Cloning and Sequence Analysis of Translational Release Factors eRFl and eRF3 of Pneumocystis carinii
Translation termination requires two polypeptide release factors–eRF1 and eRF3–in eukaryotes. eRFl plays a direct role in reading the three stop codons, while eRF3 binds guanine nucleotides and stimulates the eRFl activity by forming a stable complex with eRFl [1], Both eRFl and eRF3 are essential housekeeping proteins. The eRFl sequences are well conserved among various eukaryotic species, and many eRFl genes are functionally exchangeable, such as between human and the budding yeast, Saccharomyces cerevisiae. On the other hand, the eRF3 sequences possess a unique property. The C-terminal part of eRF3, including the G domain, is highly conservative and homologous to elongation factor EF-1α, whereas the N-terminal part is highly divergent. In S. cerevisiae, eRF3 is known to form aggregates-referred to as the [PSI+] element-that self-propagate and are transmitted cytoplasmically in the manner of the ‘protein-only’ transmission of mammalian prion diseases [2]. The N domain of eRF3 is prion-determining and contains tandem imperfect oligopeptide repeats (PQGGYQQYN) similar to those (PHGGGWGQ) of the mammalian prion determinant, PrP. The [PSI+] property of eRF3 is widely distributed in yeast species [3] with some exceptton(s) such as the fission yeast, Schizosaccharomyces pombe [4]. In this study, we cloned and analyzed the cDNAs and genomic sequences for eRFl and eRF3 from rat Pneumocystis carinii, which is classified as fungus, to examine the phylogenetic diversity and conservation of translational release factors.
MATERIALS AND METHODS
P. carinii RNA and DNA were prepared as described [5]. The eRFl and eRF3 cDNAs of P. carinii were synthesized from P. carinii RNAs by the RACE (rapid amplification of cDNA ends) method according to manufacturer's instruction (GIBCO BRL). The primers were designed from the putative eRF1 and eRF3 sequences found in the EST database (http://biology.uky.edu/Pc). The eRF1 and eRF3 genomic sequences were amplified by PCR from P. carinii DNA using primers that encode the cDNA terminal sequences. The sequences were analyzed by Wisconsin Package Version 10.2 (Genetics Computer Group).
RESULTS AND DISCUSSION
Partial sequences of putative eRFl and eRF3 genes of P. carinii were identified in the P. carinii EST database by means of homology to the overall sequence of eRF1 and to the C-terminal sequence of eRF3, respectively. The full-length cDNAs for eRFl and eRF3 were then isolated by the RACE method using these sequences, and subjected to nucleotide sequence analysis. The resulting data will appear in the DDBJ/EMBL/GenBank nucleotide sequence databases with the accession number AB052893 (eRFl) and AB052894 (eRF3).
The data indicated that P. carinii eRFl (Pc-eRFl) is composed of 432 amino acids and shares 80% and 78% sequence homology with S. cerevisiae eRFl and S. pombe eRFl, respectively. The deduced P. carinii eRF3 (Pc-eRF3) structure is, as predicted, composed of N-terminal non-conservative (196 amino acids) sequence and C-terminal conservative (433 amino acids) sequence (Fig. 1A). The latter part of Pc-eRF3, including the G-domain motifs, which is proximal to its N-terminus, is 77% homologous to S. cerevisiae and S. pombe eRF3s. On the other hand, the N-terminal part of Pc-eRF3 is rich in positive or negative charge residues (i.e., one-third amino acids are Asp, Glu, Lys and Arg; see Fig. 1B), and shares no homology with any known eRF3 sequences: hence, referred to as ‘charged N-domain’. The charged N-domain of Pc-eRF3 is slightly shorterthan those of S. cerevisiae eRF3 (253 residues) or S. pombe eRF3 (232 residues), and is free from apparent oligopeptide repeats like the prion-determining elements [3].
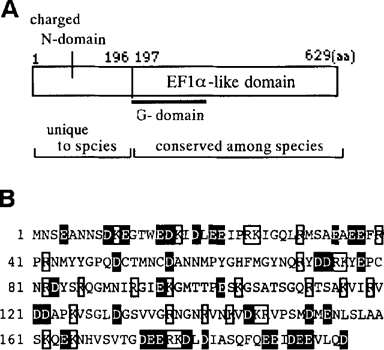
The protein property of P. carinii eRF3. (A) Two domain structures as revealed by protein sequence diversity and conservation. (B) The protein sequence of the charged N-domain. Charged residues are marked by boxes (positive) or in white on black (negative).
The genomic sequences for eRFl and eRF3 were amplified from P. carinii DNA by the PCR method using primer sequences of the cDNA end sequences. The deduced eRF1 and eRF3 genes are composed of 1884 and 2323 nucleotides, respectively. Surprisingly, when compared with their cDNAs, both genes appeared to contain unusually multiple introns, i.e., 13 (Pc-eRFl) and 10 (Pc-eRF3) introns (Fig. 2). To our knowledge, this feature is unique among phylogenetically related release factors. Most of yeast and fungus eRF3s do not encode an intron and S. pombe eRF3 may be the rare exception as it encodes one intron [4]. All introns of Pc-eRFl and Pc-eRF3 are similar in the sizes (39–52 bp) and retain the acceptor/donor site consensus sequence for splicing, GT/AG, but do not conserve the apparent branch point consensus as found in other genes [6]. Functional study of P. carinii release factors is in progress in this laboratory.
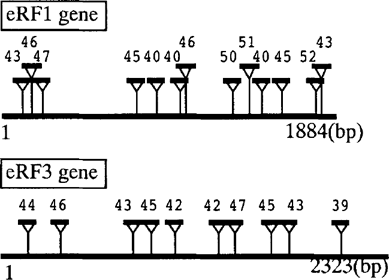
Schematic diagrams of the eRFl and eRF3 genes of P. carinii. The defined introns are marked by triangle inserts with nucleotide size.




