Molecular typing of Bartonella henselae DNA extracted from human clinical specimens and cat isolates in Japan
Editor: Ewa Sadowy
Abstract
Bartonella henselae is the causative agent of cat scratch disease (CSD). To clarify the population structure and relationship between human and cat strains of B. henselae, 55 specimens isolated in Japan, including 24 B. henselae DNA-positive clinical samples from CSD patients and 31 B. henselae isolates from domestic cats, were characterized by multilocus sequence typing (MLST) and the 16S–23S tRNA-Ala/tRNA-Ile intergenic spacer (S1) sequence, which were used previously for strain typing of B. henselae. Three different sequence types (STs) were identified by MLST, one of which was novel. Fifty-two strains (94.5%), including all strains detected in CSD patients, were assigned to ST-1. Eight S1 genotypes were observed, three of which were novel. The 52 ST-1 strains were classified into seven S1 genotypes, two of which were predominant in both human and cat strains. In addition, 5.5% of the strains (3/55) contained two different intergenic spacer S1 copies. These results indicate that the predominant B. henselae MLST ST-1 in Japan is a significantly genetically diverse population on the basis of the sequence diversity of intergenic spacer S1, and that highly prevalent S1 genotypes among cats are often involved in human infections.
Introduction
Bartonella henselae is the causative agent of cat scratch disease (CSD). Cats represent the major reservoir for B. henselae. Infected cats are usually asymptomatic and develop relapsing bacteremia for long periods (Kordick et al., 1995). Human infection usually occurs through scratches or bites by infected cats and presents as CSD, typically with localized lymphadenopathy. Occasionally, the infection may have an atypical presentation due to blood-borne spread, such as bacteremia, endocarditis, encephalopathy, neuroretinitis, or systemic CSD with hepatic and splenic granuloma (Anderson & Neuman, 1997; Murakami et al., 2002; Tsuneoka & Tsukahara, 2006). Disease symptoms depend on the immune status of the host; in immunocompromised hosts, the bacteria are often present in blood and involved in angioproliferative disorders such as bacillary angiomatosis and peliosis hepatis (Welch et al., 1992).
Isolation of B. henselae from patients is extremely difficult (La Scola & Raoult, 1999). The diagnosis of CSD relies on clinical manifestations, history of contact with cats, serology, or the detection of bacterial DNA in tissue specimens by PCR (Regnery et al., 1992; Anderson et al., 1994; Murakami et al., 2002; Woestyn et al., 2004; Tsuneoka & Tsukahara, 2006). Bartonella henselae strains are divided into two 16S rRNA (rrs) genotypes (16S type I/Houston-1 and 16S type II/Marseille), which correspond to two distinct human serotypes (Drancourt et al., 1996; La Scola et al., 2002). Although both genotypes are present worldwide, 16S type II appears to be dominant in the European cat population, whereas 16S type I is more common in Asia, including Japan (Maruyama et al., 2000; Boulouis et al., 2005).
Multilocus sequence typing (MLST) is a nucleotide sequencing-based genotyping method in which variations in approximately 450–500-bp internal fragments of housekeeping genes (generally seven) are indexed (Maiden, 2000). MLST analysis of 182 feline and human B. henselae isolates from Europe, North America, and Australia revealed that sequence type (ST)-1 was most significantly associated with human infection, but that the geographical distribution of STs was not homogenous (Arvand et al., 2007). However, the use of highly conserved housekeeping genes in MLST often fails to detect variability in closely related strains. Compared with housekeeping genes, intergenic spacers are highly variable, thus generating a clearer population structure (Li et al., 2009). In the multispacer typing (MST) scheme for B. henselae, the 16S–23S tRNA-Ala/tRNA-Ile intergenic spacer (S1) is the most variable spacer, containing a 15-bp variable number tandem repeat (VNTR) (Li et al., 2006). PCR-based genotyping methods can be applied directly to clinical specimens (Rodrick et al., 2004; Li et al., 2007). However, no data are available regarding the predominant strains causing CSD in Japan.
In this study, we examined 55 human and feline B. henselae specimens by MLST and S1 sequence to uncover the genotypic distribution and relationship between human and cat strains of B. henselae in Japan. Furthermore, we analyzed the structural diversity of ST-1 using the intergenic spacer S1 sequence to generate a clear population structure.
Materials and methods
Clinical specimens
Twenty-four human clinical specimens consisted of five lymph node specimens and 16 pus specimens from patients with typical CSD, one blood specimen from a patient with bacteremia, one liver specimen from a patient with hepatic granuloma, and one spleen specimen from a patient with splenic granuloma. The specimens were obtained from various regions of western Japan, including Yamaguchi prefecture, from 1997 to 2008.
Bacterial strains
The 31 B. henselae isolates were derived from 290 blood samples collected from domestic cats in western Japan, mainly Yamaguchi prefecture, from 2003 to 2004 (Tsuneoka et al., 2004). Primary isolates of B. henselae from cat blood samples were grown on chocolate agar plates with 5% defibrinated sheep blood at 35 °C in 5% carbon dioxide (CO2) for 2 weeks. The strains were stored at −80 °C until use. Subcultures were performed on chocolate agar plates with 5% defibrinated sheep blood at 35 °C in 5% CO2 for 5 days. A single colony of each isolate was passaged once on agar before the extraction of bacterial DNA.
DNA extraction
Total genomic DNA was extracted using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions.
Identification of B. henselae
Bartonella henselae was detected with PCR targeting 414 bp of the htrA gene and 172 bp of the Bartonella species-specific 16S–23S rRNA internal transcribed spacer region, and confirmed by partial sequencing of the 16S rRNA gene using broad-host-range primer 16SF together with 16SR, as described previously (Anderson et al., 1994; Bergmans et al., 1996; Jensen et al., 2000). No bacterial species other than B. henselae was detected in any sample.
PCR amplification and sequencing
For MLST, eight genes (rrs, batR, gltA, ftsZ, groEL, nlpD, ribC, and rpoB) were amplified and sequenced directly using MLST primers for B. henselae as described previously (Iredell et al., 2003). The intergenic spacer S1 was amplified and sequenced directly using S1 forward primer and S1 reverse primer as described previously (Li et al., 2007). When direct sequencing of spacer S1 was unsuccessful because of an atypical number of VNTRs, locus-specific PCR was performed using S1 forward primer with one of two locus-specific primers: BH12700-R (5′-ACGCCAATGTGTTATCCACTT-3′) or BH13810-R (5′-GAAACTTGTCGATGATCAGGC-3′). The PCR mixture contained 1 × Phusion HF Buffer (Finnzymes, Espoo, Finland), 0.4 U Phusion DNA polymerase (Finnzymes), 200 μM dNTP, 500 nM of each primer, 4–100 ng DNA template, and sterile-distilled water, in a final volume of 20 μL. The reaction conditions were as follows: denaturation at 98 °C for 30 s; 35–50 cycles at 98 °C for 10 s, 56–62 °C for 30 s, and 72 °C for 30–120 s; and a final extension step at 72 °C for 10 min. PCR products were purified using the High Pure PCR Product Purification Kit (Roche Diagnostics GmbH, Mannheim, Germany) and then sequenced directly using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) on both strands with a 3130 Genetic Analyzer (Applied Biosystems).
Sequencing analysis and phylogenetic analysis
The nucleotide sequences were analyzed with DNA sequencing analysis software version 5.1 (Applied Biosystems). Alleles, STs, and S1 genotypes were assigned in accordance with published data (Iredell et al., 2003; Li et al., 2006, 2007; Arvand et al., 2007). The novel allele and S1 sequence were carefully confirmed on multiple occasions, and the sequences were deposited in the DNA Data Bank of Japan (DDBJ). New S1 genotypes were deposited in the MST-Rick database (http://ifr48.timone.univ-mrs.fr/MST_BHenselae/mst). A neighbor-joining tree was reconstructed from the concatenated MLST allele sequences of 14 previously reported STs (Iredell et al., 2003; Arvand et al., 2007) and the novel ST-15 using Kimura 2-parameter distance measures as implemented in mega4 (Tamura et al., 2007).
Nucleotide sequence accession numbers
Newly encountered sequences have been submitted to DDBJ under the following accession numbers: AB525232, rpoB allele 5 (YC-073); AB525233, S1 genotype 11 (YC-053); and AB534165, S1 genotype 12 (YC-012 and YC-013).
Results and discussion
MLST analysis detected three STs in 55 human and cat B. henselae strains. Intriguingly, 94.5% (52/55) of B. henselae strains including all 24 human clinical specimens were assigned to ST-1, and only three cat isolates were assigned to other STs (Table 1). In isolate YC-073, a new allele was found in rpoB, which consisted of a single nucleotide variation (G instead of A) at position 711 784 of the B. henselae Houston-1 chromosome (accession no. BX897699). The ST containing this allele was designated as ST-15. Phylogenetic tree analysis revealed that ST-15 belongs to Group 1 and is closely related to ST-1 (Fig. 1). In the MLST analysis, ST-1 was common among cat isolates, and human clinical specimens were assigned to ST-1 at a much higher frequency (100%) than that observed in previous studies (Iredell et al., 2003; Lindroos et al., 2006; Arvand et al., 2007). However, the clonal population of ST-1 in Japan is quite different from that reported in other regions.
| ST | Allele number | No. ofisolates (n) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| rrs | batR | gltA | ftsZ | groEL | nlpD | ribC | rpoB | ||
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 28 |
| 6 | 2 | 3 | 2 | 2 | 2 | 1 | 1 | 2 | 2* |
| 15 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 5 | 1† |
- * Bartonella henselae isolates YC-012 and YC-013.
- † Bartonella henselae isolate YC-073.
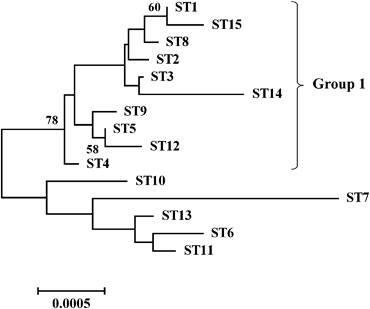
Neighbor-joining tree of concatenated MLST allele sequences of 15 Bartonella henselae STs. Concatenated MLST allele sequences representing each ST were obtained for 14 previously reported STs and the new ST-15, and a neighbor-joining tree was generated by performing bootstrap analyses (1000 replicates) using mega4. Only node values >50% are indicated in the figure.
To generate a clearer population structure, we analyzed the 55 strains using the highly variable intergenic spacer S1. We identified eight different S1 genotypes, three of which were novel (Table 2). S1 genotype 11 containing a sixfold repetition of the 15-bp sequence was found in YC-053. S1 genotype 12 with a single nucleotide deletion at position 1 412 654 of the B. henselae Houston-1 (accession no. BX89 7699) was found in YC-012 and YC-013. The third novel S1 genotype was found in three cat isolates YC-015, YC-024, and YC-073, which contained atypical numbers of VNTRs in the two spacer S1 regions (Fig. 2). The complete genome sequence of B. henselae Houston-1 (accession no. BX897699) was shown to contain two identical copies of intergenic spacer S1. Locus-specific PCR, followed by direct sequencing revealed that these isolates were assigned to S1 genotype 7+4 (Fig. 2 and Table 3).
| S1 genotype | No. of strains (n) | |
|---|---|---|
| Human | Cat | |
| 3 | 1 | 0 |
| 4 | 12 | 14 |
| 5 | 9 | 10 |
| 7 | 1 | 1 |
| 8 | 1 | 0 |
| 11 | 0 | 1 |
| 12 | 0 | 2 |
| 7+4* | 0 | 3 |
- * S1 genotype 7+4 indicates that the strain had two different copies of intergenic spacer S1 in its genome.

Representative results of direct sequencing of the 16S–23S tRNA-Ala/tRNA-Ile intergenic spacer (S1) in Bartonella henselae strain YC-015. These sequences show the 15-bp VNTR (CAATCTTTTTAGAAG) detected with the S1 forward primer. (a) The sequence, obtained from PCR using the S1 forward and reverse primers, shows the abrupt onset of ambiguous bases suggestive of overlapping sequences. (b) The sequence corresponding to the 1 412 349–1 412 683 position of BX897699, obtained from PCR using the S1 forward and BH12700-R primers, shows a double repetition of the 15-bp VNTR and belongs to S1 genotype 7. (c) The sequence corresponding to the 1 582 358–1 582 692 position of BX897699, obtained from PCR using the S1 forward and BH13810-R primers, shows a triple repetition of the 15-bp VNTR and belongs to S1 genotype 4. The same results were obtained from B. henselae strains YC-024 and YC-073.
| Spacer position on the genome* | ||
|---|---|---|
| 1 412 349–1 412 683 | 1 582 358–1 582 692 | |
| S1 genotype | 7 | 4 |
| No. of 15-bp repeat | 2 | 3 |
- * Bartonella henselae Houston-1 (accession no. BX897699).
Compared with MLST, the intergenic spacer S1 was able to generate a clearer population structure of the strains under investigation. The 52 ST-1 strains were classified into seven distinct S1 genotypes. Our results indicate that ST-1 has a high genetic diversity on the basis of the sequence diversity of intergenic spacer S1. MLST could not clearly differentiate between strains because the eight selected housekeeping genes were highly conserved and showed less sequence variability.
According to the intergenic spacer S1 sequence, two major S1 genotypes, 4 and 5, were identified in both human and cat strains (Table 2). In a previous MST study of B. henselae specimens isolated from patients in France, only S1 genotypes 3 (53.3%) and 5 (45.3%) were observed (Li et al., 2007). S1 genotype 3 was not a predominant genotype in our study, and S1 genotype 4 was not detected in France. This discrepancy may be explained by similar regional differences observed in the distribution of S1 genotypes 3 and 4 among cats in France (Li et al., 2006) and Japan, respectively. These data suggest that prevalent S1 genotypes among cats are often involved in human infections. Further studies are necessary to elucidate the association of the S1 genotype with pathogenicity.
In conclusion, we have demonstrated that the predominant B. henselae MLST in Japan, ST-1, is a significantly genetically diverse population on the basis of the sequence diversity of intergenic spacer S1, and that highly prevalent S1 genotypes among cats are often involved in human infections. These results may aid our understanding of the population structure and the relationship between human and cat strains of B. henselae.
Acknowledgements
This work was supported by Grant-in-Aid for Young Scientists (B) No. 21790538 from the Ministry of Education, Culture, Sports, Science and Technology of Japan.




