Genetic Characterization of Cryptosporidium parvum Isolates from Cattle in Portugal: Animal and Human Implications
Cryptosporidium parvum (APICOMPLEXA: COCCIDIA) is an opportunistic protozoa with a worldwide distribution. The parasite causes diarrhea in humans and in livestock -mainly neonatal diarrhea with high morbidity, variable mortality and economic losses. In calves, diagnosis is impaired by the presence of other agents such as Rotavirus, Coronavirus, E. coli K99, and Salmonella spp. Several waterborne eryptosporidiosis outbreaks have been reported [5]. C. parvum is associated with either innate or acquired imunodeficiency. Zoonotic transmission does occur and several genotypes of C. parvum are well known. PCR-RFLP is one of the techniques applied to study genetic polymorphisms. In Portugal, the prevalence of bovine eryptosporidiosis ranges between 12% in the North and 44% in the South [Pereira da Fonseca, I.M. & Cannas da Silva, J. 2000. Bovine eryptosporidiosis: epidemiological study in Portugal. Proceedings XXI Congreso Mundial de Buiatria, Punta del Este, Uruguai, 4-8 Dez., 2000]. The objectives of this work were: (i) to study the prevalence of eryptosporidiosis in cattle from several age groups at “Alentejo” and “Ribatejo e Oeste” regions of Portugal by EIA-ELISA; (ii) to detect oocysts in animal drinking water, in order to evaluate its importance as a risk factor for eryptosporidiosis; (iii) to determine the genotype of C. parvum isolates and (iv) to evaluate the importance of the obtained data in Public Health.
MATERIALS AND METHODS
Epidemiological study
A total of 553 faecal samples from cattle were collected between January 1996 and February 1998. At “Alentejo”, 456 samples were obtained and at “Ribatejo e Oeste” the number of samples was 97. Twelve age groups were defined: Group 1 (1–days old), group 2 (8-15), group 3 (16-24), group 4 (25-32), group 5 (33-66), group 6 (67-100), group 7 (101–34), group 8 (135-180), group 9 (181-270), group 10 (271-360), group 11 (361-720) and group 12 (721-1440). The EIA-ELISA was performed using the kit “Melotest Cryptosporidium Ag” from Redifarma, Spain. Positive results for C. parvum were confirmed by faecal smears stained with modified Zielh-Neelsen.
Genotyping of C. parvum isolates
Genomic DNA was extracted from 186 samples (128 positives and 58 negatives) and purified by two methods: guanidinium thiocyanate/silica and proteinase K/heat followed by ethanol precipitation. Two pairs of primers were used in the PCR [3,4]. One of the primers was directed to a 550 bp region of the Cryptosporidium oocyst wall protein (COWP) gene [Cry-15: 5′-GTA GAT AAT GGA AGA GAT TGT G-3′and Cry 9: 5′-GGA CTG AAA TAG AGO CAT TAT CTT G-3′] (94°C/50”, 55°C/30″, 72°C/45″×45 and 72°C/10′) and other to a 506 bp region of the thrombospondin-related adhesive protein of Cryptosporidium-1 (TRAP-C1) gene [Et.E: 5′-GGA TGG GTA TCA GOT AAT AAG AA-3′and Et.W: 5′-CAA TTC TCT CCC TTT ACT TC-3′] (94°C/50″, 50°C/30”, 72°C/45″×40 and 72°C/10′). The PCR-RFLP was performed with the restriction enzyme Rsal. The PCR amplified DNA and the restriction fragments were analysed on a 2% agarose electrophoresis gel stained with ethidium bromide and visualized by UV transillumination.
Oocysts detection in animal drinking water
Between May 1998 and January 2000, 11 samples of 10 litres each were collected at three dairy farms both from the tap and the calf bucket. Method 1622 of the United States Environmental Protection Agency (US EPA) was used to detect Cryptosporidium oocysts [6]. Filtration was performed with Envirocheck capsules (Gelman); for the immunomagnetic separation of the ooeysts, the kit “Dynabeads anli-Cryptosporidium (Dynal) was used. The pellet obtained was stained with monoclonal antibodies (kit Monofluo Cryptosporidium -Sanofi) and 4′,6-diamidino-2-phenylindole (DAPI) was used to evaluate oocysts viability.
RESULTS AND DISCUSSION
A global prevalence of 23.3% (129/553) was found being 21.1% (96/456) in “Alentejo” and 34.0% (33/97) in “Ribatejo e Oeste” regions using the EIA-ELISA technique. Group 3 with 47.2% (17/36) and group 2 with 41.7% (53/127) were the age groups which showed higher prevalences of eryptosporidiosis. Infections of 9.5% (2/21) and 10.8% (4/37) were detected in calves (1–7-days old) and adults, respectively. These animals were assymptomatic carriers.
The amplification results of COWP and TRAP-C1 genes by PCR in the two studied regions showed that 58.6% (14/24) of the isolates were positive for TRAP-C1 from which 71.4% were from “Ribatejo e Oeste” and 40.0% from “Alentejo”. For COWP, the genomic DNA was amplified in 25.0% (6/24) of the isolates; 14.3% were from “Ribatejo e Oeste” and 40.0% from “Alentejo”. TRAP-C1+ COWP were detected, in 16.7% (4/24) of the samples; 14.3% were from “Ribatejo e Oeste” and 20.0% were from “Alentejo”. The sample sizes were too small to allow meaningful statistical analysis on geographical differences.
After PCR-RFLP of the 24 isolates, only the cattle genotype was found (Fig. 1).
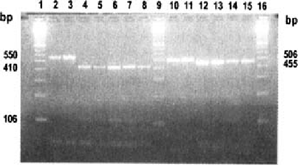
Amplification and Rsal restriction length polymorphisms of the TRAP-C1 and COWP genes. Lanes 1, 9, 16 –100 bp ladder; lanes 2 and 3 -PCR products of COWP gene: 550 bp; lanes 4 to 8 -hydrolysis fragments of COWP gene with Rsal (410, 106 and 34 bp); lanes 10 and 11 -PCR products of TRAP-Cl gene (506 bp); lanes 12 to 15 -hydrolysis fragments of TRAP-C1 gene with Rsal (455 and 51 bp).
The human genotype has different polymorphism patterns -TRAP-Cl: 314, 114 and 51 bp and COWP: 285,125,106 and 34 bp [2].
Analysis of animal drinking water showed that 6 out of 11 (55.0%) samples were positive. At farms A and B, 50.0% (2/4) of the samples had Cryptosporidium oocysts. All the samples were collected from the tap. At farm C, 66.7% (2/3) of the samples were positive; one from the calf bucket and the other from the tap. The number of Cryptosporidium oocysts detected in the water samples ringed between one to nine. Oocyst recovery rates for positive controls were 22.5% and 13.0%. The results of the viability study with DAPI on the six positive samples were negative. Cryptosporidium oocysts were detected in the animal drinking water of the three farms in every season evaluated [1].
In conclusion, the prevalence of bovine cryptosporidiosis was high at the two studied regions. The only genotype found was the bovine type, which is common to animals and man, suggesting that C. parvum transmission is not necessarily a zoonosis, although zoonotic transmission can also occurs. Bovine play an important role as reservoirs for human cryptosporidiosis and also as a source of environmental contamination. Although viability studies with DAPI were not conclusive, oocysts were observed in water used for calves. This fact can contribute for the spread of animal cryptosporidiosis, environmental contamination and public health risks. Due to the small number of oocysts found in the water samples, determination of the isolates'genotype was not possible. Cryptosporidiosis study at Portugal needs to be pursuit by genotyping more isolates from faecal, animal (drinking water and farm effluent samples using other molecular markers.
ACKNOWLEDGMENTS
Prof. Olga Matos and Dr. Margarida Alves from Instituto de Higiene e de Medicina Tropical, Lisboa, for laboratorial assistence in isolates genotyping; Dr. Leonor Falcao and Dr. Ana Cristina Almeida from Institute Nacional de Saude Dr. Ricardo Jorge, Lisboa, for their advice in the water study and Prof. Graça Ferreira-Dias for manuscript revision.
[Supported by Centra de Investigação Interdisciplinar em Sanidadc Animal (CIISA)]




