DEEP DIVERSIFICATION AND LONG-TERM PERSISTENCE IN THE SOUTH AMERICAN ‘DRY DIAGONAL’: INTEGRATING CONTINENT-WIDE PHYLOGEOGRAPHY AND DISTRIBUTION MODELING OF GECKOS
Abstract
The relative influence of Neogene geomorphological events and Quaternary climatic changes as causal mechanisms on Neotropical diversification remains largely speculative, as most divergence timing inferences are based on a single locus and have limited taxonomic or geographic sampling. To investigate these influences, we use a multilocus (two mitochondrial and 11 nuclear genes) range-wide sampling of Phyllopezus pollicaris, a gecko complex widely distributed across the poorly studied South American ‘dry diagonal’ biomes. Our approach couples traditional and model-based phylogeography with geospatial methods, and demonstrates Miocene diversification and limited influence of Pleistocene climatic fluctuations on P. pollicaris. Phylogeographic structure and distribution models highlight that persistence across multiple isolated regions shaped the diversification of this species complex. Approximate Bayesian computation supports hypotheses of allopatric and ecological/sympatric speciation between lineages that largely coincide with genetic clusters associated with Chaco, Cerrado, and Caatinga, standing for complex diversification between the ‘dry diagonal’ biomes. We recover extremely high genetic diversity and suggest that eight well-supported clades may be valid species, with direct implications for taxonomy and conservation assessments. These patterns exemplify how low-vagility species complexes, characterized by strong genetic structure and pre-Pleistocene divergence histories, represent ideal radiations to investigate broad biogeographic histories of associated biomes.
Identification of underlying mechanisms shaping the diversification of intraspecific lineages and species complexes is essential to understand speciation processes and ultimately biogeographic patterns across multiple spatial and temporal scales. Emergent approaches based on the coalescent theory coupled with geospatial methods represent exciting advances in testing for alternative models of population divergence at the species level, and historical biogeography at the landscape level (Knowles and Alvarado-Serrano 2010; Chan et al. 2011). The use of model-based inference in phylogeography (i.e., any method in which a fully specified probabilistic model describes how observed data are generated) is not free of controversy (Templeton 2010). However, as more simulation studies and empirical applications are completed, the debate declines (Bloomquist et al. 2010), and their utility as a reasonable statistical approach has now become consolidated (Beaumont et al. 2010; Bertorelle et al. 2010; Garrick et al. 2010).
Approximate Bayesian computation (ABC) is one of the most emblematic ‘likelihood-free’ model-based methods currently in use (Beaumont 2010). Briefly, ABC simulates millions of genealogies assuming different parameter and prior values under competing models, and those producing patterns of genetic variation most similar to the observed data (determined by summary statistics, SuSt) are then retained and analyzed in detail (Bertorelle et al. 2010; Csilléry et al. 2010). Recent implementations of ABC to test complex evolutionary scenarios have brought statistical rigor and power to phylogeography, enhancing the potential interfaces and outcomes of the field (Hickerson et al. 2011).
In a recent global survey, Beheregaray (2008) identified large phylogeographic knowledge gaps in most South American biomes. In the last decade or so, however, phylogeographic research has accelerated throughout much of the continent, including Amazonia (Geurgas and Rodrigues 2010; Ribas et al. 2012), Atlantic Forest (Carnaval et al. 2009; Thomé et al. 2010; Cabanne et al. 2011), Andean regions (Cadena et al. 2011; Chaves and Smith 2011), and Patagonia (Lessa et al. 2010; Breitman et al. 2011; Nuñez et al. 2011), to cite some examples. Conspicuous gaps in this emerging coverage are the highly threatened open vegetation biomes of central-eastern South America, which extend diagonally across a large latitudinal range and include the Seasonally Dry Tropical Forests (SDTFs, with the largest area, Caatinga, in northeastern Brazil), the Cerrado savanna (central Brazil), and the Chaco (southwestern South America, Fig. 1). Early studies interpreted these regions as a single complex of low-diversity open formations, the ‘diagonal of dry formations’ (Vanzolini 1963, 1976), without regionally distinct biotas, despite their strong character (reviewed by Werneck 2011). However, species diversity and endemism is high for some taxa within these biomes, including fish (Leal et al. 2003), squamate reptiles (Rodrigues 2003; Nogueira et al. 2011), Lepidoptera (Amorim et al. 2009), and plants (Oliveira and Marquis 2002; Simon et al. 2009).
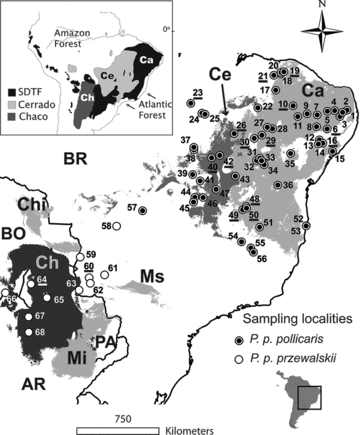
Distribution of sampled localities for Phyllopezus pollicaris relative to the refugial areas estimated through palaeomodeling for Seasonally Dry Tropical Forests (SDTFs; light gray refugia: Caatinga [Ca], Chiquitano [Chi], Missiones [Mi], and Mato Grosso do Sul [Ms][from Werneck et al. 2011]), Cerrado [Ce] (Serra Geral de Goiás refugium in medium gray [Werneck et al. in press]), and Chaco [Ch] refugium in dark gray (F. P. Werneck and G. C. Costa, unpubl. data). The insert map shows the approximate current distributions of the central-eastern South America ‘dry diagonal’ biomes. Numbers correspond to localities names in Table S1, and underlined numbers identify areas under legal preservation. Straight-line distance from locality 1 to 68 is approximately 3440 km. AR, Argentina; BO, Bolivia; BR, Brazil; PA, Paraguay. See text for details.
The combined influences of Paleogene–Neogene geological processes and Quaternary climatic and vegetational fluctuations are hypothesized to have generated and maintained the high diversity levels, and to have driven the differentiation of the open biome communities from adjacent rainforests and from each other (Werneck 2011). Recent studies have yielded mixed results concerning the primary diversification processes in some of these open biome biotas. For example, Miocene (23–5.3 Mya) geological events were interpreted as primary determinants of diversification among lizard genera (Giugliano et al. 2007) and species (Werneck et al. 2009), and within a frog species complex (Maciel et al. 2010), whereas recent adaptive shifts driven by fire dynamics (Simon et al. 2009) and Quaternary climatic changes were identified as foremost processes of some plant (Ramos et al. 2007; Caetano et al. 2008) and fruitfly (Moraes et al. 2009) groups.
The role of relatively recent Quaternary changes versus Neogene geomorphological events in the origin and diversification of high Neotropical biodiversity has been a recurrent debate (Moritz et al. 2000; Rull 2008, 2011a; Hoorn et al. 2010). Each of these hypotheses predicts very different causal mechanisms (climatic vs. geomorphological) and patterns of genetic diversification. A potential role was attributed to Quaternary glacial cycles and habitat stability (refugia), especially in the Northern Hemisphere (Hewitt 2004), but now also in the Southern Hemisphere where glaciers did not cover large areas but climate may have had considerable impacts (Carnaval et al. 2009; Pepper et al. 2011; Sérsic et al. 2011). Overall, it is expected that Quaternary stability will be reflected in higher genetic diversity and strong phylogeographic structure between refugia in opposition to unstable regions, where populations have genetic signatures of repeated distributional shifts (Hewitt 2004).
The alternative hypothesis predicts that species have been little affected by Pleistocene fluctuations when compared to previous geomorphological events that remained largely unchanged. These older events are then predicted to leave stronger genetic signatures, characterized by deep phylogeographic structure with multiple well-supported haploclades, pre-Pleistocene divergence times, and the presence of cryptic species and multiple evolutionary significant units (Moritz et al. 2000; Rull 2008; Hoorn et al. 2010). A third hypothesis, although, remains largely unexplored: that of Tertiary and Quaternary causal mechanisms leaving mutually detectable genetic signatures across different levels of sampled coalescent trees (Fujita et al. 2010; Rull 2011a). In this case, most crown clades ages would date back to the Paleogene–Neogene, whereas most extant terminals would have diverged during the Quaternary (Rull 2011a). Distinguishing among these alternative hypotheses requires the use of multiple markers spanning a wide range of coalescent times and mutation rates, coupled with dense population sampling, to estimate species trees and divergence times.
Species or species complexes broadly distributed across the ‘dry diagonal’ biomes represent ideal models to investigate subcontinental diversification patterns that span the Tertiary–Quaternary boundary. The nocturnal gecko Phyllopezus pollicaris (Phyllodactylidae; Gekkota) fits this role (Gamble et al. 2011, 2012). Phyllopezus pollicaris distribution spans three South American open biomes, with a slight break in the natural distribution near central Brazil. Current taxonomy recognizes two subspecies: P. p. pollicaris and P. p. przewalskii (Fig. 1), but a recent study indicates that P. pollicaris is composed of multiple cryptic lineages (Gamble et al. 2012).
Here, we use a range-wide dataset sampled from multiple loci to investigate the geographical and ecological factors promoting the diversification of a ‘dry diagonal’ endemic. We integrate geospatial methods with coalescent phylogeographic and population genetic analyses to test the relative contributions of Tertiary geomorphological versus Quaternary climatic events on P. pollicaris diversification. We subsequently use ABC analysis to test divergence models under alternative biogeographic scenarios relevant at the subcontinental scale. Our integrative approach represents one of the best documented examples of the complex diversification histories of South American open biome taxa, highlighting possible species limits within the group and testing biogeographic processes at the biome level.
Methods
SAMPLE COLLECTION
Samples were obtained through fieldwork led by FPW, GRC, or MTR, and through tissue loans obtained from colleagues (Table S1). Vouchers were catalogued in the Coleção Herpetológica da Universidade de Brasília (CHUNB) or Museu de Zoologia da Universidade de São Paulo (MZUSP). Phyllopezus pollicaris is a habitat specialist on rock outcrops, but at regions where these are not abundant, individuals are sometimes associated with human buildings (F. P. Werneck and M. T. Rodrigues, pers. obs.).
The complete dataset for molecular analyses includes 393 tissue samples from 68 localities, spanning the complete geographic, taxonomic, and morphological extent of variation currently known in P. pollicaris. Sampling localities are distributed within and outside modeled historical refugia for P. pollicaris and for the open biomes (Fig. 1). Phyllopezus periosus and P. maranjonensis (Gamble et al. 2012) were used as outgroups in all phylogenetic analyses.
SEQUENCE DATA COLLECTION
We sequenced partial mtDNA sequences from cytochrome b (cytb) and NADH dehydrogenase subunit 2 (ND2) genes from most individuals (some samples failed to amplify for ND2). We then obtained a maximum likelihood (ML) gene tree to direct subsampling efforts representing haplotype and geographical diversity (one or two individuals representing all haplotypes at each locality) for the nuclear loci. We screened approximately 40 rapidly evolving nuclear markers from a variety of sources, and amplified 11 variable loci within P. pollicaris (including protein coding regions and introns). Supporting Information Appendix S2 and Table S2 summarize relevant details for the 13 gene regions, primers, laboratory protocols, sequence assembling, alignment, recombination tests, and treatment of heterozygous individuals. We deposited all sequences in GenBank (accession nos. JQ825288-JQ828670) and trees in TreeBase (no. 12502).
GENE TREE ESTIMATION AND GENETIC DISTANCES
To describe the overall phylogeographic structure of P. pollicaris, we performed ML analyses on single-gene and partitioned concatenated datasets (mtDNA and nuDNA), including both gene copies in the case of phased nuclear loci, using RAxML version 7.0.0 (Stamatakis 2006). Analyses were implemented with GTR + Gamma model, 200 independent ML searches, and 1000 nonparametric bootstrap replicates to assess nodal support (Felsenstein 1985). We used Geophylobuilder version 1.0 (Kidd and Liu 2008) to construct a tridimensional representation (geophylogeny) from the mtDNA gene tree and associated geographical data, and visualized it with ArcGIS and ArcScene version 9.1 (ESRI). We calculated net among-group distances between mtDNA haploclades with MEGA version 5.05 (Tamura et al. 2011), using uncorrected and Tamura–Nei (Tamura and Nei 1993) corrected distances, and 500 bootstrap replicates to estimate standard errors.
SPECIES TREE ESTIMATION AND DIVERGENCE DATING
We estimated the P. pollicaris species tree from the multilocus trees under a coalescent model and simultaneously estimated divergence times using *BEAST version 1.6.2 (Drummond and Rambaut 2007; Heled and Drummond 2010). The lack of Phyllopezus fossils prevented rigorous divergence dating, but we used a less-ideal indirect calibration based on substitution rates to provide a first estimate of divergence dates for the major nodes relevant to testing biogeographic scenarios. We used uncorrelated relaxed clocks to allow for rate heterogeneity among lineages, a Yule prior for the species tree, and a normal prior on the global substitution rate to calibrate the estimation (mean = 0.0065 substitutions/my; SD = 0.0025 for the ucld.mean parameter) based on the mtDNA substitution rate of 0.65% changes/million years (Macey et al. 1998), widely employed in dating squamate phylogenies. For the nuclear loci, we used the default gamma prior for ucld.mean and exponential prior for ucld.stdev, with a mean of 0.5.
As with most species tree estimation methods, *BEAST requires a priori assignment of individual alleles to a species (‘traits’ file) before estimating the relationships. There is no routine procedure on how to assign individuals to species in a poorly known complex, but any approach elected should avoid underestimation of intraspecific diversity. We provisionally made assignments based on well-supported and geographically structured mtDNA haploclades. We selected at random one of the phased nuclear gene copies to represent each individual on this and subsequent analyses to avoid extremely time-consuming computations. We implemented five independent runs of 100 million generations each (total 5 × 108 generations), and assessed convergence of MCMC runs (effective sample sizes, ESS values > 200) using Tracer version 1.4.1 (Rambaut and Drummond 2007). Stationarity was reached before the 10% of the posterior samples, which was used as a conservative burn-in when pooling files from the independent runs with LogCombiner (Drummond and Rambaut 2007). We used BEAGLE version 1.0 (Ayres et al. 2012), a programing interface to accelerate analyses.
POPULATION STRUCTURE AND ASSIGNMENTS
We investigated population structure with a Bayesian probabilistic genetic clustering implemented by STRUCTURE version 2.3 (Pritchard et al. 2000) using a genotype matrix of two mtDNA and four nuDNA. This subset of nuclear loci spanned a range of nucleotide diversities and coalescent times (KIF24, MYH2, PRLR, and rpl35) in the complex, but allowed us to avoid the exceptionally time-consuming computations typical of such large datasets. We are aware that in some instances (e.g. long history of isolation), STRUCTURE can create clusters that are inconsistent with the main evolutionary divisions (Kalinowski 2011). This result seems to be forced by evaluation of an inappropriately small number of discrete genetic clusters, K (Kalinowski 2011), so we explored a large range of values by running 20 replicated analyses over a range of K from 2 to 30. Each of these 580 independent runs implemented one million generations following a burn-in of one million generations, and incorporated the possibility of mixed ancestry. We visualized the optimal K structure based on the rate of change in the log probability of data between successive K values, ΔK (Evanno et al. 2005), as calculated by Structure Harvester.
We combined the replicate analyses under the optimal K identified with CLUMPP (Jakobsson and Rosenberg 2007), and plotted results with individuals in the order of their appearance on the mtDNA gene tree, to facilitate cross-visualization of results. Because ΔK may favor smaller values of K representing basal levels of hierarchical structure (Evanno et al. 2005; Weisrock et al. 2010), we reran the STRUCTURE analyses (same set of parameters) within each of the major basal clusters identified to investigate finer level structure.
QUATERNARY PALAEOMODELING AND ANCESTRAL DISTRIBUTIONS
To estimate P. pollicaris species distribution models (SDMs) across Quaternary climatic fluctuations, we implemented the maximum entropy machine-learning algorithm MAXENT (Phillips et al. 2006). The occurrence dataset included our field GPS records, georeferenced museum or published records, and records from trusted collectors or on-line gazetteers (e.g., ACME Mapper version 2.0, and Global Gazetteer version 2.2), which were verified with Google Earth to ensure points were not located in heavily urbanized areas. Current climatic variables were downloaded from WorldClim (Hijmans et al. 2005), while Last Glacial Maximum [21 ky BP, LGM] and Holocene [6 ky BP] were based on ECHAM3 (Deutsches 1992), and Last Interglacial [130 ky BP, LIG] data were obtained from Otto-Bliesner et al. (2006). We then obtained a historical stability map by overlapping the presence/absence projections under the four scenarios. Methodology is described in detail in Werneck et al. (2011; in press), as applied to the open biomes. To estimate the ancestral distribution of the basal node of P. pollicaris, we used PhyloMapper version 1b1 (Lemmon and Lemmon 2008) with nonparametric rate-smoothing to obtain an ultrametric tree from the mtDNA gene tree based on unique haplotypes, and corresponding geographical coordinates.
GENETIC DIVERSITY AND TEST OF QUATERNARY PHYLOGEOGRAPHIC PREDICTIONS
We calculated population genetic metrics and tested general genetic predictions derived from the habitat stability (refugia) hypothesis (Table 1) based on mtDNA (Carnaval et al. 2009). We grouped samples based on the results of Structure, and into stable or unstable regions, and calculated coalescent estimators of genetic diversity with Migrate-n version 3.2.15 (Beerli and Palczewski 2010). We used two independent ML runs with 10 short chains and five long chains, for a total of 2000 and 20,000 generations, a burn-in of 20,000 steps, and starting parameters for the calculations derived from an Fst-like estimator (Beerli and Palczewski 2010). We calculated the other genetic parameters in Table 1 and respective significance levels with 10,000 coalescent simulations using DnaSP version 5 (Rozas et al. 2003). We also implemented Mantel tests with Zt (Bonnett and de Peer 2002) to investigate isolation-by-distance (IBD) patterns as characterized by correlations between geographical and genetic distances matrixes.
| Metric | Description | Expectation (stable areas) | Expectation (unstable areas) |
|---|---|---|---|
| θ | Theta diversity parameter | Higher genetic diversity | Lower genetic diversity |
| Mean Da | Average net nucleotide differences across localities | Higher differences across localities reflecting higher geographic structure within refugia | Fewer differences across localities (lower geographic structure) |
| Fay and Wu's Hs (P-value) | Test to detect population expansion signal | Lack of signature of population expansion | Signatures of population expansion |
| Mantel's correlation coefficient (P-value) | Isolation-by-distance (IBD) test | Presence of IBD patterns | No IBD patterns |
TESTS OF POPULATION DIVERGENCE AND BIOGEOGRAPHIC SCENARIOS
We used an ABC approach to test three alternative scenarios for the diversification of P. pollicaris relevant to hypothesized biogeographic scenarios (models and parameters described in Results). Analyses consisted of three steps: (1) draw parameter values from the prior distributions and simulate data under each model; (2) compute SuSt and the distance between simulated and observed datasets; and (3) use a rejection algorithm to approximate the posterior distribution of parameters by retaining the simulations closest to the observed data, based on a specified threshold, that is tolerance (Lopes and Beaumont 2010). Model choice can affect estimates and conclusions, so alternative models should represent biologically relevant hypotheses in the simplest way possible, and model fit should be explored (Carstens and Knowles 2010; Csilléry et al. 2010). We simulated 100,000 coalescent genealogies under each model with msABC (Pavlidis et al. 2010), and simulated a total of six loci (the same used for Structure analyses) (Pavlidis et al. 2010). Number of gene copies, number and length of loci, and prior distributions were chosen to reproduce observed values from the empirical dataset.
We estimated empirical relative mutation rates of loci and average theta across lineages specified in the models using Migrate-n version 3.2.15 (Beerli and Palczewski 2010) implementing the same search strategy as above, and used those parameters to convert absolute times into coalescent times (units of 4No generations), assuming one generation per year. We selected all seven global SuSt to be computed with mean and variance calculated across the loci, but excluded population-specific SuSt that cannot be compared between models with different numbers of populations. We transformed observed sequence data into ms-like files and then calculated the same global SuSt across all loci.
We estimated posterior probabilities and models support using ‘postpr’ and parameter posterior distributions using ‘abc’ functions of the R package ‘abc’, move the comma to out of the single quote implemented in R version 2.11.1 (R Development Core Team 2010). We set tolerance to 0.001 and implemented a nonlinear neural network regression, which has been shown to outperform other ABC algorithms (Camargo et al. 2012). We summarized simulated model fit to the observed data using a Principal Components Analysis (PCA): predictive plots in which the observed SuSt occurred within the cloud of simulated SuSt were interpreted as good fit. All intensive computational analyses were submitted to the BYU Fulton Supercomputing Cluster.
Results
MARKER AND DNA POLYMORPHISM
We sequenced a total of 1828 and 7477 bp of mtDNA and nuclear markers, respectively. Nucleotide diversity ranged from 0.59% (RBMX) to 16.85% (ND2); and overall haplotype diversity and average number of nucleotide differences were surprisingly high (Table 2). We found a high number of polymorphic sites for the mtDNA and most nuclear loci, including some protein coding genes (Table 2). The two mtDNA fragments were concatenated and treated as a single locus in subsequent analyses; 269 of the 388 samples analyzed represented unique haplotypes restricted to single localities. Most well-sampled localities (n≥ 4) were characterized by different haplotypes, but haplotypes were shared once between two nearby localities (Xingó[no. 12] and Poço Redondo [no. 16]), and once between two disjunct populations (Tianguá[no. 20] and Peixe [no. 39]).
| Gene | Length (bp) | N/localities | H/Hd | Pi(%)/k | S |
|---|---|---|---|---|---|
| Cytb | 942 | 393/68 | 164/0.99 | 15.65/98.4 | 344 |
| ND2 | 886 | 312/62 | 130/0.98 | 16.85/97.72 | 340 |
| KIF24 | 565 | 241/67 | 142/0.97 | 3.65/20.37 | 263 |
| ACA | 900 | 225/61 | 48/0.84 | 2.83/15.39 | 201 |
| MYH2 | 436 | 227/63 | 128/0.97 | 2.66/8.84 | 98 |
| PRLR | 563 | 265/68 | 98/0.96 | 2.65/13.70 | 174 |
| rpl35 | 617 | 206/61 | 76/0.96 | 2.15/11.91 | 151 |
| MC1R | 524 | 238/61 | 72/0.92 | 1.20/6.30 | 82 |
| MAP1Bex5 | 888 | 226/64 | 55/0.89 | 1.15/8.81 | 119 |
| MXRA5 | 919 | 258/68 | 96/0.94 | 1.09/9.75 | 166 |
| SINCAIP | 482 | 263/68 | 61/0.93 | 1.09/5.14 | 80 |
| DMXL1 | 943 | 276/68 | 69/0.85 | 0.91/8.63 | 179 |
| RBMX | 640 | 262/68 | 41/0.87 | 0.59/3.18 | 65 |
All but three nuclear loci (MC1R, MAP1B, DMXL1) had at least one fixed indel among lineages, and MYH2 and rpl35 regions had the largest number of indels. Barra do Garças (no. 57) and São Geraldo do Araguaia (no. 23) had the highest frequency of fixed indels (across five loci), followed by Babaçulândia (no. 24), Grão Mogol (no. 51), and Mucugê (no. 36), with indels across three loci.
GENE TREE ESTIMATION
The mtDNA tree identified multiple deep, moderately to strongly supported haploclades, with several localities exhibiting monophyly. We identified 44 mtDNA haploclades (A–AR, Fig. 2, Table S1), which were used as assignments for species tree estimation. We conservatively recognized some haploclades with low support, if concordant with geography, to avoid underestimating diversity in downstream analysis. Corrected mtDNA distances among haploclades ranged from 0.2% between geographically close sister-clades (Table S3) to values as high as 35% (uncorrected distances 0.2–25%, Table S4), with mean distance across the tree of 23.4% (SD = 0.9%).
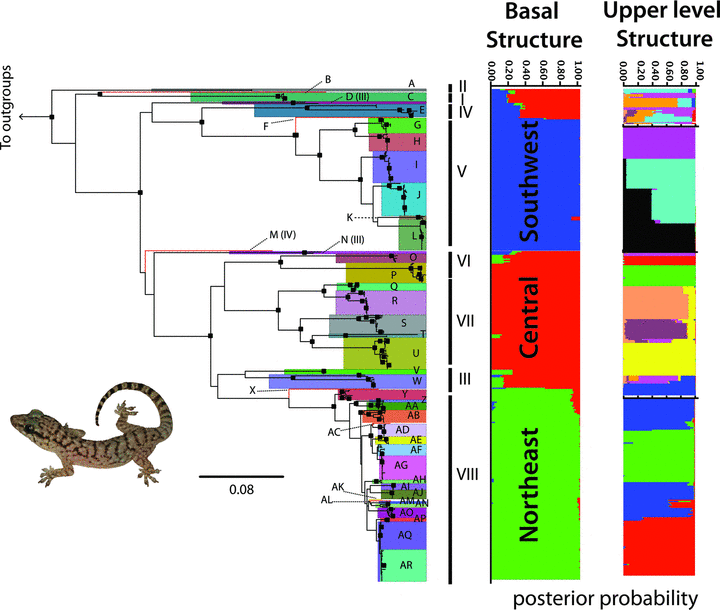
Maximum likelihood mtDNA gene tree and correspondence between terminals and STRUCTURE clusters at the two levels investigated. Black squares represent bootstrap nodal support >0.70 (terminal names are omitted for clarity, but are available in Table S1). Each population cluster is represented by a different color, with horizontal bars representing individuals and the posterior probability that a given individual is assigned to a particular cluster. Clades A–AR correspond to the 44 mtDNA haploclades identified. Vertical bars and roman numerals represent the correspondence with clades identified from the Species Tree that are interpreted as ‘candidate species’ (Fig. 4 and text for details). Photo of Phyllopezus pollicaris by MTR.
Some individual nuclear gene trees lacked resolution at the most basal levels (e.g., ACA), but most had moderate resolution and support (Fig. S1). In contrast, the concatenated nuclear ML tree recovered 12 strongly supported clades (Fig. 3). The overall genealogy of mtDNA and nuclear trees was congruent, with some relationships consistently recovered and well supported across all trees, as P. p. przewalskii (from southwestern Brazil and throughout Chaco) and the Caatinga group (Figs. 2, 3, and S1). Cerrado populations occupy three positions on the tree, two small basal lineages, including mtDNA haploclades A, B, and C, and nuclear clades (Mucugê no. 36) and (São Geraldo do Araguaia no. 23 + Barra do Garças no. 57), sister to all other lineages. A second clade, mtDNA haploclade E and nuclear clade (Serra da Mesa no. 44, Lajeado no. 37, and Palmas no. 38), is sister to the strongly supported clade comprising all P. p. przewalskii populations (Fig. 3). The third group (mtDNA haploclades Q–U and V–W) is closely related to a clade including most Caatinga localities (haploclades X–AR). With a few exceptions, populations from the Caatinga formed a well-supported clade deeply nested within the tree, but with internal relationships poorly resolved and short branches.
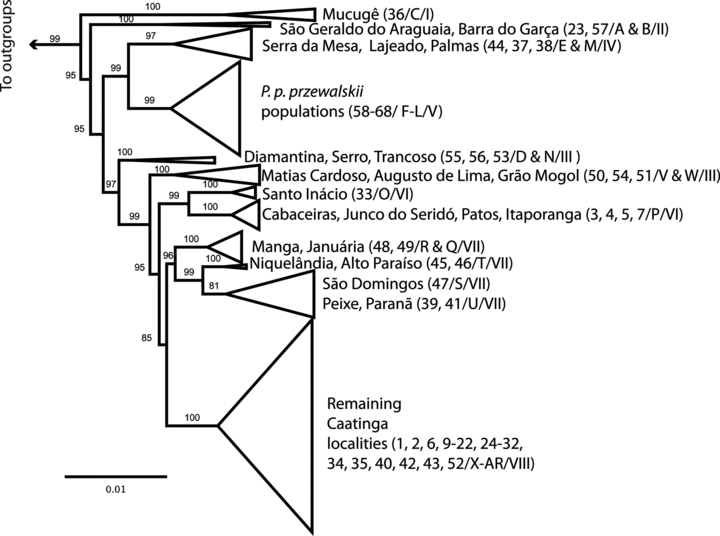
Phyllopezus pollicaris concatenated nuclear maximum likelihood tree; relationships within terminal clades are collapsed for ease of presentation and clades are labeled according to current subspecies designations or major localities. Parentheses include locality numbers (Fig. 1 and Table S1)/letters corresponding to the mtDNA haploclades (Fig. 2 and Table S1)/roman numerals corresponding to the clades recovered in the species tree (Fig. 4 and Table S1).
SPECIES TREE ESTIMATION AND DIVERGENCE DATING
We observed large ESS (> 200) and convergence for most parameters across independent and combined runs for the species tree analysis. Divergence time estimates were bound by broad confidence intervals (95% highest posterior density, HPD) and, in many instances, were not calculated at nodes of ambiguous relationships, that is, posterior probabilities lower than 0.5 (Fig. S2); thus, these are provisional conservative estimates. The species tree based on all 13 loci (Fig. 4) is mostly concordant in topology with both mtDNA and nuclear concantenated ML trees. As in previous analyses, São Geraldo do Araguaia no. 23, Barra do Garças no. 57, and Mucugê no. 36 (clades I and II) are sister to all other P. pollicaris lineages, and diverged in the mid-Miocene (∼11.5 Ma). The large clade of populations from Caatinga is deeply nested in the species tree and diverged in the Mid–Late Miocene, but internal support is low (Fig. 4, green). The strongly supported P. p. przewalskii clade was isolated approximately 8.5 Ma, and shows moderate to strong support for internal relationships (Fig. 4, blue).

Phyllopezus pollicaris species tree (maximum clade credibility tree based on all 13 loci) and divergence time estimates, derived under a coalescent model with *BEAST. Clades colors correspond to population clusters defined by STRUCTURE analyses, and the squares correspond to nodes with posterior probability >0.75. Terminals A–AR identify the 44 mtDNA haploclades, and clades highlighted by roman numerals are interpreted as ‘candidate species’, pending confirmation from ecological and morphological studies. Credibility intervals are omitted for clarity, but see Figure S2.
Divergence times estimated the origin and major crown clades splits of P. pollicaris to have occurred during the Miocene (between 11 and 5 Ma), followed by divergence within these major groups during the Pliocene–Pleistocene transition (5–1.5 Ma). These estimates are concordant with previous estimates for the origin of P. pollicaris (Gamble et al. 2011), and indicate that limited diversification occurred during Late Pleistocene, mostly among geographically close populations in the Chaco and Caatinga (Fig. 4, blue and green). Most Cerrado populations initiated and ended their diversification during the Miocene, and are characterized by long branches and older diversification dates (Fig. 4, red).
POPULATION STRUCTURE AND ASSIGNMENTS
Calculations of ΔK detected a peak at K= 3, representing the most basal hierarchical structure in the data (Evanno et al. 2005; Weisrock et al. 2010). Assignments detected at K= 3 are congruent with geographic clusters in the Southwest (SW), Central (CE), and Northeast (NE) of the P. pollicaris distribution, which are largely coincident with the distributions of the Chaco, Cerrado, and Caatinga, respectively (Figs. 5,6). One exception are samples from a few close localities in Paraíba state (Itaporanga [no. 7], Junco do Seridó[no. 4], Patos [no. 5]), assigned to the Central cluster, while surrounded by populations assigned to the Northeast cluster (Fig. 5). There is also overall concordance between identified multilocus clusters and mtDNA haploclades and geophylogeny (Fig. 5). Some individuals located at the contact regions between clusters revealed admixed assignments (Fig. 6).
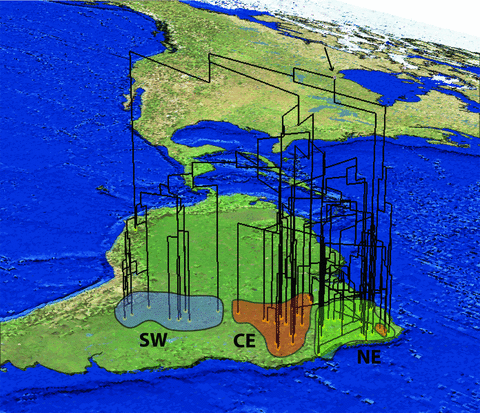
Phyllopezus pollicaris and outgroups mtDNA geophylogeny as estimated by GeoPhyloBuilder and visualized by ArcScene. Colors shapes correspond to population clusters identified with STRUCTURE and the black arrow shows the tree root.
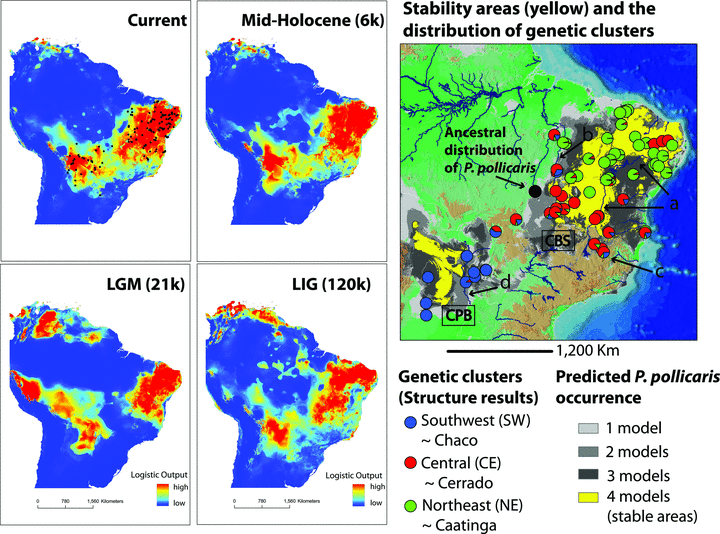
Modeled ranges of Phyllopezus pollicaris across Quaternary climatic fluctuations, and current climatic scenarios (left panels). Warmer colors of the logistic output format correspond to regions with higher probability of occurrence. Black dots in the logistic output for the Current climatic scenario (top left) represent the occurrence dataset implemented in the distribution modeling analyses, and depict the natural break in the P. pollicaris species complex distribution. On the right: Historical stability surface (gray tones and stable areas in yellow) obtained by overlapping predicted logistic outputs under the four climatic scenarios (current, 6, 21, and 120 kyr BP), and the geographical distribution of the Bayesian clustering results and ancestral distribution of P. pollicaris. All stability surfaces are plotted against a digital elevation model for South America, depicting major rivers and geomorphological features discussed in the text (brown represents higher altitudes, e.g., Central Brazilian Shield [CBS] and green represents lower altitudes, e.g., Chaco-Paraná Basin [CPB]; a, São Francisco river; b, Tocantins River; c, Espinhaço mountain range; d, Paraná River). Each genetic cluster is designated by a different color, with pie charts representing individuals and the posterior probability that a given individual is assigned to a particular cluster.
When we reran analyses within each of the major clusters, ΔK detected K= 3, K= 9, and K= 3 (total K= 15) for the SW, CE, and NE clusters, respectively, demonstrating further population structure, more pronounced in the CE populations. However, although most individual membership posterior probabilities were greater than 0.9 in the first round of analyses, coefficients are overall lower at the finer level structure, and further divide both admixed and nonadmixed sets of individuals (Fig. 2). Therefore, as we considered using population assignments to develop divergence models, and alternative models should preferentially represent biologically relevant questions in the simplest way possible, we chose to focus subsequent analyses at the basal level of clustering (K= 3).
PALAEOMODELING AND ANCESTRAL DISTRIBUTIONS
Estimated palaeomodels depict a potential distribution of P. pollicaris contracted to the southwest and northeast of the ‘dry diagonal’ during the LGM, with a break in central Brazil. These two SDMs blocks closely track the distribution of population clusters, SW to one extreme and [CE + NE] to the other (Fig. 6). However, when considering all SDMs and the stability surface, we observe that P. pollicaris potential distribution did not change drastically across Quaternary climatic fluctuations, especially from the Holocene to the present (Fig. 6). Note the extensive overlap of highly suitable areas across the four time periods (i.e., large areas of stability; Fig. 6, yellow areas). Despite minor oscillations in the species range, the P. pollicaris stability map provides a close match for the ‘dry diagonal’ biomes stability models (Werneck 2011; Werneck et al. in press). Thus, we are confident that P. pollicaris tracked changes of the open biomes and that this complex represents a suitable proxy of their biogeographic patterns at both shallow and deep temporal scales. The ancestral distribution was estimated by PhyloMapper to be located in central Brazil, nearby CE and NE current distributions, where ancient Phyllopezus lineages became extinct (Fig. 6, black circle).
GENETIC POLYMORPHISM AND TEST OF PHYLOGEOGRAPHIC PREDICTIONS
None of the groupings (or the total sample) revealed detectable signatures of population expansion or reduction (Table 3). We did detect significant IBD across all groupings, except for the SW cluster, which had overall lower diversity when compared to the other clusters (Table 3). We did not find major differences between samples from stable and unstable areas with respect to most population genetic metrics, contradicting predictions from the hypothesis of strong influence of Quaternary refugia on clade's genetic structure (Hewitt 2004; Carnaval et al. 2009). In fact, unstable areas presented higher genetic diversity than stable areas with respect to several metrics (Table 3).
| Model and parameters | Models comparisons | |||
|---|---|---|---|---|
| All models A versus B versus C i/m | No-speciation versus speciation models, pairwise | Speciation models only B versus C i/m | ||
| A versus B i/m | A versus C i/m | |||
| Model A; no-speciation | 0.089/0.169* | 0.158/0.159 | 0.08/0.169 | NA |
| Model B; early speciation | 0.098/0.168* | 0.842/0.841 | NA | 0.007/0.286 |
| Model C; speciation in two clades (SW [NE+CE]) | 0.813/0.663* | NA | 0.84/0.831 | 0.993/0.714 |
- *No resolution at tolerance rate = 0.001; results shown at an increased tolerance rate (tol = 0.1).
| Region | N | θ | θsites (95% confidence interval) | Pi | k | Mean Da (min.; max.) | Hs (P-value) | Mantel's correlation coefficient (P-value) |
|---|---|---|---|---|---|---|---|---|
| Population cluster | ||||||||
| Central (CE) | 131 | 185.23 | 0.120 (0.046–0.165) | 0.15680 | 200.6 | 0.174 (0.010; 0.226) | 0.029 (0.330) | 0.410 (0.002) |
| Northeast (NE) | 151 | 96.29 | 0.194 (0.083–0.364) | 0.03736 | 64.478 | 0.037 (0.00006; 0.06132) | −0.065 (0.333) | 0.303 (0.003) |
| Southwest (SW) | 105 | 91.38 | 0.058 (0.025–0.078) | 0.07096 | 121.9 | 0.076 (0.001; 0.118) | −0.082 (0.332) | 0.065 (0.439) |
| Stability surface | ||||||||
| Stable | 217 | 151.54 | 0.136 (0.114–0.165) | 0.1405 | 202.3 | 0.118 (0.005; 0.216) | 0.229 (0.329) | 0.507 (<0.0001) |
| Unstable | 170 | 169.89 | 0.084 (0.067–0.104) | 0.1565 | 233.8 | 0.165 (0.001; 0.225) | 1.520 (0.323) | 0.616 (<0.0001) |
| Total | 387 | 194.08 | 0.123 (0.108–0.142) | 0.155 | 219.1 | 0.147 (0.001; 0.224) | 0.048 (0.331) | 0.575 (<0.0001) |
TESTS OF POPULATION DIVERGENCE AND BIOGEOGRAPHIC SCENARIOS: ABC
We developed three models representing alternative biogeographic scenarios for the diversification of P. pollicaris across the ‘dry diagonal’ based on (1) speciation expectations from estimated genealogies, population structure, and palaeomodels; and (2) available geological information with relevant times that fall within the dated chronogram from *BEAST. The use of external information (e.g., dates of geological events) is crucial to narrow the universe of possible histories (Garrick et al. 2010). The first model (A) represents a no-speciation null model and reflects views from early studies that the open biomes had a shared history (Vanzolini 1976); it was modeled as a large population, with no substructure and free migration. Model B hypothesizes an approximately simultaneous early divergence from the ancestral area (as estimated by PhyloMapper) into three clades (SW, CE, and NE, as recovered by Structure), and represents a geologically older speciation scenario. Model C represents an early diversification of the southwest (SW) group from the other groups (CE + NE), followed by a more recent divergence between these last two. This hypothesis would correspond to two speciation events within P. pollicaris, and it predicts a (SW [CE + NE]) topology (Fig. 7).
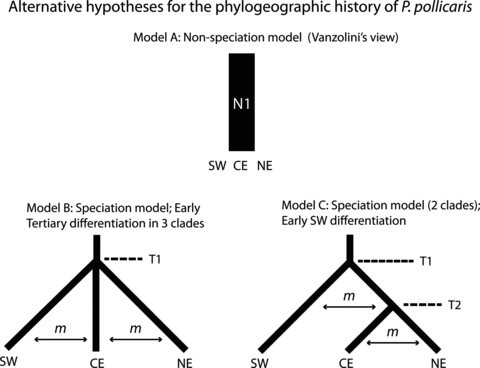
Alternative models for the phylogeographic history of P. pollicaris tested with multilocus ABC. Each model consisted of branching order and divergence times, which were converted from absolute times into coalescent times (units of 4No generations). Models B and C were also tested including migration rates as an additional parameter. Although the early divergence event in both speciation models (B and C) is referred to as T1 (because they are attributed to the same event) and the more recent divergence time at model C as T2, the coalescent simulations are preceded backwards on time (i.e., for model C, T2 is simulated before T1). See text for details.
We attribute the early divergence time (T1) to the final uplift of the Central Brazilian Plateau that took place 7–5 Ma, an important event hypothesized to have driven divergence across the open biomes (Werneck 2011). We hypothesize the more recent divergence time (T2) to represent the final phase of the ecological expansion of grass-dominated vegetation and increased fire frequency (5–3 Mya), which may have promoted additional ecological diversification of the Cerrado from the adjacent open biomes (Beerling and Osborne 2006; Simon et al. 2009; Edwards et al. 2010). Both divergence times fall within estimated divergence times for P. pollicaris lineages (Fig. 4). Although the use of three subgroups estimated by Structure does not fully corroborate the phylogenetic results (which recover a paraphyletic CE group), the models were intended to capture the broad substructure and biogeographic patterns supported by available geological data for the ‘dry diagonal’. Because divergence with some degree of gene flow may be common (Nosil 2008), we also considered the possibility of migration in the diversification models B and C. Model A included the per locus mutation parameter θ (= 4Noμ) for a single lineage across multiple loci, whereas models B and C included additional parameters: the divergence times in coalescent units (τ, in units of 4No generations) and the migration rate between divergent lineages (m, prior varied from 0, no migration, to 1, high migration). We assumed stable population sizes in all models.
When compared simultaneously, ABC could not attain resolution between the three models, unless the tolerance rate was increased (Table 4). ABC favored the speciation scenarios over the no-speciation scenario in all pairwise comparisons. More specifically, diversification of P. pollicaris into two large groups, SW and CE + NE (model C), was selected in every comparison that included this model (Table 4). Posterior probabilities were overall lower for comparisons that included migration, relative to equivalent comparisons without migration, reflecting increased difficulty in distinguishing between speciation and no-speciation models when migration is present (Nielsen and Wakeley 2001). Bayes factors of pairwise comparisons to the preferred models attained moderate values (up to ∼12). The observed SuSt occurred within the bounds of the prior sample of simulated SuSt at PCA predictive plots, confirming good model fitting (Fig. S3).
Discussion
The degree to which phylogeographic approaches can resolve broader biogeographic processes is directly dependent on the coverage of geographical sampling, the biology of the study taxa, and the nature of markers used. Single-locus (mtDNA) phylogeography has been profusely criticized (Brito and Edwards 2009), because it ignores coalescent stochasticity and tends to provide biased and overly simplistic inferences (Knowles and Maddison 2002). Regarding sampling design, robust inferences require a trade-off between number of loci and individuals, but generally an increased number of individuals within species/populations improve species tree accuracy (Maddison and Knowles 2006). However, most phylogeographic studies for Neotropical taxa are still based mainly on mtDNA and limited population and individual sampling, so their broader biogeographic implications are limited.
In this article, we present a densely sampled case study for a broadly distributed Neotropical habitat-specialist species complex. Our findings revealed a very deep mtDNA structure typical of low-vagility species, unprecedented levels of cryptic genetic diversity, and diversification dating back to at least the Neogene with persistence across Quaternary fluctuations. Ultimately, we tested alternative divergence scenarios, which proved useful to infer major biogeographic patterns across the ‘dry diagonal’.
DNA POLYMORPHISM, GENETIC DISTANCES, AND POPULATION STRUCTURE
Patterns of DNA polymorphism, gene and species trees are clearly consistent with the occurrence of a complex of cryptic species within P. pollicaris, as hypothesized by Gamble et al. (2011, 2012). The large mtDNA genetic distances among clades found here exceed typical values ranging from 5% to 11% reported among gecko sister species (Rocha et al. 2009; Fujita et al. 2010), and for gecko complexes with likewise high hidden genetic diversity and low haplotype sharing between localities (Geurgas and Rodrigues 2010).
The P. pollicaris species complex has a strong population structure in which the three major clusters at the deepest levels are concordant with the geographic limits of the Chaco, Cerrado, and Caatinga biomes; and also approximately congruent in the gene and species trees and geophylogeny, implying that these ‘dry diagonal’ biomes do not share a single diversification history. Although the SW cluster is more isolated from the others, the CE and NE are intermingled over several regions of overlap, as revealed by admixture assignments and lower diversity in the SW Chaco cluster. The CE cluster is characterized by higher population subdivision, genetic diversity, and nucleotide differences across localities than the other two clusters. This cluster is mostly confined to the Cerrado, which has a landscape compartmentalized between ancient plateaus and young valleys (Cole 1986). This heterogeneity at local and regional scales potentially enhances environmental opportunity for ecological divergence and speciation (Werneck 2011), if these changes result in reduced gene flow and isolation between lineages (Rundell and Price 2009). Geomorphological complexity and heterogeneity are then likely to have shaped the genetic structure among CE populations.
MAJOR RELATIONSHIPS AND DIVERGENCE TIMES
Analyses of mtDNA and nuclear data showed overall congruence in the major relationships, differing mostly in their branch support at deeper levels (higher for most nuclear loci). We found higher support for the nuclear concatenated analysis relative to species trees, which have been suggested to be an artifact of uninformative loci for short branches, and still comprise a challenge for species-tree methods for higher level phylogenies (Townsend et al. 2011). As uninformative nuclear loci are more frequent at the lower levels, we expected such an artifact to have stronger influences on species-trees estimated from phylogeographic datasets. Indeed, McCormack et al. (2011) also reported slightly lower topological support for the species tree in Aphelocoma jays, although their study included only three nuclear markers.
Instances of topological discordance are evident at internal nodes of more recent lineages, and at the admixture zones between CE and NE phylogeographic groups. For example, relationships within the NE group are poorly resolved. This pattern can be attributed to the reduced topographic complexity of the Caatinga that is disrupted by few prominent landforms, such as elevated plateaus (e.g., Chapadas do Araripe, Apodi, and Borborema, Serra do Espinhaço), and relictual rainforest enclaves (Sampaio 1995). Fewer geological barriers in the Caatinga likely enhanced opportunity for gene flow, yielding less-resolved diversification histories.
A few samples are characterized by discordant placement among datasets or methods. For example, the clade D (Diamantina no. 55 + Serro no. 56) occupying a basal position in the mtDNA (Fig. 2) and the species trees (Fig. 4), but a more nested position in the nuclear concatenated tree (Fig. 3). Similarly, Matias Cardoso no. 50 + Augusto de Lima no. 54 and Grão Mogol no. 51 (clades V and W) are basal in the species tree but deeply nested in both concatenated trees, and these samples show admixed structures (Fig. 6). These cases show the confounding effect of contact zones between adjacent groups on species tree inference, and the importance of multilocus coalescence methods for estimating species trees in instances of discordance (Leaché 2009). In these cases, we favor the species tree hypothesis that places some of the northern Minas Gerais populations closer to the base of the tree (V, W, and D), and remaining samples (Januária [Q, no. 49] and Manga [R, no. 48]) as sister to the large clade of Caatinga populations (Fig. 4).
The most interesting outcome of this hypothesis comes from the disjunct placement of Manga (clade R, no. 48) and Matias Cardoso (clade V, no. 50) localities that are less than 15 km apart, but are separated by the São Francisco River (Fig. 6, letter a, SFR). The SFR is recognized as a barrier that has driven speciation at high taxonomic levels, with several endemic genera and species pairs isolated on opposite sides (Rodrigues 1996). More recently, the SFR was suggested as an important phylogeographic barrier to Atlantic Forest taxa distributed closer to the delta (Pellegrino et al. 2005; Carnaval and Moritz 2008). Recent divergence estimates in the lizard genera Eurolophosaurus (Passoni et al. 2008) and Calyptommatus (Siedchlag et al. 2010) suggest that the SFR probably formed a vicariant barrier during the Late Miocene, concordant with P. pollicaris divergence times. The SFR also likely prevented secondary contact between previously diverged P. pollicaris lineages, reinforcing its importance as a singular center of diversification. Our results also suggest a role for the Tocantins River as barrier between CE and NE clusters (Fig. 6, letter b). Denser sampling and investigation of other taxa distributed in the region are, however, needed to further explore this hypothesis.
São Geraldo do Araguaia (no. 23/A), Barra do Garças (no. 57/B), and Mucugê (no. 36/C) on the other hand were recovered as highly distinct and early differentiated populations in all analyses. Mucugê together with other populations (Diamantina [no. 55/D] and Serro [no. 56/D]) are located in the high altitude “campos rupestres” in the Serra do Espinhaço mountain range (Fig. 6, letter c), a region well known for its high levels of endemism, including squamates (Cassimiro and Rodrigues 2009; Nogueira et al. 2011). Establishment of the Espinhaço range was a regional event that potentially influenced P. pollicaris divergence (see below).
Divergence within the P. pollicaris complex proceeded in a nearly continuous fashion since about 11.5 Ma, but with crown clades originating in the Mid–Late Miocene. Phyllopezus pollicaris przewalskii from Chaco is strongly supported by gene and species tree, and diverged approximately 8.5 Ma. Pleistocene diversification is recovered among some geographically close Caatinga populations; a time correlating with geological evidence for the exposition of formerly submerged granite surfaces (lajedos) where the species occur. These times suggest a major influence of divergence/geographical barriers during the Miocene, followed by a nearly continuous diversification, with no clear bursts of Quaternary diversifications (Rull 2011b). Long-term diversification with combined Neogene and Pleistocene causal mechanisms is expected to be apparent at higher taxonomic levels (Rull 2011a), whereas the prolonged complex history of ‘dry diagonal’ biomes is here evident within a single complex.
Likewise, ancient divergence dates have been reported in other gecko complexes (Pellegrino et al. 2005; Geurgas and Rodrigues 2010; Oliver et al. 2010), and these are commonly older than codistributed taxonomic groups. Patterns of New World gecko diversification exemplify most of the scenarios described for terrestrial vertebrates, including ancient vicariance, trans-Atlantic rafting and temporary land bridge dispersal, and human introductions (Gamble et al. 2011), and reflect their ancient Gondwanan origin (Kluge 1987). Positive correlations between clade age and species diversity described for geckos (Gamble et al. 2011) can account for some of the patterns we report for P. pollicaris complex (ancient divergence times and high diversity).
MODEL-BASED POPULATION DIVERGENCE: BIOGEOGRAPHIC IMPLICATIONS
We are aware that our models represent simplifications (e.g., no substructure within major clusters, use of combined splitting times, and use of a nearly monophyletic group [CE]), but they do reflect phylogeographic patterns within P. pollicaris that correlate with major biogeographic events across the ‘dry diagonal’. ABC favored the speciation scenarios over the no-speciation model, and we found stronger support for model C, representing an early diversification (∼7–5 Ma) of the southwest (SW) from the (CE + NE) group, followed by a more recent divergence between these last two (∼5–3 Ma). Comparisons of model C under isolation and migration suggest that divergence may have occurred with some gene flow. Due to the high correspondence between groups tested and the ‘dry diagonal’ biomes, we can infer geomorphological events potentially responsible for an early diversification of Chaco lineages followed by divergence of Cerrado and Caatinga lineages, and their biogeographic implications.
Key Neogene events often emphasized as causal mechanisms for Neotropical diversification (e.g., closure of the Panama Isthmus, Andean uplift, change of drainage patterns with the cessation of marine incursions into the Amazon basin; Hoorn et al. 2010; Rull 2011a) almost certainly impacted indirectly the ‘dry diagonal’ biota diversification, which is largely confined to the Brazilian Shield (Werneck 2011). However, other correlated, but less-understood, events were more relevant for this region and for the diversification of P. pollicaris complex. From the hypothesized ancestral area in central Brazil, some ancestral population dispersed across the ‘dry diagonal’ before being subject of the following suggested biogeographic events that generated the three major genetic lineages.
Marine transgressions during the Mid–Late Miocene (∼10–5 Ma), supported by sedimentology and marine fossil records, covered large portions of the Chaco-Paraná Basin (CPB), and formed the second flooding pulse of the Paranaense Sea, isolating large areas of the Chaco depression and associated biota (Hernández et al. 2005; Ruskin et al. 2011). Isolation of southwest populations of P. pollicaris initiated by these marine transgressions was most likely enhanced by the Chaco subsidence due to an intense uplift phase of the Andes and the final uplift of the Central Brazilian Shield during the Late Miocene–Early Pliocene transition, 7–5 Mya (Gubbles et al. 1993; Uba et al. 2006; Mulch et al. 2010). We hypothesize that these events, in concert, drove allopatric speciation of the P. p. przewalskii clade (SW). The resulting altitudinal break acted as a geographical barrier and prevented secondary contact, with the lowland populations isolated on the SW side (CPB and Paraguay River basin) and the higher elevation Brazilian Plateaus populations to the CE–NE side (Fig. 6). Strong isolation coupled with low densities of P. pollicaris preferred habitat (rock outcrops) in the Chaco, may have limited population sizes, and thus explain the reduced molecular variation in the SW clade.
We also hypothesize that subsequent regional tectonic events responsible for the final establishment of the Brazilian Plateau and enclosed regional mountain ranges (e.g., Serras do Espinhaço, do Mar, da Mantiqueira, 2–4 Mya) coupled with ecological events such as expansion of grasslands and increased fire frequency (Beerling and Osborne 2006; Simon et al. 2009) would promote differentiation between Cerrado and Caatinga clades. However, because CE and NE lineages are not reciprocally monophyletic and show some admixture, they may have diverged through ecological speciation along environmental gradients, without physical barriers to prevent gene flow (Nosil 2008). The alternative of sympatric speciation is supported by an increasing body of evidence in marine vertebrates (Foote et al. 2009; Crow et al. 2010), where obvious barriers are less evident, but is also documented for terrestrial vertebrates (Niemiller et al. 2008; VanderWerf et al. 2010), and must also be considered as a possibility here.
Although the exact mechanisms are not clear, events at the Tertiary–Quaternary boundary that significantly remodeled eastern Brazil's landscapes may have confounded the genetic signatures of the earlier isolating events, including vegetation shifts (Werneck et al. 2011, in press), and neotectonic activity (Riccomini and Assumpção 1999). We suggest that the less dramatic differentiation between Cerrado and Caatinga lineages could be due to a combination of more recent divergence and gene flow, and represent a possible example of ecological or sympatric speciation (Vanzolini 1976).
PAST DISTRIBUTIONS AND TESTS OF QUATERNARY PHYLOGEOGRAPHIC PREDICTIONS
Our results suggest a limited influence for Quaternary climatic fluctuations on the phylogeographic history and SDMs of P. pollicaris. Patterns of genetic variation and structure reflect the impact of earlier influences and a Miocene origin of the crown clades followed by persistence through the Pleistocene without major demographic changes. Our modeling suggests a slight range reduction of P. pollicaris during the LGM with a break in central Brazil. This “two LGM refugia” scenario reflects very distinct SDMs between the SW and the [CE + NE] clusters, and these ecological niche differences may have promoted additional diversification between these two groups.
Although Pleistocene distributional shifts shaped strong genetic signatures in many groups (Carnaval et al. 2009; Knowles and Alvarado-Serrano 2010; Lessa et al. 2010), there are counter-examples in which phylogeographic signatures reflect events that have not been overwritten by Pleistocene climate dynamics (Leavitt et al. 2007; Bell et al. 2010; Thomé et al. 2010; Hoskin et al. 2011). For P. pollicaris specifically, the influence of climatic fluctuations seems minor when compared to geographical and topographical influences that remained largely unchanged since the Miocene, however, they can not be neglected, and likely played a relevant role to promote further differentiation between and within previously diverged lineages.
TAXONOMIC IMPLICATIONS AND PROVISIONAL SPECIES LIMITS
Phyllopezus pollicaris should clearly be treated as polytypic, and because the consistently recovered clades are deeply divergent and strongly supported in all analyses, they are almost certainly valid species. Here, we comment briefly on species limits and taxonomic implications within P. pollicaris complex.
The P. p. przewalskii clade is strongly supported across all analyses, and it has a distinct structure and a singular SDM. Although morphological uniformity generally characterizes the P. pollicaris complex, P. p. przewalskii is also morphologically diagnosable by the lack of postcloacal tubercles and slightly lower ventral scales count (Vanzolini 1953), and in its karyotype (Pellegrino et al. 1997). Under a general lineage species concept (de Queiroz 2007) and our empirical evidence for both ecological and genetic nonexchangeability (Crandall et al. 2000), P. p. przewalskii should be elevated to species status. The nested placement of P. p. przewalskii within P. pollicaris renders the other subspecies, P. p. pollicaris, paraphyletic. We hypothesize that the following clades (Roman numerals in Fig. 4 and Table S1) should be treated as ‘candidate species’: Clades I (no. 38) and II (no. 7 and no. 54), which are the sister groups to all other P. pollicaris, should be treated as two candidate species, along with these six remaining clades: Clade III (6 localities); Clade IV (3 localities); Clade V (P. przewaslkii, 11 localities); Clade VI (6 localities); Clade VII (7 localities), and the Clade VIII (the 32 remaining Caatinga localities). See Figure S4 for the geographic distribution of the eight ‘candidate species’. Aside from P. przewaslkii, we do not suggest formal names for any of these lineages, but as ‘candidate species’ (Morando et al. 2003) they can guide biogeography-based conservation planning because they register broad evolutionary patterns without the necessity of formal names (Bickford et al. 2007; Riddle et al. 2011).
IMPLICATIONS FOR ‘DRY DIAGONAL’ CONSERVATION
Recent studies show that current knowledge of amphibian and reptile species diversity in the Neotropics has been underestimated (Fouquet et al. 2007; Geurgas and Rodrigues 2010; Gamble et al. 2011; Funk et al. 2012). The cryptic genetic diversity disclosed here reveals that the South American ‘dry diagonal’ biomes are no exception. With the management task of preserving adaptive diversity, evolutionary processes, and natural genetic connections across the range of species complexes (Crandall et al. 2000; Davis et al. 2008), we can propose some conservation implications of our results.
Our assessment of the P. pollicaris complex shows that protection of the eight candidate species is necessary to preserve the evolutionary processes that generated them, especially if these geographic regions have fostered codivergence in other taxa (Davis et al. 2008). Protection of long-isolated populations should be augmented by protection of admixture zones to maximize adaptive diversity and evolutionary potential (Crandall et al. 2000; Smith et al. 2001; Moritz et al. 2009). This can be accomplished by placing reserves at the core of the three major clusters, and several are established and were included in our sampling (e.g., no. 6, 10, 16, 19, 40, 42, 48, 49, 50, 64; Fig. 1). Fortunately, reserves have also been established in the transition zones between CE and NE clusters (no. 23 and 26), but additional reserves in this region (e.g., no. 36, 37, 38, 51, 54, 55, 56,) and between the SW populations and the other lineages (e.g. no. 57, 58, 61) should be considered in regional conservation planning.
Persistence through Quaternary fluctuations and high diversity within the P. pollicaris complex suggests that it has the potential to tolerate adverse scenarios and to adapt to new conditions (Davis et al. 2005), as long as rock outcrops persist. These patterns provide important insights about responses to future environmental changes and long-term population viability, a critical variable for establishing efficient conservation strategies (Diniz-Filho et al. 2008). However, other taxa associated with ‘dry diagonal’ features more susceptible to oscillations may be more susceptible to extinction due to climate change. Allocation of conservation resources will then be more effective if comparative studies can provide evolutionary histories of a diverse array of codistributed ‘dry diagonal’ endemics.
Conclusions
This study highlights how phylogeographic patterns can be studied in a spatio-temporal framework to investigate broader historical biogeographic processes through the coupling of SDMs, multilocus genetic data, and model-based approaches (Knowles 2009; Knowles and Alvarado-Serrano 2010). For species complexes such as P. pollicaris, whose current and historical distributions overlap closely those of the relevant biomes, major phylogeographic breaks may reflect influential historical biogeographic processes. These characteristics make such complexes important evolutionary radiations to study the deep history of associated biomes. This approach is particularly relevant for poorly studied regions, such as the South American ‘dry diagonal’ biomes where threats are escalating.
In this perspective, the deeply divergent lineages of P. pollicaris, which are geographically structured along the southwest–northeast ‘dry diagonal’, reflect complex diversification scenarios and the primary influence of geologically old processes on the dry biomes diversification. Long-term isolation is not essential to generate significant genetic differentiation (Knowles and Alvarado-Serrano 2010), but this is the case for the P. pollicaris complex, whose distributions are characterized by substantial stability and genetic signatures of deep historical events that were not erased by Pleistocene climate shifts. Recent environmental instability thus does not appear to be the primary parameter influencing the genetic outcomes of the diversification process in this taxon.
Each biome has unique population clusters and lineages; and our model-based approach yielded stronger support for the diversification models, contradicting early views that the ‘dry diagonal’ biomes would have a shared diversification history. More precisely, the central portion of South America appears to have served as important diversification center over the last several million years, as a response to geomorphological rearrangements of the landscape. These results have important implications for identification of areas to maximize the protection of rich lineages unique to the region, which evolved in concert with the diversification of central-eastern South American ‘dry diagonal’ biomes.
Associate Editor: B. Fitzpatrick
ACKNOWLEDGMENTS
FPW was supported by fellowships from CAPES/Fulbright (no. 15073722–2697/06–8) and by a graduate research assistantship from the BYU Department of Biology. This work was funded by the National Geographic Society (NGS, no. 8642–09), National Science Foundation DDIG award (DEB-1210346), Society of Systematic Biologists (SSB), Neotropical Grassland Conservancy (NGC), Society for the Study of Amphibians and Reptiles (SSAR), Idea Wild, Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), and Fundação de Apoio à Pesquisa do Distrito Federal (FAPDF). The work in the Sites lab was also supported by a BYU Mentoring Environment Grant (2011), and National Science Foundation (NSF) awards EF 0334966 (“Deep Scaly” project) and OISE 0530267 (“PIRE – Patagonia” project); this last project to the following institutions (listed alphabetically): Brigham Young University, Centro Nacional Patagónico (AR), Dalhousie University, Instituto Botánico Darwinion (AR), Universidad Austral de Chile, Universidad de Concepción, Universidad Nacional del Comahue, Universidad Nacional de Córdoba, and University of Nebraska. FPW thanks the Instituto Chico Mendes de Conservação da Biodiversidade (ICMBio; no. 16381–2) and Instituto Estadual de Florestas (IEF-MG; no. 066–070/09) for issuing collecting permits. We thank the following researchers: D. Loebman, I. J. Roberto, F. S. Rodrigues, M. Pérsio, V. H. G. L. Cavalcante, P. Peloso, A. Prudente, D. M. Borges-Nojosa, J. Cassimiro, M. Teixeira, Jr., R. Recoder, F. F. Curcio, D. Pavan, J. Roscito, R. M. L. Santos, R. Villela, P. H. Valdujo, F. L. Souza, M. Uetanabaro, C. Cândido, D. O. Mesquita, F. R. Delfim, R. G. Faria, C. B. Carvalho, R. A. Santos, H. Zaher, F. Graziotin, C. C. Mello D’Horta, V. L. Ferreira, C. Strüssmann, and curators of the following institutions for providing tissue samples for this study: CHUNB, MZUSP, Coleção Herpetológica da Universidade Federal do Ceará (CHUFC), Laboratório de Zoologia da Universidade Federal do Piauí (LZUFPI), Museu Paraense Emílio Goeldi (MPEG), Departamento de Zoologia da Universidade Federal do Mato Grosso (UFMT), Coleção Herpetológica da Universidade Federal do Mato Grosso do Sul (UFMS/CH), Museo Nacional de Historia Natural del Paraguay (MNHNP), Zoologisches Forschungsinstitut und Museum Alexander Koenig (ZFMK), Museum für Tierkunde Dresden (MTD), Museo Nacional de Ciencias Naturales (MNCN), Herpetological collection of L. J. Avila and M. Morando (LJAMM) of the Centro Nacional Patagónico (CENPAT–CONICET). FPW thanks A. H. S. B. Soares, R. Leite, D. C. de Paula, M. M. Espírito-Santo, M. M. Caixeta, R. Recoder, S. F. Balbino, A. F. Béda, V. L. Ferreira, N. Rufino, F. C. Villela, L. J. Avila, N. Feltrin, N. Frutos, M. Motte, N. J. Scott, P. Cacciali, G. H. C. Vieira, D. O. Mesquita, S. Vieira, S. C. Ribeiro for their invaluable assistance in the field expeditions. This manuscript benefitted from discussions of the analyses with A. Camargo, R. N. Leite, P. Pavlidis, J. E. McCormack, J. Breinholt, and G. C. Costa; and from helpful comments from two anonymous reviewers.




