Gene expression analysis of an epidermolysis bullosa simplex Dowling-Meara cell line by subtractive hybridization: recapitulation of cellular differentiation, migration and wound healing
Abstract
Abstract: An intact keratin 5/keratin 14 intermediate filament cytoskeleton is vital for the integrity of basal keratinocytes and for the development and maintenance of epidermal structures. In patients with epidermolysis bullosa simplex Dowling-Meara (EBS-DM), heterozygous mutations in the keratin 14 gene in keratinocytes cause a cytoskeletal collapse leading to fragile cells susceptible to cellular stress. The primary aim of this work was to extend analysis of differentially expressed genes in an EBS-DM model cell line to obtain insights into the molecular consequences resulting from the keratin 14 mutation. In a first step, suppression subtractive hybridization (SSH), a powerful technology to enrich for differentially expressed genes, was used to identify genes whose up-regulation may be a direct or indirect result of the keratin 14 mutation, R125P. We discovered 55 candidate genes (SSH genes) that were further analysed by RTq-PCR. Of the 55 SSH genes, 14 (25.45%) were found to be congruently up-regulated. Bioinformatic analysis revealed significant enrichment of genes regulating epidermal development, migration, apoptosis and wound healing.
Introduction
Epidermolysis bullosa simplex (EBS) is an inherited autosomal dominant genodermatosis characterized by loss of tissue integrity resulting in intraepithelial blisters within the basal epidermal layer in response to comparatively mild mechanical trauma. Phenotypes in EBS are primarily the consequence of mutations in type 2 keratin 5 (K5) or type 1 keratin 14 (K14) (1–11). Co-expression of K5 and K14 is a hallmark of highly mitotic basal layer epidermal keratinocytes (12–15). K5 and K14 usually form heterodimers, which build up the structure of the cytoskeletal intermediate filament (IF) in these cells, guaranteeing mechanical resilience sufficient to overcome physical stress but sufficiently pliable to maintain proliferative capacity of the cells (16,17). On closer inspection of keratin structure, the molecules all exhibit a non-α-helical N-terminal head domain and a C-terminal tail domain flanking an α-helical rod domain, segmented into highly conserved subhelices 1A, 1B, 2A and 2B. In particular, the helix initiation peptide in region 1A and the helix termination peptide in the 2B domain are important for heterodimerization of compatible type 1 and type 2 keratins (18,19). The phenotypes of the three major subtypes of EBS, EBS Dowling-Meara (EBS-DM), localized EBS (EBS-loc) and generalized other EBS are the consequence of mutations at different residues in K5 or K14 genes; mutations in the critical 1A and 2B regions lead to the most severe form, EBS-DM (20).
To better understand the mechanisms accounting for the phenotypic characteristics and to be able to estimate unwanted side effects of different therapeutic approaches, we analysed the gene expression profile in the EBS-DM keratinocyte model KEB7 relative to wild-type NEB1 cells. KEB7 cells carry an autosomal dominant mutation R125P in the 1A segment of the rod domain of the K14 molecule, which triggers a cytoskeletal collapse. We expected the gene expression profile of these cells to be influenced by this collapse.
Relatively few gene expression profiling studies relevant to EBS simplex have been accomplished. Microarray experiments have been performed by Liovic et al. (21) to investigate the response of the EBS keratinocyte cell lines KEB4 (mild EBS-loc phenotype, K14 mutation V270M) and KEB7 (severe EBS-DM phenotype, K14 mutation R125P) to hypo-osmotic stress in comparison with the wild-type control NEB1. Liovic et al. (22) focused on screening for differences in the expression profile of cytoskeletal components in NEB1, KEB4, KEB7 and KEB1 (K5 mutation E475G) cells and isogenic pathomimetic cell lines. In 2007, Lu et al. (23) investigated the expression profile in the epidermis of a K5−/− EBS mouse model, concentrating on the regulation of inflammatory cytokines. Currently available information regarding differentially expressed genes in EBS is summarized in Table S1.
To extend the list of differentially expressed genes and potentially to confirm previously reported regulated genes in EBS-DM, we chose to perform suppression subtractive hybridization (SSH). SSH is an ‘open system,’ which means that the method is not predefined by a finite list of candidate genes available on a microarray chip nor does it need any gene annotation or information to search for differentially expressed genes. The method, therefore, permits finding even currently unidentified/unannotated new genes and splice variants (24–26). Another advantage of SSH is that it permits the detection of low-abundance transcripts (27), which are often not detected in microarray analyses (28–32).
Materials and methods
Cell lines and culture conditions
The immortalized keratinocyte cell lines NEB1 (wild-type control cell line) and KEB7 (EBS-DM phenotype caused by the severe R125P mutation) (33) were cultured in DMEM with 25% Ham’s F12 and 10% fetal bovine serum plus the additional agents adenine (1.8 × 10−4 m), hydrocortisone (0.4 μg/ml), transferrin (5 μg/ml), liothyronine (2 × 10−11 m), insulin (5 μg/ml) and EGF (10 μg/ml). The cell lines were grown to a confluency of approximately 90% in plastic tissue culture grade flasks at 37°C, 5% CO2 without fibroblast feeder cells.
Isolation of total RNA
After a minimum of seven days in culture, total RNA from cells growing in the same passage at approximately 90% confluency was isolated after trypsinization using the RNeasy Mini Kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer’s protocol. Concentration of RNA was determined by optical density measurement, and quality was examined by 1% agarose 3-(N-morpholino) propanesulfonic acid RNA gel electrophoresis. Total RNA samples were stored at −80°C.
Construction of a subtracted cDNA library using suppression subtractive hybridization
Suppression subtractive hybridization was accomplished using the NEB1 RNA as driver and the KEB7 cDNA as tester via the PCR-Select cDNA Subtraction Kit (Takara Bio Europe/Clontech, Saint-Germain-en-Laye, France) following the manufacturer’s instructions, except that total RNA was used instead of purified poly A+ RNA, and purification of double-strand cDNA in steps D7-19 was replaced by purification using the Illustra GFX PCR DNA and Gel Band Purification Kit (GE Healthcare Europe GmbH, Vienna, Austria). Subtracted KEB7 cDNA fragments were subsequently cloned via the StrataClone PCR Cloning Kit (Agilent Technologies Austria GmbH, Vienna, Austria) and checked for positive cloning by colony PCR using the M13(-20) forward and M13 reverse primers. Plasmids were isolated using the PureYield Plasmid Miniprep System (Promega GmbH, Mannheim, Germany) and sequenced with the M13 reverse primer.
Real-time quantitative PCR
Pools of total RNA extracted from five consecutive NEB1 and KEB7 passages at approximately 90% confluency each were used as templates for cDNA synthesis employing the iScript cDNA Synthesis Kit (Bio-Rad Laboratories Ges.m.b.H., Vienna, Austria) according to the instructions of the supplier. Aliquots of cDNA (1 μl) were used for each RTq-PCR performed in combination with the iQ SYBR Green Supermix (Bio-Rad Laboratories, Inc) in a total reaction volume of 25 μl. Three PCR runs were performed for each cell line including biological and technical triplicates of every gene. Seven housekeeping genes (B2M, HPRT1, RPL13A, GAPDH, ACTB, ANXA1 and TUBB) served as internal controls for normalization and for evaluation of experimental variance, which provided an experimental estimate of the fold-change cut-off. Probes for KRT14 intron 1 and BEST1 intron 2 were included to assess genomic DNA contamination.
Results
As the K5/K14 IF system represents the major cytoskeletal component in basal keratinocytes, providing flexibility plus mechanical resilience, we expected the R125P mutation in K14 to alter the expression of a specific set of genes based on the observation that many signalling pathways are dependent on intact cellular integrity (34–36). The essential features of our experimental design for investigating alterations in gene expression patterns in a patient cell line compared to a wild-type (WT) control cell line under normal growth conditions are depicted in Fig. 1. Beginning with RNA purification from the EBS-DM cells and a wild-type control cell line and quality control of the isolates, we proceeded with an SSH assay screening for up-regulated genes in the patient cell line. Taking advantage of the DAVID functional annotation tool, we were able to assign identified SSH genes to biologically enriched functional groups (37,38). As a last step, the up-regulation of these genes was confirmed by RTq-PCR.
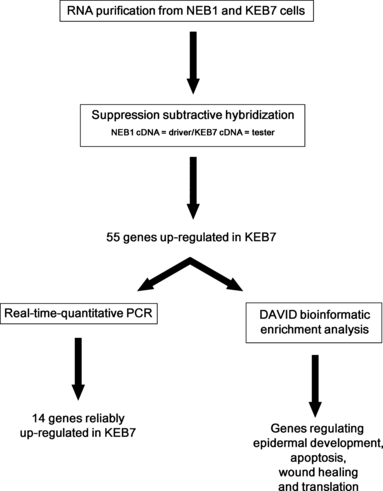
Schematic depiction of the experimental design used for the identification of genes up-regulated in the patient epidermolysis bullosa simplex Dowling-Meara KEB7 cell line. Expression of 55 genes involved in epidermal development, apoptosis, wound healing and translation was found to be increased by suppression subtractive hybridization. Of these, 14 were confirmed by RTq-PCR.
Fifty-five differentially expressed genes were identified in the EBS-DM cell line
To study and understand the regulation of a variety of molecular processes, it is vital to have knowledge of the expression patterns of genes forming the basis of those mechanisms. Subtractive hybridization (SH) techniques are one way to comparatively identify differentially expressed genes in two cell populations. The outstanding advantages of these systems, compared to other expression profiling technologies, are that there are no limitations to the number of genes to be investigated, and differentially expressed cDNAs can easily be identified by sequencing and can be isolated and cloned for various downstream applications (24–27). However, in addition to being labour intensive with multiple subtraction steps, SH techniques also hold several disadvantages including a requirement for large amounts of poly(A)+ RNA (more than 20 μg) and inefficiency in obtaining low-abundance transcripts. To overcome these problems, we chose a PCR-based SSH method described by Diatchenko et al. (27) in which amplification of non-target cDNAs is suppressed, whereas differentially expressed target fragments are selectively amplified. Total RNA from the wild-type control cell line NEB1 was used to produce driver cDNA, and cDNA derived from total RNA of the EBS-DM patient cell line KEB7 served as tester. A total of 119 clones were analysed, and following the removal of redundancies, 55 different genes could be identified as being up-regulated in KEB7 (Table S3). On closer examination of the SSH data, two large enriched gene groups become obvious: keratins (approximately 11%) and ribosomal proteins (approximately 29%). Among the keratin group, several type 1 keratins including K14, K16 and K17 and the type 2 keratins K5, K6A and K6B were identified.
Bioinformatic analysis of the SSH library reveals enrichment of genes regulating epidermal development, apoptosis, wound healing and translation
To obtain a more detailed look at this expression profile, we subjected our SSH data set to gene ontology (GO) and enrichment analysis with the DAVID web tool (37,38). The functional annotation clustering tool, grouping highly related and redundant annotation terms, provided by this suite was used to assign the genes to significant functional groups displayed in Table S2. Significance is indicated by the enrichment score (ES), which is the geometric mean of the enrichment P-values (P) of each GO term held in the cluster. As the ES represents a relative score, it is transformed to a negative logarithm; thus, clusters with scores above 1.3 were considered to be significant and were investigated more closely. Notably, the most enriched cluster (ES = 13.9) comprises annotation terms for translational regulation and ribosomal structure owing to the overrepresentation of 16 ribosomal proteins (approximately one-third) in the subtracted gene library. Other interesting and significant enriched clusters highlight an influence of the K14 R125P mutation on epidermal development/differentiation (ES = 4.27) and regulation of apoptosis/wound healing (ES = 2.37).
The DAVID suite also provides a disease chart compiling annotations from the Online Mendelian Inheritance in Man genetic disorders database, which permits assigning the enriched genes to diseases. The top diseases shown by DAVID (P ≥ 0.0076 for all) included the other two forms of EBS (EBS-loc and generalized other EBS), pachyonychia congenita (Jackson-Lawler and Jadasson-Lewandowsky types), as well as EBS-DM investigated in this study.
RTq-PCR analysis confirms the up-regulation of cell junction proteins, K14, the basal cell carcinoma (BCC)-associated K17 and wound healing–related K16 in EBS-DM
Although the 55 candidates were found by subtractive hybridization to be up-regulated in the EBS-DM cell line, it was necessary to confirm their up-regulation by a complementary gene expression profiling method. Therefore, RTq-PCR experiments based on forward and reverse oligonucleotide primer pairs for all 55 candidates were performed on NEB1 and KEB7 cDNA. To guarantee the reliability of our real-time data, each experiment was independently performed three times. An experimental fold-change cut-off of 1.3 was experimentally validated using seven housekeeping genes (for each biological replicate) for normalization. To ensure conservative results, only genes with a fold change above 1.5 were further examined. Significance of differences in the mean fold-change values of NEB1 and KEB7 were assessed by Student’s t-test. P ≤ 0.05 was interpreted as indicating significance.
Of the 55 genes that were up-regulated in the EBS-DM cell line by SSH, 14 (25.45%) were confirmed by RTq-PCR and are presented in Fig. 2a. The list of genes can be broken down into two subcategories: (i) Twelve genes were moderately up-regulated, exhibiting a fold change <3.0 (twice the fold cut-off of 1.5). These were desmoglein 3 (DSG3), tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide (YWHAZ), annexin A8 (ANXA8), keratin 6B (K6B), thioredoxin interacting protein (TXNIP), ubiquitin-conjugating enzyme E2K (UBE2K), DNA-damage-inducible transcript 4 (DDIT4), coagulation factor 3 (F3), desmoplakin (DSP), metastasis-associated lung adenocarcinoma transcript 1 (non-protein coding) (MALAT1), keratin 17 (K17) and the TP53 apoptosis effector (PERP). (ii) Two genes, keratin 16 (K16) and keratin 14 (K14), were highly up-regulated, exhibiting a fold change >3.0.
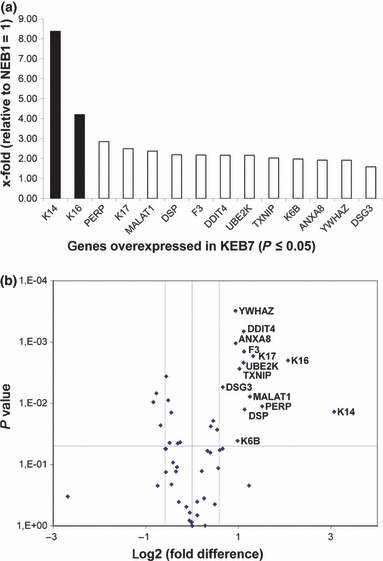
RTq-PCR-based examination of KEB7 up-regulated genes revealed by SSH. A. Two highly up-regulated genes (K14, K16) depicted by black bars and 12 moderately up-regulated genes (PERP, K17, MALAT1, DSP, F3, DDIT4, UBE2K, TXNIP, K6B, ANXA8, YWHAZ, DSG3) depicted by white bars, not exceeding 100% above the fold cut-off of 1.5, were confirmed to be increased significantly in KEB7 cells. B. Volcano plot shows the overall outcome of the RTq-PCR experiment depicting the 14 significantly up-regulated genes localized below the P-value of 0.05 (blue line) and above the fold-change cut-off of 1.5 shown as log2 (right pink line).
A more detailed look at the overall outcome of the RTq-PCR study depicted in the volcano plot (Fig. 2b), which examines the presumptively up-regulated KEB7 SSH genes, reveals 38 genes to be unregulated and 3 to be significantly and unexpectedly down-regulated. Therefore, these 41 genes number among the SHH candidates whose up-regulation could not be confirmed by RTq-PCR. As noted above and in Fig. 2a, 14 genes were significantly up-regulated in the patient cell line compared to the WT control cells.
As demonstrated by these results, the cytoskeletal keratin group identified by SSH screening, consisting of K6B, K14, K16 and K17, was confirmed by RTq-PCR to be up-regulated in KEB7. K5, the heterodimerization partner of K14, was the only cytoskeletal keratin gene not significantly up-regulated (P = 0.055). In addition, a group of cell junction proteins, DSG3, DSP and PERP, were confirmed by RT-PCR to be up-regulated in the patient cell line.
Discussion
The primary aim of this work was to analyse differential gene expression in the patient cell line KEB7 compared to the wild-type cell line NEB1 to identify candidate genes potentially implicated in keratin-associated stress signalling that might account for the pathophysiological consequences observed in K14-defective EBS-DM keratinocytes. Furthermore, using a complementary subtractive hybridization analysis, we planned to extend the library of gene expression data obtained in earlier DNA microarray–based mRNA profiling studies.
In a first step, PCR-based SSH was used to identify genes whose up-regulation might be a result of the K14 mutation R125P. The identification of such genes through mRNA profiling has the potential to contribute to the elucidation of mechanisms responsible for the severe phenotype of EBS-DM. Through SSH analysis, we identified 55 candidate genes that were further subjected to RTq-PCR analysis. Of these 55 SSH candidate genes, 14 (25.45%) were confirmed by RTq-PCR analysis to be up-regulated. Methodologically, the lack of a higher consensus between SSH and RTq-PCR results (up-regulation of 41 SSH genes was not confirmed) may be attributed to false-positive candidates obtained by SSH. Factors that may have influenced the false-positive rate include the use of total RNA rather than purified poly A+ RNA, lack of optimization of first- and second-step hybridization times or use of a suboptimal driver/tester cDNA ratio (39). In this regard, the set of ribosomal proteins, representing a group of high-abundance proteins, could be an artifact generated during the hybridization steps of the SSH procedure. Importantly, despite the identification of false-positive candidates, 14 presumably true positive candidates were identified.
Massive dysregulation of genes involved in wound healing could be associated with a state of activated wound healing and cytoskeletal flexibility in EBS-DM cells
An interesting group of genes found to be up-regulated in KEB7 consisted of the keratins K6B, K14, K16 and K17. Surprisingly, the K14 gene, which carries the heterozygous R125P mutation in the KEB7 cell line, turned out to be the most highly up-regulated gene among our 14 confirmed candidate genes. A possible explanation for this phenomenon could be the influence of active stress signalling, including the SAPK/JNK pathway, in KEB7 cells (40). As a consequence of active stress signalling, AP-1 transcription factor family members are activated (41) and bind the K14 promoter (41–43), thereby potentially leading to the enhanced expression of K14 in KEB7 cells. This expression profile in general indicates that in addition to impairment of K5/K14 IF dynamics triggered by the K14 R125P mutation in basal keratinocytes (44), IF composition and architecture might also be changed. In wound healing and hyperproliferative tissue, K6, K16 and K17 are transiently co-expressed. Especially in wound repair of adult skin (13), psoriatic adult epidermis (45) and skin development (46), K16 is thought to engage in keratinocyte activation and is expected to positively influence the migration of cells.
Not to be overlooked is the ability of K16 to replace K14 in the K5/K14 heterodimer, a consequence of the high homology of K14 and K16 primary structures. In this regard, Paladini et al. were able to partially mitigate skin blistering in keratin 14 null mice, which would die shortly after birth, by targeted expression of K16 (47,48). Possibly, EBS-DM keratinocytes benefit from up-regulation of K16 and K14, which stoichiometrically compete for K5 binding and prevent the mutant K14 from sequestering K5, which could otherwise lead to assembly of erroneous K5/K14 IFs or K5/K14 heterodimers unable to assemble IFs.
We hypothesize that the recurrent bullous lesions occurring in EBS-DM induce an activated ‘wound healing state’ of keratinocytes. This would be guaranteed by the up-regulation of both K6 and K16. We also hypothesize that different heterodimeric combinations of the type 2 keratins K5 and K6 and the type 1 keratins K14, K16 and K17 are used to compensate for of the impaired resistance to mechanical trauma resulting from the K14 R125P mutation.
Another gene in the KEB7 up-regulated candidate list attracted our attention: the coagulation factor 3 or tissue factor (F3). In synergy with coagulation factor 7 (F7), F3 is primarily known to participate in the initiation of thrombus formation in the extrinsic coagulation cascade (49,50), an important step in wound healing. F3 is also expressed in skin keratinocytes and was found by Xu et al. (51) to be crucial for positively influencing keratinocyte migration, re-epithelialization, leucocyte migration and inflammatory signalling in keratinocytes during wound healing. This characteristic also matches the observation of Morley et al. (33) who showed in a scratch wound assay that re-epithelialization occurred significantly faster in EBS cells than in the control cell lines, which again fits our model of an activated ‘wound healing state’ of EBS-DM keratinocytes.
Also connected to wound healing, cell migration and adhesion is the YWHAZ, also known as 14-3-3zeta. YWHAZ has been reported to interact with the ADAM (a disintegrin and metalloproteinase) family protein, ADAM 22 (52), which is primarily expressed in brain (53). Members of this family are membrane-anchored glycoproteins, and based on its molecular structure, ADAM 22 is thought to play a role in cell–cell adhesion, functioning as an integrin ligand. Interaction of YWHAZ with the cytoplasmic tail of ADAM 22 was found to be crucial for the promotion of cell adhesion and spreading (52). Kligys et al. (54,55) have investigated the role of YWHAZ in migrating keratinocytes. They found that a heterodimer of YWHAZ and 14-3-3tau regulated slingshot 1 (SSH1) phosphatase, thereby influencing migration of the cells (54).
Relationship between basal cell carcinoma and EBS-DM could be linked by K17
In the previous studies, up-regulation of K17 has been implicated in BCC (56,57). A striking characteristic exclusively of EBS, particularly EBS-DM, is the elevated risk of developing BCC with increasing age and long-term exposure to sunlight. Within the white American population, the estimated lifetime risk is 28–33% (58) contrasted with a remarkable risk of 44% in patients suffering from EBS-DM (59). The role of K17 in BCC was further indicated by the observation that genetic ablation of K17 delayed growth and tumor initiation of basaloid follicular hamartoma, a basaloid skin tumor similar to BCC, in the epidermis of mice with constitutive Hedgehog signaling (56).
Increased levels of K17 have been shown to compensate for the ablation of K14 in recessive EBS (60), possibly by reducing the dominant negative effect of mutated K14 by copolymerizing with K5 in vivo (56). In aggregate, these findings suggest that a negative side effect of the increased levels of K17 in EBS-DM found in our study could be a factor responsible for the more frequent occurrence of BCC in EBS-DM.
Activation of the ubiquitin-proteasome pathway (UPP) in EBS-DM
A K14 mutation in keratinocytes on amino acid 125 causes severe misfolding of the protein and leads to the formation of keratin aggregates and ring-like structures in the cell periphery (61,62). These aggregates are dynamic structures requiring permanent de novo protein synthesis and turnover (44). This mutant keratin turnover is regulated by the UPP (63). The UPP system requires the successive action of an E1 ubiquitin-activating enzyme, an E2-conjugating enzyme and an E3 ubiquitin ligase, potentially providing for substrate specificity (64,65). A recent study of Löffek et al. more closely explores this UPP/keratin turnover relationship, finding the stress-induced chaperones HSP70 and HSP90 to be up-regulated in neonatal K5−/− mice and, upon proteasome inhibition, to be increased in a cell culture model of EBS-DM (MCF-7 cells expressing K14 R125C). Furthermore, overexpression of the chaperone-associated E3 ligase CHIP (carboxyl terminus of Hsc/p70 interacting protein) in this cell model reduced keratin aggregates. In addition, CHIP is credited with a central role in keratin quality control as a facilitator of mutant keratin turnover (61). Interestingly, by SSH, we were also able to identify a HSP90 variant, HSP90AB1, to be up-regulated in KEB7 cells. Moreover, the ubiquitin-conjugating enzyme E2K (UBE2K) was reliably identified to be more abundant in KEB7 than in NEB1 confirming an involvement of the UPP in EBS-DM. The exact role of UBE2K in EBS remains uncertain. UBE2K is known to interact with huntingtin, the gene product for Huntington’s disease (66), and to regulate the expression and activation of caspase-12 in ER stress-mediated amyloid-β peptide (Aβ) neurotoxicity, assumed to contribute to the pathogenesis of Alzheimer’s disease (67), both are protein conformational disorders where protein aggregation is observed (68,69).
Increased expression of cell junction proteins in KEB7
Our observation that expression of the cell junction proteins PERP, DSG3 and DSP was increased in KEB7 relative to NEB1 is contrary to the findings of Liovic et al., who found DSG3 and DSP to be down-regulated in KEB7. Furthermore, studies on isogenic pathomimetic cell lines expressing either WT K14 or mutant K14 (R125P) showed reduced DSG3 expression by RTq-PCR and decreased levels of DSG3 and DSP by western blot analysis in the isogenic mutant cell line. Down-regulation of DSG3 and DSP was also seen in EBS-DM KEB1 cells carrying a heterozygous dominant mutation in K5 (E475G) and in parallel isogenic cell lines (22).
Desmoglein 3 and DSP are both components of desmosomes, which provide a platform for intercellular junctions and are most abundant in tissues with a need to withstand mechanical stresses, such as the epidermis (70,71). Proteins comprising desmosomal structure are derived from three major gene families: (i) the cadherins, represented by desmoglein (DSG) and desmocollin (DSC), transmembrane proteins forming the adhesive interface between cells (70–75), (ii) the armadillo family members plakoglobin, plakophilins and p0071, which interact with the cytoplasmic tails of the cadherins (70,71,76,77) and (iii) the plakin protein family, with its member desmoplakin (DSP) linking the desmosomal plaque to the IF cytoskeleton (70,71,78).
Additionally, the PERP, also a target of the master regulator for stratified epithelial development p63, is suggested to play a key role in the promotion of the robust assembly of desmosomal adhesive units and was shown to colocalize to desmosomal structures. Also noteworthy is the fact that PERP−/− mice show dramatic blistering in stratified epithelia and die shortly after birth (79).
We interpret the up-regulation of DSG3, DSP and PERP observed in our study as an attempt by the KEB7 cells to overcome the negative effects of disturbed K5/K14 IF cytoskeleton by potentially increasing the number and internal integrity of desmosomes. A possible explanation for the discrepancy between our study and the systematic work of Liovic et al. might be an effect of differing composition of agents and growth factors added to the culture medium, different cell confluencies and different elapsed times following trypsinization. The two last factors have been shown by D’Alessandro et al. (40) to influence the sensitivity of NEB1 and KEB7 cells to hypo-osmotic shock. The extent to which this is also true for the different gene expression patterns reported here is uncertain and remains to be resolved by further investigation.
Perturbations of genes involved in epithelial differentiation
The thioredoxin interacting protein (TXNIP), also known as vitamin D-up-regulated protein 1 (VDUP1), and ANXA8 were also found to be increased in KEB7 cells. TXNIP interacts with sciellin, a precursor of the cornified envelope, and is assumed to have a role in regulating the transition of transient amplifying cells to suprabasal differentiating cells in epithelial tissues (80). As ANXA8 also has been found to be most abundant in the stratified epithelia of the epidermis, the forestomach, the oesophagus, the tongue and the cornea, it has been suggested that it plays a role in epithelial differentiation (81). ANXA8 was proposed to have an anticoagulatory activity in lung endothelial cells (82,83) and to participate in the interaction between the plasma membrane and the cytoskeleton owing to its high specificity binding to phosphatidylinositol (4,5)-bisphosphate and interaction with F-actin in HeLa cells (84).
Finally, DNA-damage-inducible transcript 4 (DDIT4) and metastasis-associated lung adenocarcinoma transcript 1 (non-protein coding) (MALAT1) showed heightened expression in KEB7 cells. The connection of DDIT4 to keratinocytes is substantiated by the findings of Ellisen et al. (85), which revealed a role of DDIT4 in keratinocyte differentiation. DDIT4 and p63, which have an essential role in the formation of epithelial structures including the skin (86), are coordinately down-regulated in differentiating primary keratinocytes and are suggested to contribute to the maintenance of an undifferentiated state. The mechanism is thought to involve reactive oxygen species (ROS) because of their modulation by DDIT4 (85) and the fact that delicate shifts in intracellular ROS levels impact tyrosine kinase growth factor receptors (87–89).
The only gene identified in our study that cannot yet be implicated in EBS is MALAT1. MALAT1 is a non-coding RNA that has been shown to be associated with metastasis in lung adenocarcinoma (90).
The extent to which these proteins account for the phenotypes observed in EBS-DM keratinocytes remains subject to future clarifications. Analysis of current literature indicates that regulation of the majority of the genes identified by SSH and corroborated by RTq-PCR is consistent with a chronic wound healing state in the EBS keratinocyte cell line and is in line with the pathogenesis of EBS. However, this study was primarily intended to amend the list of differentially regulated genes in EBS initiated by Lu et al. (23), Liovic et al. (21) and Liovic et al. (22) (Table S1) and offers new, thought-provoking insights for functional studies and novel routes for therapeutic approaches.
Acknowledgements
We gratefully thank Prof. Birgit Lane for kindly supplying the wild-type NEB1 and the patient KEB7 keratinocyte cell lines. This work was supported by the Dystrophic Epidermolysis Bullosa Research Association (DEBRA), Austria.
Author contributions
KO designed the original concept of the study supervised the study and contributed to writing the manuscript; MW carried out all experiments, evaluated the data and wrote the manuscript; and HH and JWB gave scientific support and critically revised the manuscript. All authors read and approved the manuscript.
Conflict of interests
The authors have declared no conflicting interest.




