Immunohistochemistry Based Assay to Determine the Effects of Treatments on Cryptosporidium parvunt Viability
SUMMARY
Cell culture infectivity assays can provide an accurate means of detecting viable Cryptosporidium parvum oocysts from environmental samples or to test the effects of various treatments on oocyst infectivity. Cell culture assays can also be used to test candidate chemotherapeutic agents. The use of a human cell line provides a situation close to human infection. The present assay uses an anti-Cryptospordium primary antibody, combined with a biotinylated secondary antibody, and an immunoperoxidase detection system. Cryptosporidium parvum oocysts excysted in vitro when placed on monolayers of HCT-8 cells and developmental stages including schizonts and merozoites were visualized using light microscopy of the immunoperoxidase stained slides and by transmission electron microscopy of infected HCT-8 cell cultures. Because the immunoperoxidase system used gives a permanent preparation, the cell cultures can be retained and examined later. Dose titration of oocysts indicated that as few as 50 inoculated oocysts could be detected. The activity of paromomycin was evaluated in this system and 500 μg/ml produced a 97.8% reduction in infection.
Cryptosporidium parvum is an Apicomplexan protozoan parasite that is a cause of waterborne and foodborne outbreaks of diarrheal disease [2–10,14]. Infection occurs by ingestion of the thick-walled oocysts. CHnical disease caused by C. parvum is manifested by gastrointestinal illness lasting 1–2 weeks in previously healthy individuals or lasting indefinitely in immunocompromised individuals. Currently, there is no uniformly successful treatment for cryptosporidosis. An effective in vitro assay is essential for assessing the efficacy of potential therapies. Viability assays using vital dyes and oocyst excystation are not reliable and the use of animal models is often expensive and time-consuming [1,11,12]. Infectivity for cell cultures has been determined by immunofluorescence, but may have high background staining and slides cannot be held indefinitely and observed at a later date. The present study describes a reliable infectivity assay using cell culture and an immunoperoxidase staining procedure to rapidly detect C. parvum developmental stages.
MATERIALS AND METHODS
Human illeocecal adenocarcinoma cells (HCT-8 cells) (ATCC CCL244) were maintained in RPMI 1640 media (Mediatech Cellgro) supplemented with L-glutamine (300 mg/L), and HEPES (25 mM), 5% fetal bovine serum for maintenance and 10% fetal bovine serum for infection [13]. Stock HCT-8 cells were maintained in 75-cm2 tissue culture flasks in 5% CO2 atmosphere, at 37°C, and passaged every 3–5 days. Collected cells were grown on sterile glass coverslips in 6-well plates at 1 × 106 cells per well and grown to ∼95% confluency (48 hours). Cryptosporidium parvum oocysts (Beltsville isolate) were treated with 0.525% sodium hypochlorite at 4°C, washed twice with Hanks Balanced Salt Solution (HBSS), incubated with the treatment of interest, and washed twice with HBSS.
Cells were then incubated with treated or non-treated (positive control) oocysts for 90–120 minutes and washed twice with HBSS. Cells were then grown for 48 hours with maintenance media.
Coverslips were fixed with 100% methano! for 20–30 minutes. Coverslips were then processed using a rabbit anti-Cryptosporidium parvum primary antibody (courtesy of C. Dykstra, Auburn University) and a biotinylated anti-rabbit secondary antibody (Vectastain ABC Kit, Vector Laboratories). Life stages were visualized with an immunoperoxidase stain using hydrogen peroxide, diaminobenzidine tetrahydrochloride (DAB, Sigma), with hematoxylin used as a counter stain.
As a negative control, oocysts were frozen in liquid nitrogen for two hours, prior to host cell inoculation. Selected cultures were fixed in 3% gluteraldehyde in 0.1M sodium phosphate buffer 2 days after inoculation of oocysts, and processed for transmission electron microscopy (TEM).
Paromomycin (Sigma, St. Louis. MO) was used as a positive anti-C parvum drug to determine the ability of our assay to detect reduction in developmental stages in treated cell cultures. Infected cell monolayers were incubated with media containing 5% fetal bovine serum and 500 μg/ml paromomycin. Paromomycin-treated cultures were compared to infected non-treated cultures. Treatment effectiveness was determined by counting the number of positive fields out of one hundred total fields. Percent reduction compared to control untreated oocysts was determined using the following equation: [(Control-Treated)/Control] × 100.
RESULTS AND DISCUSSION
Immunoperoxidase staining of HCT-8 cell cultures inoculated with viable oocysts demonstrated dark brown stained developmental stages of C. parvum against a light blue host cell cytoplasm background produced by the hematoxylin counter stain. Developmental stages were usually observed in clusters indicating that development occurred at the site of excystation. No brown staining stages were observed in HCT-8 cells inoculated with frozen oocysts and processed using the immunoperoxidase procedure. This indicates that residual nonexcysted oocysts or nonviable oocysts are washed off in our procedure and not detected by the immunoperoxidase stain. Based on examination of one hundred 40× microscopic fields, infection by 1 × 106 untreated oocysts consistently produced between 90–98% infection. Detection is possible with an initial infection of as few as 50 oocysts (1% infection) or 100 oocysts (1%-20% infection), but detection is enhanced when ≥10,000 oocysts are used. Experiments and treatments were evaluated a minimum of three times.
Schizonts and merozoites were observed 48 hr after infection using TEM (Fig. 1). They were located in a parasitophorous vacuole within the microvilli of HCT-8 cells. The TEM results clearly demonstrate that development of C. parvum occurred in the HCT-8 cells.
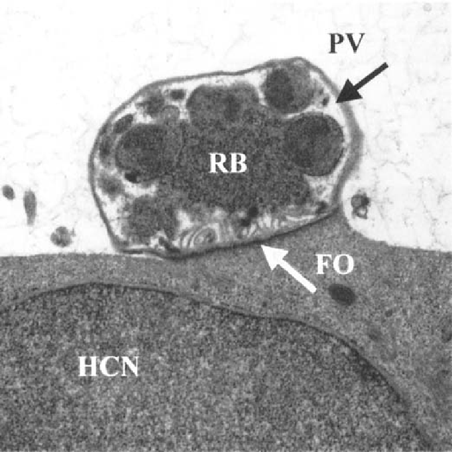
Transmission electron microscopy of a Cryptosporidium parvum schizont growing in an HCT-8 cell. Parasitophorous vacuole (PV), feeder organelle (FO), residual body (RB), and host cell nucleus (HCN) are indicated
This assay demonstrated that treatment with 500 μg/ml paromomycin produced a 97.8% reduction in developmental stages based on five experiments. Treatment with paromomycin serves to validate the assay.
This method provides a relatively simple and reliable means for determining the effects of treatments on Cryptosporidium parvum viability in an in vitro assay. This method provides permanent slide preparations that can be examined independently and in a blinded fashion by a single or several evaluators.
ACKNOWLEDGMENTS
Supported in part by CSREES USDA special food safety grant No. 98–34382–6916 and VPI & SU HATCH project #135563.




