Extraction of high-quality epidermal RNA after ammonium thiocyanate-induced dermo-epidermal separation of 4 mm human skin biopsies
Abstract
Abstract: To obtain a separation of the epidermal and dermal compartments to examine compartment specific biological mechanisms in the skin, we incubated 4 mm human skin punch biopsies in ammonium thiocyanate. We wanted to test (i) the histological quality of the dermo-epidermal separation obtained by different incubation times; (ii) the amount and quality of extractable epidermal RNA and (iii) its impact on sample RNA expression profiles assessed by large-scale gene expression microarray analysis in both normal and inflamed skin. At 30-min incubation, the split between dermis and epidermis was not always histologically well-defined (i.e. occurred partly intra-epidermally), but also varied between subjects. Consequently, curettage along the dermal surface of the biopsy was added to the procedure. This modified method resulted in an almost perfect separation of the epidermal and dermal compartments, and satisfactory amounts of high-quality RNA were obtained. Hybridization to Affymetrix HG_U133A 2.0 GeneChips showed that ammonium thiocyanate incubation had a minute effect on gene expression resulting in only one significantly downregulated gene (cystatin E/M). We conclude that epidermis can be reproducibly and almost completely separated from the dermis of 4 mm skin biopsies by 30 min incubation in 3.8% ammonium thiocyanate combined with curettage of the dermal surface, producing high-quality RNA suitable for transcriptional analysis. Our refined method of dermo-epidermal separation will undoubtedly prove valuable in the many different settings, where the epidermal and dermal compartments need to be evaluated separately.
Introduction
Studying cutaneous biology at a transcriptional level under physiological or pathological conditions requires reproducible isolation of high-quality RNA from skin samples. As the epidermis constitutes an entirely different cellular compartment than the underlying dermis, various separation techniques have been used to obtain layer-specific skin sampling prior to RNA extraction.
Recently, Trost et al. (1) tested different dermo-epidermal separation methods on large (200–300 mg) human foreskin samples and concluded that incubation for 20 min in 3.8% ammonium thiocyanate was sufficient to produce a split localized within the lamina lucida of the basement membrane zone (BMZ) and that the isolated epidermal RNA would be of superior quality compared with a range of other separation methods. While a 200 mg sample may be difficult to obtain in a clinical setting, skin punch biopsies are performed on a routine basis. The disadvantage to such biopsies is the limited amount of RNA that can be isolated (2).
We wanted to test and optimize the ammonium thiocyanate method for layer-specific isolation of RNA from 4 mm human skin punch biopsies. As any delay in RNA-preserving procedures may have a detrimental effect on RNA integrity as well as a confounding effect on the analysis of gene expression (3,4), we also wanted to examine the impact of 30 min ammonium thiocyanate incubation on skin gene expression by the use of microarrays.
Methods
The study was performed in accordance with the Declaration of Helsinki and approved by the local ethics committee (Videnskabsetisk komité for Region Syddanmark; S-20070012). Written informed consent was given by the participants.
Optimization of dermo-epidermal separation using ammonium thiocyanate
Normal skin specimens from breast surgery were received within 45 min after excision. Four millimetre punch biopsies comprising epidermis and dermis were placed directly in 3.8% ammonium thiocyanate (VWR-Bie&Berntsen, Herlev, Denmark) in phosphate-buffered saline (PBS), pH 7.4. Incubation time in ammonium thiocyanate varied from 20 min to 24 h at room temperature (RT). The splitting was performed with a watch-maker’s forceps with or without subsequent curettage of the denuded dermal surface of the biopsy. Curettage was performed by scraping with the sharpened edge of a 16 gauge syringe until the exposed dermal surface appeared white and shiny with no epidermal remnants left. The curetted cells were pooled with the epidermal disc. Epidermal and dermal fractions were formalin fixed, paraffin embedded, stained routinely with haematoxylin and eosin, and in addition, to define the dermo-epidermal split, immunohistochemically with antibodies to cytokeratin 13 and laminin (Sigma-Aldrich, Brondby, Denmark), cytokeratin 10 (Dako, Glostrup, Denmark) and collagen IV (Chemicon-Millipore, Copenhagen, Denmark).
Determination of RNA degradation after different ammonium thiocyanate incubation times
Four millimetre punches direct from surgically excised skin were either directly frozen in fluid nitrogen (control) or were incubated in ammonium thiocyanate at RT or 4°C for ½, 1 or 2 h and separated as described above. Epidermal discs were transferred to RNAlater (Applied Biosystems/Ambion, Nærum, Denmark). RNA from epidermis and full skin was extracted following the Qiagen RNeasy Mini Kit (Qiagen, Copenhagen, Denmark) protocol for normal and fibrous tissue respectively.
Skin biopsies from healthy volunteers
Four millimetre punch biopsies were taken from the volar forearm (n = 36) or upper buttocks (n = 19) skin of healthy human volunteers and were immediately padded dry on a sterile cloth to remove significant amounts of blood from the sample. Further processing of the biopsies varied according to the different designs.
RNA quantification and quality assessment of epidermal and dermal samples
Forearm biopsies from 36 volunteers were placed directly in 3.8% ammonium thiocyanate in PBS, pH 7.4. After 30 min incubation at RT, the biopsy was split and the dermal surface curetted as described above. The epidermal and dermal fractions were stored separately in RNAlater in sterile 2.0 ml tubes (Eppendorf). Samples were stored at 4°C for 48 h and then at −20°C until processing.
Samples were thawed for 30 min and carefully transferred to new 2.0 mL microtubes containing one stainless steel bead (Qiagen, Copenhagen, Denmark) and 350 ml of buffer RLT (supplied with the RNeasy Micro Kit) according to the manufacturer’s instructions.
Next, the new microtubes were placed in a TissueLyser® (Qiagen) bead beater to homogenize the tissue for 2 × 2 min (epidermis) or 2 × 3 min (dermis) at 30 Hz. RNA was extracted using the RNeasy Micro Kit (Qiagen) following the manufacturer’s instructions. For dermal samples, a protein digestion step (proteinase K; Qiagen) was added as recommended in the fibrous tissue protocol. RNA quantity was measured on a NanoDrop 1000 (Thermo Scientific, Copenhagen, Denmark) spectrophotometer, while RNA integrity was assessed on a Agilent Bioanalyzer (Agilent Technologies, Nærum, Denmark).
Determination of ammonium thiocyanate influence on gene expression in normal and irritated skin
Large Finn® chambers (Ø = 16 mm) containing filter paper discs saturated with 25 μl sodium lauryl sulphate (SLS; Sigma-Aldrich) 2% (aq.) or SLS 5% (aq.) were placed on the upper buttock skin in four healthy volunteers. After 24-h exposure, the patches were removed and visual scoring was performed. Equally irritated sites with a 1+ score were selected for biopsy.
For each volunteer, four biopsies were obtained: two from inflamed skin and two from adjacent normal skin. One irritated and one normal skin sample was placed directly in RNAlater. The remaining two samples were incubated in ammonium thiocyanate for 30 min at RT and then placed in RNAlater without performing any separation of the dermal and epidermal layers. Additionally, a biopsy was obtained from another irritated site in three of the four subjects and separated as described above (30 min ammonium thiocyanate incubation plus curettage). Dermal and epidermal fractions were formalin fixed separately to histologically examine the actual plane of separation in freshly harvested specimens as well as the possible influence of the inflammation on the separation. All samples for RNA extraction were homogenized in the TissueLyser as described above, and the RNeasy Fibrous Tissue Mini Kit (Qiagen) was used to extract RNA.
Hybridization of extracted RNA to microarrays
For the microarray experiment, RNA from 16 buttocks full thickness skin samples and 13 epidermal samples were amplified with the MessageAmp II Enhanced kit (Applied biosystems/Ambion), following the manufacturer’s instructions for single-round amplification using 500 ng total RNA. cRNA from full skin samples was hybridized to Affymetrix® GeneChip (Affymetrix, Santa Clara, CA, USA) HG_U133A 2.0 and cRNA from epidermal samples was hybridized to HG_U133 plus 2.0 following the Affymetrix manual.
Statistical analysis
Raw data have been deposited at NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE15101 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE15101).
Gene expression values were generated from CEL-files using perfect match probes in dChip software (http://biosun1.harvard.edu/complab/dchip/). Data from full skin samples performed with HG_U133A 2.0 chips were normalized using quantile method in dChip. For comparison of full skin and epidermis, data from identical probe sets from HG_U133A 2.0 and HG_U133 plus 2.0 were extracted and normalized as one data set using quantile method. Hierarchical clustering of full skin samples as well as the combined data set was performed in r environment (http://cran.r-project.org/) using all genes in data sets. The ANOVA model was introduced to detect the day as well as the labelling effects on gene expression variation for each gene. The false discovery rate (FDR) was calculated to adjust for multiple testing. These calculations were performed in r.
Results
Completeness of the dermo-epidermal split increases with incubation time and varies between subjects
Optimization series showed that the epidermis is more prone to separate from the dermis at the BMZ when the time in the ammonium thiocyanate solution is prolonged. An incubation time for 2 h or longer induces a well-defined split between the epidermal and dermal compartments, leaving only a few basal cells with the dermal compartment focally (Fig. 1a). Shorter incubation periods may induce less predictable and ill-defined splits. Incubation times of 20–40 min resulted in either an incomplete (partially intra-epidermal, Fig. 1b) or an almost complete separation of the two compartments. Typically, the remaining epidermal cells were situated at the periphery of the biopsy (Fig. 1c).
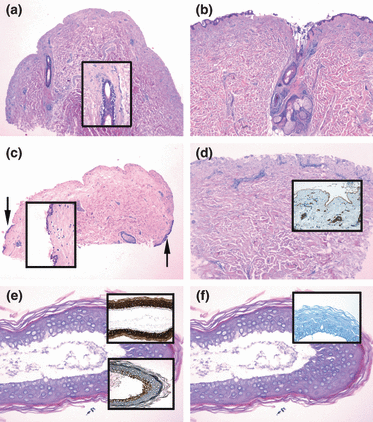
Separation of 4 mm biopsies resulting from different ammonium thiocyanate incubation times with or without curettage. Punch biopsies incubated in ammonium thiocyanate for 120 (a) or 30 (b,c) min showing varying completeness of dermo-epidermal split. (a) Complete detachment of the epidermis with a well demarcated dermal split without epidermal remnants. The follicular epithelium is partly falling apart (inset); (b) intra-epidermal and focally subepidermal split with the epidermis only partially detached; (c) almost complete epidermal-dermal separation, but focally epidermal cells are left with the dermal compartment (arrows and inset); (d) 30 min ammonium thiocyanate incubation and subsequent curettage showing a well-preserved contour of the dermal papillae without epidermal remnants. The basement membrane zone appears intact as demonstrated by an unbroken band of laminin (inset); (e) epidermal sheet separated from a punch biopsy incubated in ammonium thiocyanate for 120 min. A preserved basal cell layer is demonstrated by the negative staining with cytokeratin 10 (CK10, upper inset) and the positive staining with cytokeratin 13 (CK13, lower inset); (f) epidermal sheet treated like (e): the negative staining with collagen IV (inset) confirms that no dermal tissue is left with the epidermal compartment.
No differences in separation were observed comparing 20, 30 and 40 min of incubation. Neither did 24 h incubation result in a more complete separation compared with 2 h incubation (not shown). Preincubation of punches in RNAlater for 24 h was tested; however, the tissue changed into a rubber-like substance making subsequent separation by ammonium thiocyanate technically impossible.
Epidermal cells left with the dermal compartment can be removed by adding curettage to the procedure
Unless appropriate measures are taken, RNA degradation progresses with time which was confirmed for breast skin samples incubated in ammonium thiocyanate for up to 2 h with or without cooling of ammonium thiocyanate to 4°C (not shown). To optimize the compartment separation while maintaining a short incubation time, a curettage of the remaining epidermal cells was performed after ammonium thiocyanate incubation. Microscopic control confirmed that the dermal compartment was intact but with a few basal cells left. Along the line of separation, the architecture of the dermal compartment was well preserved without signs of injury and with an intact contour of the dermal papillae. Immunohistochemical staining for collagen IV (not shown) and laminin (Fig. 1d) confirmed that the lamina densa was intact. There was no difference between normal and inflamed skin (not shown). Immunohistochemical staining showed that the epidermal sheet was intact including the basal cells visualized by positive staining with cytokeratin 13 and negative staining with cytokeratin 10 (Fig. 1e). Accordingly, a negative collagen IV staining of the basal cells confirmed that no dermal components were left with the epidermal sheet (Fig. 1f).
After 30-min ammonium thiocyanate incubation, RNA of high quality but variable quantity can be extracted
Amount, purity and integrity of RNA extracted from epidermal and dermal fractions obtained from healthy volunteer skin biopsies are summarized in Fig. 2. The variance in RNA yield is considerable ranging from 250 to 3200 ng (epidermis) and from 260 to 1600 ng (dermis). Usable RNA was extracted from all samples and RNA quality was comparable for the two compartments.
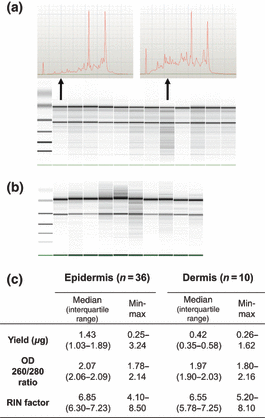
Quantity, purity and integrity of RNA obtained from human epidermal and dermal compartments from 4 mm biopsies. (a) Agilent 2100 Bioanalyzer pseudogel output showing 12 representative RNA-profiles extracted from normal human epidermal samples. Two select samples are shown as electropherograms as examples of very well-preserved (upper left) and less well-preserved (upper right) RNA. (b) Bioanalyzer gel result for RNA extracted from 10 dermal samples. (c) Yield and OD values obtained using the NanoDrop spectrophotometer and RIN values obtained using the Agilent Bioanalyzer 2100. Samples are considered of high purity with OD 260/280 ratios ≥2.0 and high integrity with RIN values ≥6.0. OD, optical density; RIN, RNA integrity number.
30-min incubation in ammonium thiocyanate has minute effect on the overall RNA expression pattern
Postsampling biological processes may continue for an unspecified period of time resulting in altered gene expression as previously reported for surgical specimens (3,4). Hypothetically, 30-min incubation in ammonium thiocyanate may also affect gene expression. Gene clustering of eight pairs of skin biopsies, exposed and non-exposed to ammonium thiocyanate, is shown in Fig. 3a. The effect of SLS is the most powerful factor on the gene expression as defined by the two main clusters. The second most powerful factor is the variation between individuals. No clustering in favour of an ammonium thiocyanate-induced influence on the gene expression is seen because a and c samples then should have clustered together. This picture was seen irrespective of samples being from normal or inflamed skin.
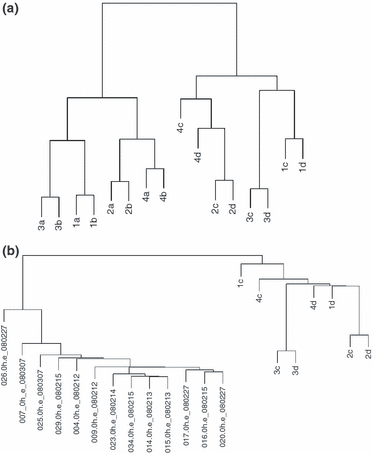
Hierarchical clustering of full skin and split skin samples. (a) Cluster analysis performed using all 22 277 probes on the Affymetrix HG_U133A 2.0 GeneChip. RNA was isolated from 16 full thickness skin biopsies from four individuals (1–4). Samples with suffix ‘a’ and ‘b’ were in vivo SLS exposed; ‘c’ and ‘d’ were non-exposed control skin; ‘a’ and ‘c’ were incubated for 30 min. in ammonium thiocyanate ex vivo and subsequently placed in RNAlater; whereas ‘b’ and ‘d’ were placed directly in RNAlater. (b) Clustering of 13 normal epidermal (left main cluster) and 8 normal full thickness biopsies (right main cluster) using all 22 277 probes represented on the Affymetrix HG_U133A 2.0 GeneChip. Epidermal samples were hybridized to the Affymetrix HG_U133 plus 2.0 platform, whereas full skin samples were hybridized to the Affymetrix HG_U133A 2.0 platform. Suffixes ‘c’ and ‘d’ are the same as described above.
Clustering samples using all genes is a crude analysis giving no information about the genes that are influenced by the treatment. Consequently, an ANOVA was performed to test for up- or downregulated genes as a result of (i) in vivo skin exposure to SLS and (ii) ex vivo skin biopsy incubation in ammonium thiocyanate. A total of 3997 probes were significantly (FDR < 0.05) regulated (1792 up and 2205 down) in response to SLS irritation. No probes were upregulated, and one probe (cystatin E/M; gene symbol CST6) was significantly downregulated caused by the effect of ammonium thiocyanate incubation (FDR = 0.003). The second most regulated (down) probe was SPINK5. However, following a correction for multiple testing, this proved non-significant (FDR = 0.301).
Epidermal sheets are transcriptionally different from whole skin biopsies and show less a priori variation between samples
A cluster analysis was performed on normal skin from different individuals comparing full thickness biopsies (n = 8) with epidermal sheets (n = 13). Two main clusters corresponding to the two sample types are formed (Fig. 3b). Thus, as expected, the cluster dendrogram confirms that the two types of samples are transcriptionally distinct. Notably, the interindividual variance between epidermal sheets appears to be smaller than between full thickness biopsies as demonstrated by the generally smaller vertical distance between epidermal samples.
Discussion
We have developed a method to completely separate the epidermal and dermal compartments and to reliably obtain fair amounts of high-quality RNA from epidermal sheets of 4 mm skin punch biopsies. This method implies incubation in ammonium thiocyanate at RT and subsequent curettage of the exposed dermal surface of the biopsy. We have demonstrated that the method can be applied to biopsies collected from both normal and inflamed skin, although only eczematous skin and not other dermatoses with a more pronounced acanthotic pattern (e.g. psoriasis) was tested.
First described by Juhlin and Shelley (5), ammonium thiocyanate has been used for murine epidermal sheet preparation (6–8) and has also been applied to small-sized (down to 20 mm2) human biopsies (9). However, in the latter study incubation was at 37°C and sheets were prepared for immunostaining rather than for RNA extraction.
Using large human foreskin samples, Trost et al. (1) convincingly argued for the use of ammonium thiocyanate compared with other separation methods. While they found the split to occur within the lamina lucida of the BMZ visualized by immunofluorescence, our experience with the method from microscopically controlled punch biopsies has been somewhat different: short incubation times (20–30 min) in ammonium thiocyanate may be enough to reliably separate the epidermis from the dermis within the lamina lucida in some subjects, whereas longer incubation times are needed to obtain perfect splits for all subjects (Fig. 1). However, when RNA analysis is requested, incubation times of 2 h or longer are unacceptable because of the ongoing RNA degradation. This was especially true because no RNA-protective effect of ammonium thiocyanate could be demonstrated (data not shown). A strategy of short incubation combined with curettage secured maximal amounts of epidermal cells in the epidermal fraction which was crucial not only for the amount of cells contributing to the overall RNA yield, but also for inclusion of basal keratinocytes. In fact, the selective pathogenic significance of the basal cells in certain inflammatory skin conditions has previously been demonstrated (10).
Epidermal sheets obtained by the present method is not a pure sample of keratinocytes but rather a mixture of keratinocytes, Langerhans cells, melanocytes and, in inflamed skin samples, additional inflammatory cells. Obtaining keratinocyte-specific RNA, including basal- and spinous-cell specificity, from biopsy specimens may require a more sophisticated technique like laser microdissection.
Importantly, our results indicate that the a priori interindividual variation is smaller for the RNA material obtained from epidermal sheets compared with full thickness biopsies (Fig. 3b). This is probably caused by the smaller repertoire of cells present in the epidermal fraction and advocates further for the use of dermo-epidermal separation of skin biopsies before extracting RNA for large-scale transcriptional analysis. While we cannot entirely rule out that the formation of two main clusters (dermal and epidermal samples) results from the comparison of two different Affymetrix platforms, the smaller variation between individuals could not be explained by this technical deviation.
It can be concluded from the microarray experiment that some postsampling gene regulation takes place. This is consistent with the previous findings (3,4). The significantly downregulated gene CST6 encodes cystatin E/M, a cysteine protease inhibitor that, through its inhibitory effect on cathepsins V and L, plays a regulatory role in epidermal differentiation, cornification and desquamation processes (11). Interestingly, of all 22 277 probes, the second most downregulated probe was SPINK5 which encodes the serine protease inhibitor LEKTI that is well-known for its causative role in Netherton syndrome and has inhibitory effect on skin kallekreins (12). Thus, two distinct pathways in epidermal proteolysis regulation show loss of function in response to ex vivo ammonium thiocyanate incubation. Speculatively, the downregulation of these genes may result from the action of ammonium thiocyanate on desmosome cohesive proteins.
In conclusion, short-time ammonium thiocyanate incubation combined with curettage along the dermal surface of the biopsy can be safely applied to small-sized punch biopsies to reproducibly obtain epidermis-specific RNA of high quality for transcriptional analysis. The curettage secures high specificity/purity of the epidermal and dermal fractions of all collected samples, but despite the addition of this optimization step few epidermal cells may still be left with the dermal compartment.
Acknowledgements
The authors thank Charlotte Skoubo, Department of Biochemistry, Pharmacology and Genetics, Odense University Hospital, Jette Krapalis and Kirsten Hammond Andersen, both Department of Dermatology, Odense University Hospital for technical assistance and Katrine Jacobsen, Medical Biotechnology Center, University of Southern Denmark for lending the NanoDrop Spectrophotometer.




