The Identification of Ingested Dandelion Juice in Gastric Contents of a Deceased Person by Direct Sequencing and GC–MS Methods*
Supported by research and development expenditures of National Institute of Scientific Investigation.
Abstract
Abstract: DNA and chemical analysis of gastric contents of a deceased person were handled in this work. The body of the victim was discovered in his car, submerged in a lake. We were asked to determine whether or not the gastric contents of the victim harbored drugs and dandelion material. It was suspected that the victim had been murdered by poisoning with an excess amount of sleeping medication (doxylamine), which had been homogenized with dandelion. The concentrations of 11.4 and 27.5 mg/kg of doxylamine detected from spleen and liver of the victim were far higher than the assumed therapeutic concentration. Via gas chromatography–mass spectrometry (GC–MS) analysis and direct sequencing analysis of plant genetic markers such as intergenic transcribed spacer, 18S ribosomal RNA (rRNA), rbcL and trnLF, it was confirmed that the gastric contents of the victim contained taraxasterol, which is one of the marker compounds for dandelion and contained dandelion species-specific rbcL and trnL-trnF IGS (trnLF) sequences. The initial PCR of the genomic DNA isolated from the gastric contents showed insufficient quantity, and the second PCR, of which the template was a portion of the initial PCR products, exhibited a sufficient quantity for direct sequencing. rbcL and trnLF located in the cpDNA resulted in the successful determination of dandelion DNA in a decedent’s stomach contents. GC–MS identifies the actual presence of a taraxasterol at 28.4 min. Raw dandelion was assumed to be used as a masking vehicle for excess sleeping drug (doxylamine).
Fragmentary plant evidence tends to make traditional morphological investigation difficult, and is often ignored by many investigators for this reason. DNA analysis, as well as drug or toxin analyses of stomach contents, can constitute valuable evidence for providing investigative leads in investigations of death. Currently, the identification of ingested vegetative samples relies on microscopical and morphological examinations, but this sometimes proves problematic. DNA technology has been used extensively in human profiling (1,2) and in animal studies for forensic purposes (3), and has been gradually applied to the analysis of forensic botanical materials (4,5).
The sequencing method of specific loci or markers with different base substitution rates has been generally adopted for plant phylogenetic studies (6–14). Intergenic transcribed spacer (ITS) and 18S ribosomal RNA (rRNA) are located in the genomic nucleolar organizing region. ITS allows for the appreciation of phylogeny of a variety of monocot and dicot plants (7,8). 18S rRNA organizes small subunits of ribosome and is used on inferring angiosperm phylogeny (9). Chloroplasts are organelles that are found in plant cells and eukaryotic algae, and are the primary organelles involved in photosynthesis. Chloroplast DNA (cpDNA) has been used extensively to infer plant phylogenies at differing taxonomic levels (10–14). Molecular markers of rbcL and trnLF are located in the DNA of chloroplasts. rbcL encodes for the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase, which catalyzes carbon fixation in chloroplasts, and exhibits a low evolutionary rate that renders it useful for evolutionary studies at higher taxonomic levels (11,14). trnLF includes the 3′ partial sequence of trnL gene and a noncoding region that is the intergenic spacer between two tRNA genes, trnL(UAA) and trnF(GAA). Noncoding regions tend to evolve more rapidly than coding regions, and have therefore proven quite useful for evolutionary studies of related species, and probably also for populations of the same species (12,14).
After 17 days of disappearance, the victim in this case was found dead in his car, which had been submerged in a lake at hot summer (missing time: 23 July 2006, body recovery date: 9 August 2006). It was suspected that the victim had been murdered by poisoning with an excess amount of sleeping medication, which had been homogenized with dandelion. As the corpse was much decomposed and blood specimens were nonexistent, we analyzed spleen and liver instead of blood for the detection of drug substances. The concentrations of doxylamine and zolpidem detected from the victim’s internal organs were toxic and therapeutic levels, respectively.
The victim had cultivated dandelions, and routinely drank dandelion juice to ameliorate his liver trouble at 10 pm every night. One half-cup of dandelion juice in the morning or evening, taken on an empty stomach, is popular in Korea as an effective treatment of liver disorders and diseases. The victim had recently taken out mobile insurance when he was subjected to a traffic accident (in 2003) and when his wife became a beneficiary (US$40,000). Since then, he was suffering from trauma and insomnia symptoms and was prescribed with a nervous sedative and a sleeping drug (zolpidem). It was assumed that the suspect had mixed homogenized dandelion and sleeping drugs (doxylamine) to avoid detection by the victim. Thus, it was speculated that the suspect had compelled the victim to ingest a great deal of sleeping medication without being noticed because homogenized dandelion had a bitter taste itself. After the patient had drunk the dandelion juice on the night in question, he might have lost consciousness. It was assumed that the suspect might have positioned him in the car, and deposited the car and victim together into the lake, in an attempt to simulate a suicide. The police asked the forensic medicine doctor to autopsy the decedent as a component of the investigation into the death. The investigator suspected the victim’s wife of his death. The investigator also obtained the cellular phone records of the victim’s wife, and determined that she had been in constant telephone communication with someone on the evening prior to the victim’s death. When her husband died, she received 100 million won (US$100,000) in a life insurance payout. The couple had been in fairly strained financial circumstances and had barely been able to make the life insurance payments.
For the identification of dandelion substances, the stomach contents of the victim were analyzed by chemical and DNA analysis. In the chemical analysis, taraxasterol, one of the marker compounds in dandelions, was verified. As taraxasterol exists in various plants belonging to the Asteraceae family, DNA analysis is keenly needed. In the DNA analysis, the dandelion-specific sequences of rbcL and trnLF were confirmed. Out of four genetic markers such as ITS, 5′ partial region (about 411bp) of 18S rRNA, 5′ partial region (446 bp) of rbcL and trnLF, two markers of rbcL and trnLF included in cpDNA supported the presence of dandelion in the stomach of the victim.
Materials and Methods
DNA Analysis
Sample Collection— During the medicolegal autopsy, gastric content samples were collected as a component of the investigation into the victim’s death. Photomicrographs of the plant tissues in gastric content samples and grinded dandelion control tissues are shown in Fig. 1 (light microscopy, ×100). Gastric content samples were collected in three different forms such as green granule, pellet, and supernatant. At first, the police requested three control samples of the dandelions cultivated in the victim’s garden for comparison with the substances in the victim’s stomach (Fig. 2). Eight supplementary leaves of dandelion controls which were randomly collected from the victim’s garden are shown in Fig. 2.
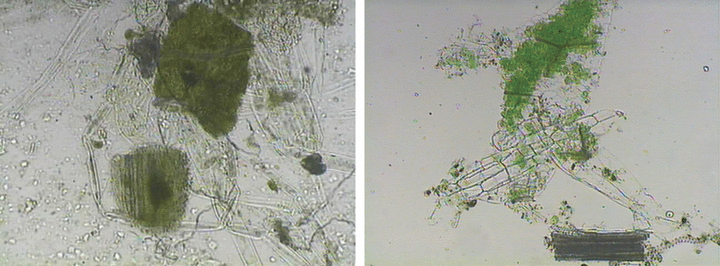
—Plant tissues in gastric contents (×100) are shown in the left panel and dandelion control tissues (×100) are shown in the right panel.

—First three control samples which were collected from the victim’s house are shown in the left panel and supplementary eight leaves are shown in the right panel.
DNA Extraction— Gastric samples were washed several times with sterilized distilled water and homogenized in a 1.5 mL tube using a plastic tool with a conical head in order to prevent sample loss. DNA was extracted from crushed green granule samples using a DNeasy Plant Mini Kit (QIAGEN Corp., GmbH, Hilden, Germany). DNA was extracted from crushed pellet samples and supernatant samples cooperatively using DNA IQ systems and Wizard Genomic DNA Purification Kit (PROMEGA Corp., Madison, WI). The isolated DNA was first quantified by agarose gel electrophoresis but the quantity of DNA could not be calculated because of its small amount. The isolated DNA might have the DNA of the epithelial cells of the victim which were collected unintentionally and might make it difficult to quantify only plant DNA by gel electrophoresis and spectrophotometer as these methods were not human-specific.
Molecular Markers and PCR Amplification— The universal primers for the species identification of land plants (terrestrial plants) and algae were employed in the amplification of ITS (6). Modified “e” primer (5′-AAAATCGTGAAGGTTCAAGTC-3′) and modified “f” primer (5′-GATTTGAACTGGTGACACGAG-3′) for the amplification of trnLF were used (12–14). The modified “e” primer was designed by Sang et al. (13) and was ten nucleotides farther away from the 3′ end of the trnL 3′ exon than the primer “e” of Taberlet et al. (12) in order to read sequences closer to the 5′ end of the intergenic spacer. The modified “f” primer had one more nucleotide at the 5′ end of the primer “f” of Taberlet et al. (12). The primer pairs were utilized in the amplification of the segment of rbcL (446 bp), which could be read from the translation start codon to 420 base pair. The forward primer for partial rbcL was 5′-AATTCATGAGTTGTAGGGAGGGA-3′ (bound to -26, designed by the first author, Eun-jung Lee), the reverse primer (making 446 bp) was 5′-AATTCGCAGATCCTCCAGACGT-3′ (designed by Eun-jung Lee) and other reverse primer (making 209 bp) was 5′-AGATTCCGCAGCCACTGCAGCCCCTGCTTC-3′ (rbc19 in Ref. 15). The forward primer for 5′ partial 18S rRNA was 5′-TGCAGTTAAAAAGCTCGTAG-3′ (Angio1F in Ref. 15), the reverse primer making about 411 bp was 5′-GGTTGAGACTAGGACGGTATCTG-3′ (designed by Eun-jung Lee) and other reverse primer making 157 bp was 5′-GCACTCTAATTTCTTCAAAG-3′ (Angio2R in Ref. 15). PCR amplification was conducted in a 50 μL total volume containing 5 μL of Gold 10× buffer (500 mM KCl, 100 mM Tris–HCl pH8.3, 15 mM MgCl2, 1% Triton X-100, 1600 μg/mL BSA, 2 mM dNTP pH8.2–8.4, PROMEGA Corp.), 2 μL of 10 μM forward and 10 μM reverse primers (synthesized by BIONEER Corp., Yu-sung, Daejon, South Korea), 2.5 units of AmpliTaq Gold® DNA polymerase, 2.5 μL of template DNA isolated from the gastric contents and sterile distilled water. The PCR mixture was subjected to dwelling 11 min at 95°C and 28 cycles of 1 min each at 95°C, 1 min at 50°C, 1 min at 72°C, and 7 min at 72°C using an ABI 9700 system (manufactured by Applied Biosystems, Foster City, CA). The second PCR, in which a component of the initial PCR products was used as a template (template was 1 μL of first PCR products), was applied. In the second amplification of rbcL and 18S rRNA, nested primers (15) were used for the survey of PCR specificity. The template of positive control for PCR is Papaver somniferum L. (opium poppy). The PCR reactions of DNA from dandelion reference leaves were conducted after the acquisition of genetic information from gastric contents.
DNA Sequencing— A 1/10 volume of the amplification products was assessed via electrophoresis on 2% agarose gels stained with SYBR™ Green I Nucleic Acid Stain (Cambrex Corp., East Rutherford, NJ). The remnants were then purified using a QIAquick PCR Purification Kit (QIAGEN). The purified PCR products were then sequenced directly using forward and reverse primers with an ABI PRISM BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). The unincorporated fluorescent dyes were eliminated using a DyeEX™ 2.0 Spin Kit (QIAGEN) and the purified sequencing products were dried approximately 1 h at 55°C. The sequencing products were then resuspended in 25 μL of Hi-Di formamide (Applied Biosystems). DNA sequencing was conducted using an ABI 310 sequencer and analyzed with DNA Sequencing Analysis Software 3.7 (Applied Biosystems).
Alignments and BLAST Query— The sequences of the molecular markers obtained from the samples were arranged using BioEdit (16), MEGA software (17) and ClustalX software (18) to determine whether they were identical or not. In case of the query of our rbcL segment to NCBI (National Center for Biotechnology Information), the 5′ region including the forward primer-binding region and some bases prior to the ATG start codon and the 3′ region including the reverse primer-binding region were excluded in order to improve the sequence specificity. A comparison was made between our secured sequences and the NCBI using BLASTn (nucleotide–nucleotide The Basic Local Alignment Search Tool) (19). For BLASTn, the Jukes-Cantor method is used to calculate the distances in NCBI. BLAST computes a pairwise alignment between a query and the database sequences and an implicit alignment between the database sequences is constructed for purposes of tree presentation of a query sequence (distance tree of results). Namely, genetic trees were simultaneously produced using BLAST pairwise alignments. Deferring to our options on BLAST of NCBI, the genetic tree method is the Neighbor-joining method, sequence label is taxonomic name, and maximum sequence difference is 0.75. Open dot represents eudicot (true dicotyledons) and black filled dot means unknown (our sequence) in the genetic tree (3, 4).
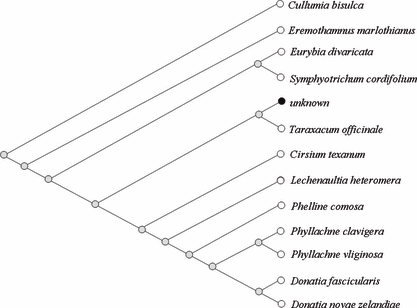
—Part of the genetic tree acquired from BLASTn which aligns our query data of rbcL (400 bp) sequences with database in NCBI. Our query data are named as “unknown” and topology is slanted type (tree method: Neighbor-joining method, sequence label: taxonomic name, maximum sequence difference: 0.75. Open dot represents eudicot and black filled dot unknown).
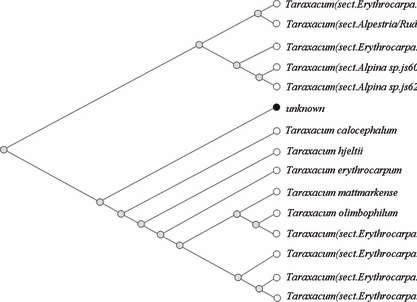
—Part of the genetic tree acquired from BLASTn which aligns our query data of trnL-trnF IGS sequences obtained from gastric contents with database in NCBI. Our query data are named as “unknown” and topology is slanted type (tree method: Neighbor-joining method, sequence label: taxonomic name, maximum sequence difference: 0.75. Open dot represents eudicot and black filled dot unknown).
Chemical Analysis
Extraction Procedure— Quantitative Analysis—2 g of spleen and liver were homogenized with 5 mL H2O and dextromethorphan (I.S., 100 mg/L) was added. The sample was mixed with 3 mL 10% trichloroacetic acid for its denaturation. After centrifugation at 1150 × g for 5 min, upper layer was transferred to a new tube. The solution was basified with 0.2 mL of 6N aqueous sodium hydroxide and was extracted with 10 mL of ethylacetate. After phase separation by centrifugation, the organic layer was transferred and evaporated to dryness. The residue was injected to gas chromatography–mass spectrometry (GC–MS). The comparison for identification of taraxasterol was made with the Wiley library and the retention times of standard samples.
For target drugs, the quantitation was calculated on the ratio of peak areas of doxylamine and zolpidem to those of internal standard (dextromethorphan). Doxylamine, zolpidem, and dextromethorphan were extracted at 58, 235, and 271 m/z respectively for quantitation. Three concentration points (5, 10, and 50 mg/L) were used for the calibration. The coefficient (r2) of calibration determination curve was 0.99.
Qualitative Analysis—In case of gastric contents, except denaturation step, qualitative analysis was followed upper sequence.
GC–MS System and Condition— The GC–MS system consisted of an Agilent series 6890 GC interfaced with a model 5973 Agilent mass selective detector. GC–MS analysis was conducted using a HP-5 MS column (cross-linked 5% methyl phenylsilicone, 30 m long, 0.25 mm i.d., 0.25 mm film-thickness). The column oven temperature was programmed to operate at an initial temperature of 100°C for 0.5 min, increased to 280°C at a heating rate of 20°C/min, and then maintained at 280°C for 10 min. The total run time of this method was 35 min. The temperatures of the injector port and the interface were set at 270°C and 280°C, respectively. The flow rate of the carrier gas (helium) was set at 1 mL/min, the ionization energy was set to 70 eV. Mass spectra were collected via scanning from m/z 50 to 550, at 2 sec intervals.
Results and Discussion
The plant tissue particles were extracted from the gastric contents of the victim. Nuclear genomic DNA and chloroplast genomic DNA were prepared simultaneously from the plant tissues. The genomic DNA concentration was insufficient for a verification of its intensity value via agarose gel electophoresis and it was difficult to discriminate plant DNA from the victim’s human DNA. There was some chance that stomach acids remained in the isolated DNA solution, and so a few template DNAs (2.5 μL) were employed in the PCR reaction. Primer pairs of ITS, 18S rRNA, rbcL, and trnLF were utilized for the amplification of each gene, but proved to be unproductive. In order to increase the magnitude of the amplification products, the second PCR method, in which a portion of the initial PCR products was used as a template, was applied. One microliter of the initial PCR products was used as the template for the secondary PCR reaction. Nested reverse primers of rbcL and 18S rRNA were used for the survey of the second PCR specificity. The reverse primer referred to in a previous paper by Rollo et al. (rbc19 and Angio2R in Ref. 15), which makes an rbcL of 209 bp and makes an 18S rRNA of 157bp, made amplicons successfully from the first PCR product of rbcL and 18S rRNA (Fig. 5). First PCR products of 209 bp-sized rbcL and 157bp-sized 18S rRNA appeared more abundant than longer amplicons did. Sequence comparisons of ≤200 bp-sized amplicons might be inefficient in species identification, due to their small sizes because small sizes harbor little information. The results of the second PCR proved adequate for the accomplishment of the next sequencing reactions. The outcomes of the second PCR of rbcL from all G, P, and S samples (in Fig. 5) and of trnLF from only P sample were successfully sequenced, but those of ITS and 18S rRNA were not. Sequences of rbcL and trnLF were queried as to whether NCBI possessed sequences related to them (19, Table 1).
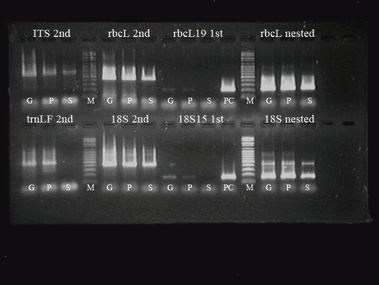
—The results of second PCR (ITS, rbcL, trnLF and 18S rRNA) and nested PCR (rbcL and 18S rRNA) of gastric samples (G: green granules, P: pellets, S: supernatant). First means the first PCR and second means the second PCR. “rbcL19 first” means the first PCR products of 209 bp-sized rbcL.
| Gene (Segment Size) | NCBI Accession # | Taxa | Identities (Homology, %) |
|---|---|---|---|
| rbcL (380 bp) | AY395562.1 | Taraxacum officinale (dandelion) | 380/380 (100) |
| rbcL (400 bp) | AY874432.1 | Ainsliaea acerifolia (Danpoong-Chwi) | 397/400 (99) |
| rbcL (446 bp) | DQ383816.1 | Lactuca sativa (lettuce) | 439/445 (98) |
- All trimmed segments showed highest homology with different taxa mutually dependent on their compared sizes.
In rbcL, sequences of rbcL from all G, P, and S samples were identical among them. Our 446 bp sequences including both primer-binding sites exhibited 98% homology with rbcL of Lactuca sativa (439/445 bp, Table 1) and 400 bp sequences exhibited 99% homology with that of Ainsliaea acerifolia (397/400 bp). Every species exhibiting sufficiently high degrees of homology with our 400 bp rbcL in the NCBI were members of the Asterales order. In particular, six species such as Cullumia bisulca, Eremothamnus marlothianus, Eurybia divaricata, Symphyotrichum cordifolium, Taraxacum officinale, and Cirsium texanum were included in the Asteraceae family (Fig. 3). It has been estimated that the rbcL gene can be used to determine the order or family of unknown plant evidence and this study proves this assumption.
In trnLF, all species exhibiting higher levels of homology with our trnLF in the NCBI database were found to be members of the Taraxacum genus (Fig. 4). These facts indicate that the study of rbcL is not as advanced as the study of trnLF, or that registered sequences in NCBI as the same gene are read from different PCR primer pairs by various researchers, or possibly that rbcL, which shows a low evolutionary rate, cannot be used for the discrimination of genus or species. However trnLF, which has a rapid evolutionary rate, was capable of discriminating between species and intraspecies in this study. Different PCR primer pairs that bind at various sites in the same gene make it difficult to compare uniform nucleotides, hence the lead researcher to obscure directions. If our 446bp rbcL sequence was trimmed away 46 and 20 nucleotides at 5′ and 3′ region, respectively, it showed 100% homology with rbcL of T. officinale [AY395562] (Table 1). The reason for these phenomena is thought to be that sequences which showed higher homologies with our queried sequences in BLAST of NCBI are ranked in due order according to their compared sequence sizes.
After securing the sequences of rbcL and trnLF from the stomach contents (about 3 days later), we conducted control experiments in order to compare the sequences of the stomach contents with the sequences of three dandelion reference samples cultivated within the victim’s house. These time-differential experiments were to prevent the contamination of case samples by control samples. In rbcL, the sequences from stomach contents showed 100% homology with the sequences from three dandelion references, and showed no nucleotide length polymorphisms, such as insertions and/or deletions (data not shown). However, in trnLF, the sequences from the stomach contents did not manifest 100% homology with the sequences from the three dandelion references. Control #1 exhibited a 98.7% homology with the stomach sequences (1 bp insertion at 354 bp, 4 transitions/1 transversion, Fig. 6) but controls #2 and #3 showed differences in size (77 bp shorter than stomach sequences) as well as in sequences (1 bp insertion at 354 bp, 7 transitions/4 transversions). Seventy-three base pairs of the trnLF 3′ end regions from controls #2 and #3 were repeated next to the trnF gene of the stomach sequences and control #1 (Fig. 6). They differed primarily as the result of a polymorphic tRNA pseudogene located next to the trnF gene (20).
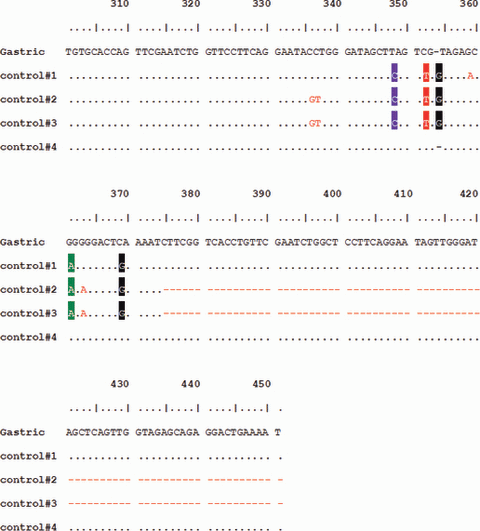
—Nucleotide sequences of trnL-trnF IGS of plant tissues from the gastric contents (named as “Gastric”) and of control dandelions.
Looking for the same sequences with the trnLF from the stomach contents, we asked the police to collect more dandelion samples cultivated within the victim’s house. Many leaves were collected piece by piece, and eight randomly chosen leaves (Fig. 2) were subjected to the same experiments. Among eight samples, only one had the same sequence with the trnLF from the stomach contents (Table 2, Con #4), another six samples displayed the same sequence with control #1 (Con #5, 6, 7, 8, 10, 11), and the last one (Con #9) showed the same sequence with control #2, #3. Needless to say, the rbcL sequences of the eight additional control samples showed no sequence variations with gastric sequences. According to the opinion of the plant morphologist, the species of dandelion cultivated within the victim’s house was T. officinale.
| Amplicon | GC | Con. #1 | Con. #2 | Con. #3 | Con. #4 | Con. #5 | Con. #6 | Con. #7 | Con. #8 | Con. #9 | Con. #10 | Con. #11 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rbcL | 446 | 446 | 446 | 446 | 446 | 446 | 446 | 446 | 446 | 446 | 446 | 446 |
| trnL-trnF | 492 | 493 | 416 | 416 | 492 | 493 | 493 | 493 | 493 | 416 | 493 | 493 |
- Detection frequency of 492bp trnL-trnF IGS is 16.7%.
The segment of rbcL gene can be used to determine the family or subfamily of unknown plant evidence. Discrepant results from the query of our rbcL (380 bp or 400 bp or 446 bp, Table 1) and from the query of our trnLF to NCBI might be caused by the lack of international agreement about plant DNA barcodes which can standardize PCR primer pairs of the same gene. Nucleotide sequences read by different primer pairs for the same gene manage to guide us to the discrimination of the family or genus of unknown samples.
Universal markers including ITS (intergenic spacer region) and 18S rRNA are powerful with regard to the resolution of the phylogenetic relationships between plants (7–9), but were unsuitable for the DNA analysis of gastric contents, because their primers could bind to the ITS and 18S rRNA of humans (refer to NCBI accession number U13369) (21,22). A few of the gastric epithelial cells from the victim remained mixed with the plant cells, which made it difficult to determine the ITS and 18S rRNA sequence of the plant evidence via direct sequencing. From this study, markers on cpDNA are recommended in attempts to identify gastric contents and to identify other plant evidence from human cell mixed samples.
Gastric content, spleen, and liver of the victim were submitted to analyze whether they had drug or toxic compounds. Doxylamine and zolpidem, kinds of hypnotics, were detected in gastric content (data not shown). At quantitative analysis in spleen and liver, we found that the concentrations of doxylamine were 11.4 and 27.5 mg/kg, respectively and the concentration of zolpidem was <0.10 mg/kg in the spleen and liver. It was reported that the assumed therapeutic concentration of doxylamine in liver ranged from 0.2 to 1.7 mg/kg which were calculated from postmortem specimens in nonintoxication cases (25,26). The concentration of doxylamine in victim specimens was far higher than the assumed therapeutic concentration in liver and was evaluated as a toxic level. In the case of zolpidem, it had been reported that the therapeutic concentration in blood was from 0.06 to 0.27 mg/L (27). The concentration of zolpidem in victim specimens was lower than therapeutic concentration in blood and so was evaluated as therapeutic level.
The mass spectrum from the GC–MS analysis of the gastric contents is shown in Fig. 7. The comparison for identification was made with the Wiley library and the retention times of standard samples. Sisosterol and stigmasterol substances of dandelion were also detected from the gastric juice of the victim by GC–MS analysis (data not shown). Although taraxasterol is a marker compound of dandelion, many plants such as Daucus carota (carrot), Glycine max (soybean), Vitis vinifera (grape), Helianthus annuus (sunflower), Latuca sativa (lettuce), etc., have taraxasterol (23).
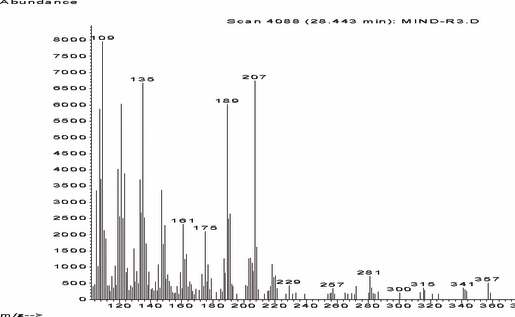
—Mass spectrum of taraxasterol from the gastric contents. The comparison for identification was made with the Wiley library and the retention times of standard samples.
From these DNA and GC–MS results, it was assumed that raw dandelion had been used as a masking vehicle for excess sleeping drugs. Food remains in the stomach for approximately 2–6 h in what amounts to an acid bath, in which mechanical churning and grinding occurs. Some factors affecting the rate with which the stomach empties include caloric content, food acidity, meal volume, gender, etc. (24). The combination of the victim’s empty stomach and hypochlorhydria symptom that was chronic with the victim and the unripe juice, in this case, allowed for the effective isolation of the dandelion DNA.
Acknowledgments
We want to thank Mu Yeol Kim (the professor in Chonbuk National University) for the morphological species identification of dandelion samples cultivated at the victim’s house.




