in landscape plantings at nearby sites in Hillsborough, and more distant sites in Manatee and Sarasota counties, was undertaken during which true date palm (P. dactylifera), Canary Island date palm (P. canariensis) and Mexican fan palm (Washingtonia robusta Wendl.) with decline-type symptoms were identified. We report the detection and characterisation of three distinct coconut LY group (16SrIV) phytoplasma strains, occurring either as single or as mixed infections, in palms with symptoms in the tri-county region.
Materials and methods
Palm samples and DNA extraction
A total of 12 palms with pronounced foliar discolouration symptoms in production groves at a nursery in Hillsborough county were selected for study. These consisted of eight P. sylvestris, one P. sylvestris ×dactylifera hybrid, one P. sylvestris ×canariensis hybrid and two Queen palms (S. romanozoffiana). Two symptomless, seemingly healthy, P. sylvestris were also included for comparative purposes. Samples (3–6 g), each consisting of interior tissues, were removed from the lower trunks (stems) of the palms using a portable electric drill or brace and bit (Harrison et al., 2002a,c). Tissues were collected directly into clean, sealable plastic bags and shipped by overnight courier to the University of Florida’s Fort Lauderdale Research and Education Center (FLREC) for further analysis. Subsequently, 26 additional palms displaying similar decline-type symptoms were observed in landscape plantings at nine other sites by disease survey work (Table 1). These included representatives of four palm species, namely P. canariensis, P. dactylifera, S. romanozoffiana and W. robusta. Stem tissue samples were collected from each palm and shipped to the FLREC, as previously described.
| Sample Identity | Palm Species | Location | Phytoplasma Strain Identity | |
|---|---|---|---|---|
| City/Town | County | |||
| PDR | Phoenix dactylifera | Ruskin | Hillsborough | 16SrIV-D |
| PC1 | Phoenix canariensis | Apollo Beach | Hillsborough | 16SrIV-D |
| PC2 | P. canariensis | Apollo Beach | Hillsborough | 16SrIV-D |
| PC3 | P. canariensis | Apollo Beach | Hillsborough | 16SrIV-D |
| SL1G | P. canariensis | Thonotosassa | Hillsborough | 16SrIV-D |
| SL2G | P. canariensis | Thonotosassa | Hillsborough | 16SrIV-D |
| SEG | P. canariensis | Brandon | Hillsborough | 16SrIV-D |
| RPA | P. dactylifera | Brandon | Hillsborough | 16SrIV-D |
| WP | P. canariensis | Tampa | Hillsborough | 16SrIV-D |
| RMco | P. canariensis | Bradenton | Manatee | 16SrIV-A |
| SA1 | P. canariensis | Bradenton | Manatee | 16SrIV-A |
| SA4 | P. canariensis | Bradenton | Manatee | 16SrIV-D |
| EPO | P. canariensis | Ellenton | Manatee | 16SrIV-D |
| S1-QP | Syagrus romanozoffiana | Bradenton | Manatee | 16SrIV-D |
| S2-PD | P. dactylifera | Bradenton | Manatee | 16SrIV-D |
| S4-PS | P. sylvestris | Bradenton | Manatee | 16SrIV-D |
| S5-PS | P. sylvestris | Bradenton | Manatee | 16SrIV-D |
| S6-PS | P. sylvestris | Bradenton | Manatee | 16SrIV-D |
| WTL1 | P. canariensis | Bradenton | Manatee | 16SrIV-D |
| WTL2 | P. canariensis | Bradenton | Manatee | 16SrIV-D |
| CID1 | P. canariensis | Sarasota | Sarasota | 16SrIV-D |
| CID2 | P. canariensis | Sarasota | Sarasota | 16SrIV-D |
| CID3 | P. canariensis | Sarasota | Sarasota | 16SrIV-D |
| VW | P. dactylifera | Sarasota | Sarasota | 16SrIV-A and 16SrIV-F |
| BCT | P. dactylifera | Venice | Sarasota | 16SrIV-A and 16SrIV-F |
| FP | Washingtonia robusta | Venice | Sarasota | 16SrIV-F |
Total nucleic acids were obtained from stem tissue samples (3 g) by the procedure of Doyle & Doyle (1990) using a 2% hexadecyltrimethylammonium bromide (CTAB) extraction buffer augmented with 1% polyvinylpyrrolidone (PVP-40). After final precipitation with 95% ethanol, nucleic acid extracts were recovered by centrifugation at 12 000 g for 15 min, dried briefly in vacuo and resuspended in 150 or 200 μL of TE [10 mM Tris, 1 mM ethylenediaminetetraacetic acid (EDTA), pH 8] buffer. Aliquots (50 μL) of each extract were purified separately on spin columns (Wizard DNA Clean-Up System; Promega, Madison, WI, USA) and eluted in 50 μL of sterile TE buffer. DNAs derived from LY-diseased P. sylvestris (LYPS) and healthy coconut palm in Broward county, southeastern Florida, were used in this study as known positive and negative experimental controls, respectively.
Polymerase chain reaction assays
In preliminary tests, palm samples were evaluated for phytoplasma DNA by a PCR assay (35 cycles) employing rRNA operon primer pair P1 (Deng & Hiruki, 1991) and P7 (Smart et al., 1996). Resulting amplification products were diluted 1:10 with sterile deionised water, and 2 μL of each dilution was used for PCR (35 cycles) with nested 16S rRNA gene primer pairs R16F2n/R16R2 (Gundersen & Lee, 1996) or LY16Sf/LY16Sr (Harrison et al., 2002c). To enhance detection specificity, primers P1m (5′-TCC TGG CTC AGG ATT AAC-3′) and LY16-23Sr (Harrison et al., 2002b) followed by LY16Sf2 (5′-AAC GGG TGA GTA ACA CGT AAG-3′) and LY16-23Sr2 (5′-TTA GAC TGG TGG GCC TAA ATG-3′) were adopted for all subsequent nested PCR assays. Primers P1m and LY16Sf2, designed during this study, corresponded to base positions 8–25 and 84–104, respectively, of an LY phytoplasma 16S rDNA sequence (accession no. U18747), whereas 16SrIV group-specific primer LY16-23Sr2 corresponded to positions 193–213 of an LY phytoplasma 16-23S intergenic spacer region (ISR) sequence (AF024639) archived in the GenBank database.
Restriction fragment length polymorphism analysis of polymerase chain reaction products
Products (2–3 μL) of nested PCRs were digested separately with restriction endonucleases AluI, BstUI, DraI, EcoRI, HaeIII, HhaI, HinfI, MspI, MseI or RsaI (New England BioLabs, Waverley, MA, USA) at 37°C for a minimum of 16 h. Digests with TaqI (Promega) were performed at 65°C. Products of digests were separated by electrophoresis through 8% nondenaturing polyacrylamide gels with TBE (90 mM Tris–borate, 2 mM EDTA) as running buffer. DNA fragment profiles in gels were visualised and recorded as described above.
Cloning and sequencing of rDNA products
Nested PCR products amplified with primer pair LY16Sf2/LY16-23Sr2 were purified separately on spin columns (Wizard PCR Preps Purification System; Promega), eluted in 40 μL of sterile ultrapure water and cloned in vector pGEM-T (Promega) and Escherichia coli DH5-α cells (Invitrogen Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions. Sequencing of representative clones was performed with automated equipment at the University of Florida’s Core DNA Sequencing Service Laboratory.
Analysis of phytoplasma rDNA
Sequences of cloned rDNA were assembled using SeqMan software (Lasergene™ 7.1; DNASTAR, Madison, WI, USA). A database search of homologous sequences was performed by Blast analysis at the National Centre for Biotechnology (NCBI) website (http://ncbi.nlm.nih.gov/BLAST). Pairwise similarities between sequences were calculated using sequence distance option in MegAlign (Lasergene). Putative restriction sites in sequences were identified using pDRAW32 software (AcaClone, http://www.acaclone.com). Phylogenetic interrelationships among palm-associated phytoplasmas, representatives of other phytoplasma groups and Acholeplasma palmae were assessed based on partial 16S rRNA gene sequences corresponding to the F2n/R2 portion (Gundersen & Lee, 1996) of the gene. Sequences were aligned using ClustalW (Thompson et al., 1994) with the following parameters: gap opening penalty – 5.0, gap extension penalty – 0.2, delay divergent cutoff – 30% and DNA transition weight – 0.5. A phylogenetic distance tree was constructed from the alignment by the neighbour-joining method and visualised using MEGA 4.0 software (Tamura et al., 2007). A. palmae was used as the outgroup to root the tree. Bootstrapping (100 replicates) was performed to estimate the stability of the inferred subclades.
Results
Phytoplasma detection by polymerase chain reaction
When palm samples were first assessed for phytoplasma infection by nested PCRs incorporating primer pair P1/P7 followed by either R16F2n/R16R2 or LY16Sf/LY16Sr, an rDNA product of either 1.2 or 1.4 kb, respectively, was amplified from diseased palms only. However, additional smaller products were consistently co-amplified from both diseased and symptomless Phoenix palms during these assays despite adjustments to thermal cycling conditions when assays were repeated (data not shown). Although the origin of these products was not determined, past aetiological investigations of diseases affecting Phoenix palms (Harrison et al., 2002c) as well as other plant species (Lee et al., 2002; Fránová, et al., 2007) have reported amplification of rDNA products from nontarget gram-positive bacteria by PCR assays employing phytoplasma universal rRNA gene primer pairs. Unambiguous detection of phytoplasma DNA was achieved only after primer pair P1m/LY16-23Sr followed by LY16Sf2/LY16-23Sr2 were adopted for all subsequent PCRs. Using this alternative assay, positive detections, indicated by weak to moderate amplification of an rDNA product of about 1.6 kb, were obtained for all 12 diseased palms (8 P. sylvestris, 2 P. sylvestris hybrids and 2 S. romanozoffiana) from a field nursery in Hillsborough county and for LY phytoplasma DNA included as a known positive experimental control. No discernible PCR products were obtained from two symptomless P. sylvestris palms included for comparative purposes or from healthy coconut DNA and reactions devoid of template DNA, which served as negative experimental controls (data not shown).
The aetiological investigation of declining palms begun at a Hillsborough county palm nursery was expanded to include additional palms with similar symptoms that were found at other sites in Hillsborough, Manatee and Sarasota counties by disease survey work. As a result, phytoplasmas were detected by nested PCRs in 26 additional landscape palms. Affected palms included 17 P. canariensis, 3 P. sylvestris, 4 P. dactylifera, 1 S. romanozoffiana and 1 W. robusta in plantings at nine additional sites within the tri-county region.
Phytoplasma characterisation by restriction fragment length polymorphism analysis
Products of nested PCRs primed by LY16Sf2/LY16-23Sr2 from P. sylvestris, P. sylvestris hybrids and S. romanozoffiana at a palm nursery in Hillsborough county, as well as from P. dactylifera and P. canariensis palms in nearby Apollo Beach, were digested separately with AluI, DraI, HaeIII, HinfI (data not shown), BstUI, HhaI, MseI, MspI, RsaI or TaqI endonuclease. Representative patterns resulting from digests are illustrated in Fig. 1. No differences in restriction fragment profiles were evident among palms, indicating that they all contained very similar or possibly the same phytoplasma strain. However, profiles revealed by HhaI (Fig. 1B) or MseI (Fig. 1E) digests clearly differentiated these strains from LY phytoplasma (LYPS)-positive control. Based on these results, HhaI and MseI were chosen for a comparative analysis of rDNA products from declining landscape palms subsequently identified by disease survey work (Fig. 2). MseI digests (Fig. 2A) revealed two dissimilar profiles one of which [e.g. P. canariensis (SEG)] was associated with most (80.8%) of diseased palms throughout the tri-county survey region. This profile was also identical to that of palm-associated phytoplasmas previously obtained from nursery-grown palms. The other profile was associated with five palms (19.2%) only, including a solitary P. canariensis (RMCo) in Manatee county as well as P. canariensis (SAI), P. dactylifera palms (VW and BCT) and W. robusta (FP) in Sarasota county. The latter profile was indistinguishable from that of LY phytoplasma (LYPS). By comparison, four distinct fragment profiles were resolved among these same landscape palms by HhaI digests (Fig. 2B). Of these, one profile (e.g. sample SEG) was associated with 21 palms that included 7 P. canariensis and 2 P. dactylifera in Hillsborough county, 4 P. canariensis, 3 P. sylvestris, 1 P. dactylifera and 1 S. romanozoffiana in Manatee county and 3 P. canariensis in Sarasota county. This most prevalent rDNA fragment profile matched that of phytoplasmas initially identified in nursery-grown palms (Fig. 1B). A second profile limited to solitary P. canariensis palms (SA1 and RMCo) in Manatee county corresponded to that of LY phytoplasma. A third profile was associated with two P. dactylifera palms (VW and BCT), while a fourth unique profile was obtained from a solitary W. robusta (FP) palm. The latter three palms were all sampled in Sarasota county.
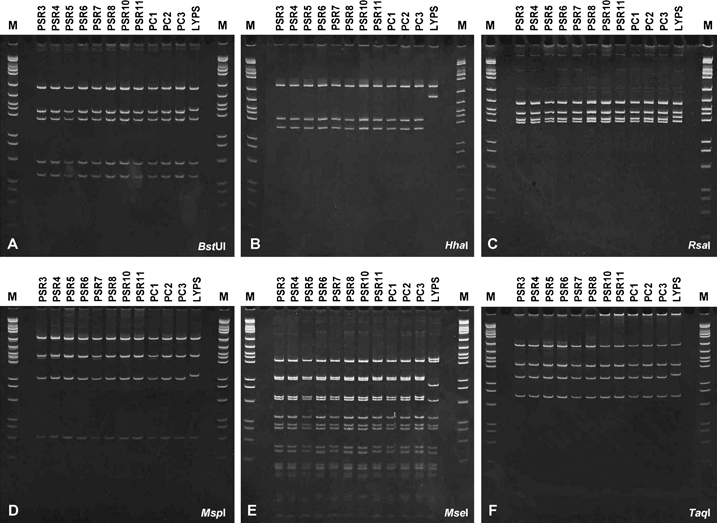
Representative restriction fragment length polymorphism (RFLP) profiles of phytoplasma rDNA (1.6 kb) amplified from symptomatic Phoenix palms by a nested PCR assay incorporating RNA gene operon primer pairs P1m/LY16–23Sr followed by LY16Sf2/LY16–23Sr2. PCR products were derived from Phoenix sylvestris (PSR), P. sylvestris ×dactylifera (PSR10) hybrid, P. sylvestris ×canariensis (PSR11) hybrid, Phoenix canariensis (PC) and lethal yellowing diseased P. sylvestris (LYPS). Products were digested with: A, BstUI; B, HhaI; C, RsaI; D, MspI; E, MseI and F, TaqI. pGEM, molecular size (bp) markers in descending order: 2465, 1605, 1198, 676, 517, 460, 396, 350, 222, 179, 126, 75, 65, 51 and 36.
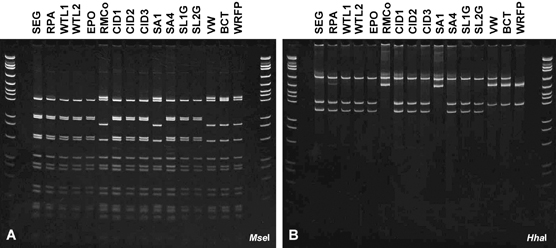
Comparison of representative fragment profiles following endonuclease digestion of phytoplasma rDNA products (1.6 kb) amplified from symptomatic Phoenix palms by nested PCR with rRNA gene operon primer pairs P1m/LY16–23Sr and then LY16Sf2/LY16–23Sr2. PCR products were derived from edible date palm (Phoenix dactylifera), palms (RPA, VW and BCT) and Mexican fan palm (Washingtonia robusta) palm (WRFP), while all other products were from Canary Island date (Phoenix canariensis) palms. pGEM, molecular size (bp) markers in descending order: 2465, 1605, 1198, 676, 517, 460, 396, 350, 222, 179, 126, 75, 65, 51 and 36.
Upon closer inspection, rDNA fragment profiles associated with P. dactylifera palms (VW and BCT) appeared to be a composite of two dissimilar patterns as each contained fragments in common with profiles from P. canariensis palms (RMCo or SA1) and the unique profile associated with W. robusta (FP). Composite RFLP profiles have been observed during past aetiological investigations of phytoplasma diseases and attributed to presence of two heterogeneous copies of the 16S rRNA gene within causal phytoplasmas (Liefting et al., 1996; Jomantiene et al., 2002; Davis et al., 2003) or to mixed infections involving combinations of two or more strains belonging to the same 16S rDNA RFLP group or different groups (Alma et al., 1996; Staniulis et al., 2000; Lee et al., 2003). The 16S rDNA interoperon sequence heterogeneity within LY phytoplasma (i.e. subgroup 16SrIV-A) strains from palms in southern Florida has been demonstrated (Harrison et al., 2002a) and may occur in other subgroup 16SrIV strains also (Cordova et al., 2000). However, in the present study, mixed infections composed of two mutually distinct phytoplasmas seemed the most likely explanation to account for RFLP patterns associated with P. dactylifera palms (VW and BCT). Evidence to support this interpretation was obtained after nested PCR products from P. dactylifera (BCT) and W. robusta (FP) were cloned separately in vector pGEM-T, re-amplified by LY16Sf2/LY16-23Sr2-primed PCR and digested with HhaI. For P. dactylifera (BCT), digests revealed two distinct rDNA fragment profiles among eight clones that were examined by this means. One profile derived from each of the four clones was identical to that of LY phytoplasma, while a second profile associated with remaining clones matched that of W. robusta (FP). By comparison, identical fragment profiles were obtained from all W. robusta (FP) rDNA clones examined by this means indicating that this palm harboured one phytoplasma strain only.
Sequence similarities and phylogenetic analysis
Eight rDNA products amplified by nested PCRs from five affected palm species were cloned and sequenced. Blast analysis verified all eight sequences to be of phytoplasma origin and that each was composed of 16S rDNA and a 16-23S ISR. Sequences were either 1631 or 1644 bp and were a result of differences in length (190 vs 203 bp) of the 16-23S ISR. Pairwise similarity comparison between rDNA sequences of palm-associated phytoplasmas in west central Florida yielded values ranging from 98.1% to 100% or from 98.2% to 100% when 16S rRNA gene sequence alone was considered. These value ranges indicated that all palms harboured group 16SrIV phytoplasma strains as was inferred by prior results from RFLP analysis.
Further comparative analysis revealed phytoplasma rDNA sequences from P. sylvestris (PSR3) (accession no. EU096499), P. dactylifera (PDR) (EU241518) and S. romanozoffiana (SR1) (EU241513) to be co-identical and indistinguishable from those of Texas Phoenix decline (TPD) phytoplasma, a previously characterised subgroup 16SrIV-D strain (Harrison et al., 2002c). Sequences from P. canariensis (RMCo) (EU241517) and P. dactylifera (BCT) (EU241514) matched those of LY phytoplasma in P. sylvestris (LYPS) (EU241516), a finding that extends the spread of LY disease beyond subtropical southern Florida (hardiness zone 10) to include Sarasota and Manatee counties (hardiness zone 9). Two co-identical sequences from W. robusta (FP) (EU241512) and P. dactylifera (BCT) (EU241515) differed from all known sequences of group 16SrIV phytoplasmas currently archived in NCBI’s GenBank database. Delineated primarily on the basis of their actual HhaI restriction profile, analysis of putative restriction sites in rDNA sequences confirmed that these strains possess a unique HhaI site located at position 655 in the 16S rRNA gene. On this basis, we propose that these strains be classified as members of a new subgroup, 16SrIV-F, sensu Lee et al. (1993, 1998, 2000).
A phylogenetic tree constructed from 16S rDNA sequences (Fig. 3) confirmed that all eight phytoplasma strains examined in this study clustered closely together with other strains (Cordova et al., 2000; Harrison et al., 2002b; Martinez et al., 2007; Wei et al., 2007) composing the coconut lethal yellows phytoplasma subclade (Gundersen et al., 1994). Tree branching patterns also determined that strains most frequently associated with diseased Phoenix palm species and newly recognised host S. romanozoffiana in west central Florida were part of an existing lineage of 16SrIV-D phytoplasma strains that included TPD phytoplasma (Fig. 3). Furthermore, phytoplasmas infecting W. robusta (FP) and P. dactylifera (BCT) gave rise to a new branch not included in any previous phylogenetic trees, indicating that these strains represent a new lineage (i.e. subgroup) within the coconut lethal yellows phytoplasma group (Gundersen et al., 1994; IRPCM, 2004).
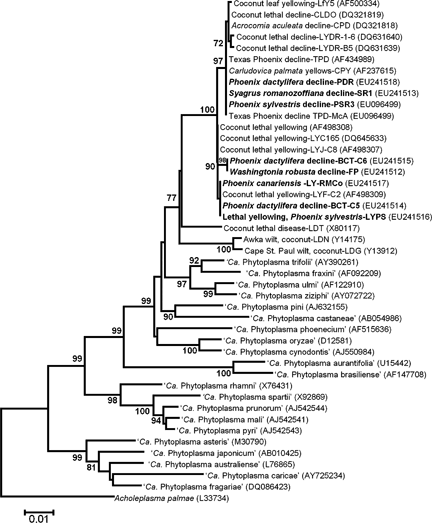
Phylogenetic tree of 16S rRNA gene sequences from representative phytoplasmas in the coconut lethal yellow groups and other phytoplasma groups constructed by the neighbour-joining method. Phytoplasma strains sequenced in this study are indicated in bold type. Boot strap values (an indicator of the reliability of the analysis) >70% are shown on branches.
Discussion
All palms displaying symptoms of decline suggestive of LY disease in west central Florida contained phytoplasmas. This finding was unusual as there have been no prior reports of palm mortality to LY in this region. Rather, spread of the disease has been confined to the southern subtropical tier of the state, a region where M. crudus, the cixiid vector of LY is most abundant (Howard et al., 1982, 1984) and coexists together with the greatest density and diversity of susceptible palm species in landscape and amenity plantings (McCoy et a.l, 1983; Howard & Barrant, 1989).
In Hillsborough county, where dying P. sylvestris palms were first identified, this species along with P. canariensis and P. dactylifera were most affected by disease. All three are known to be susceptible to LY (McCoy et al., 1983; Howard, 1992; Harrison & Jones, 2004); thus, this disease could not be entirely discounted as the underlying cause of palm mortality. However, observation of similarly affected S. romanozoffiana, a widely planted species that does not contract LY (McCoy et al., 1983; Meerow, 2006), suggested that an aetiological agent other than LY phytoplasma, a subgroup 16SrIV-A strain (Lee et al., 2000; Harrison et al., 2002a), might be involved. This possibility was confirmed after phytoplasma rDNA sequences associated with each of these four host species were found to be indistinguishable from those of TPD phytoplasma. The latter subgroup 16SrIV-D strain was previously identified as the probable cause of a lethal disease of P. canariensis in Corpus Christi, Texas (Harrison et al., 2002b). Since this earlier report, we have identified subgroup 16SrIV-D strains (EF042899) in P. canariensis in the Rio Grande Valley of southern Texas (unpublished observations), indicating that TPD is more widespread than was first thought, although, until the present study, it was not known to occur in Florida.
In the tri-county region encompassed by survey work, a decreasing disease incidence from north to south implicated Hillsborough County as the probable epicentre of infestation by subgroup 16SrIV-D phytoplasmas. Although the origin of these strains is uncertain, when strain identity, common palm hosts and geographic proximity were all considered, it seemed reasonable that they could have spread to Florida from southern Texas. That this event occurred by importation of infected palms is doubtful because none of the affected palm species identified in the present study are produced in Texas for sale in Florida. Thus, an influx of inoculative vector insects is the most likely explanation for the new disease outbreak. Presently, vector(s) of subgroup 16SrIV-D strains are unknown, although the cixiid Oliarus acicus (Caldwell) was the most abundant potential vector associated with P. canariensis in the Rio Grande Valley (Meyerdirk & Hart, 1982) at a time when losses of this species and P. dactylifera to a disease, believed to be LY, were underway (McCoy et al., 1980a,b). Whether this species also occurs in west central Florida warrants further investigation.
While successively fewer diseased palms were located in Manatee and Sarasota counties most harboured subgroup 16SrIV-D strains. Notably, these included P. canariensis palms (CID1, CID2 and CID3) that, according to Sarasota city records, had been established there on the same site for at least 50 years. This particular observation lends support to the hypothesis that these strains are a recent introduction into Florida. Palms containing subgroup 16SrIV-A phytoplasmas were also identified in both counties, each with no prior history of LY. Presumably, further, more northerly, distribution of LY indicates that ecological conditions conducive to supporting and sustaining vector M. crudus populations now prevail in these counties. The onset of favourable conditions has coincided with an increased abundance of landscape palms because of new residential growth and development. Furthermore, mild winters accompanying growth and development activity over the past decade have encouraged the planting of nontraditional, cold-sensitive palms. Evidence of this trend is coconut palm, an LY-susceptible species, which can now be found in coastal landscape plantings as far north as Tampa in Hillsborough county.
The occurrence of a third, previously undocumented subgroup 16SrIV-F phytoplasma in declining P. dactylifera and in W. robusta in Sarasota county was unexpected. As with Queen palm (S. romanozoffiana), Mexican fan palm is among the most widely used of all palms in both urban and residential landscapes due largely to this species having remained free of LY despite >35 years of exposure to the disease. Although sudden localised die offs of W. robusta have increased in southern Florida in recent years, none have been attributed to phytoplasma disease. While little is known about this new subgroup 16SrIV-F strain, its close similarity (99.6%) and occurrence as mixed infections with LY phytoplasma in P. dactylifera may be of epidemiological and evolutionary significance as it suggests that both strains are vectored by M. crudus.
The emergence of new lethal diseases of palms attributed to 16SrIV group phytoplasmas besides the LY agent in Florida has closely followed similar developments in other geographic locations where LY is known to occur. These include reports of novel 16SrIV strains associated with recent outbreaks of disease among both coyol (Acrocomia aculeata) and coconut palms in central Honduras (Roca et al., 2006) and among coconut in southern Dominican Republic (Martinez et al., 2007). In Jamaica, a resurgence of LY on coconut (Lebrun et al., 2008) has been accompanied by the appearance of new diseases among unrelated understorey dicot plants (Brown et al., 2007) which together with Cedusa sp. leafhoppers (Brown et al., 2006) have provided additional sources of novel 16SrIV group strains whose subgroup status remains to be fully determined.
Acknowledgements
We thank Jim Mertely, Randy Frazier, Andrew Cushman, Barry Troutman, Mary Beth Henry, Margaret Dessaint and Don Rainey for their help in providing samples from diseased palms for aetiological investigation in this study.