Prevalence of type 2b ‘Malmö/New York’ von Willebrand disease in Italy: the role of von Willebrand factor gene conversion
von Willebrand disease type 2B (VWD2B) is due to a unique gain-of-function variant of von Willebrand factor (VWF) that spontaneously interacts with circulating platelets, usually resulting in loss of VWF high molecular weight multimers (HMWM) in plasma and, in most cases, low platelet counts, especially after stress situations [1,2]. Diagnosis of VWD2B is based on heightened ristocetin-induced platelet aggregation (RIPA) in platelet-rich plasma (PRP). VWD2B ‘Malmö [3] or New York [4]’, previously reported as type I, is associated with increased RIPA at low concentrations of ristocetin but normal HMWM and no thrombocytopenia after stress situations. This peculiar VWD2B variant is caused by the mutation 3797C>T (P1266L) [5] in the VWF gene [6]. However, the majority of previously reported patients carrying the mutation P1266L show more than one nucleotide substitution [5,7–10], all matching the published VWF pseudogene sequence [11]. This finding, first described in VWD by Eikenboom et al. [7], has been explained by a mechanism of gene conversion between the VWF gene and its pseudogene.
The aims of this study were to determine the prevalence of this mutation in a large cohort of VWD2B patients enrolled in the Italian Registry of VWD and to evaluate whether or not gene conversions play a role in generating the mutations identified. Criteria for VWD2B were those recommended by the International Society on Thrombosis and Haemostasis Scientific and Standardization Committee VWF Subcommittee (ISTH-SSC on VWF) [2]. RIPA and multimeric analyses were performed as previously reported [12,13]. Patients’ bleeding severity score was obtained using a detailed questionnaire [14]. Platelet counts had been obtained over 2 years at baseline and after physiologic (pregnancy) or pathological (infections, surgeries) situations or by the use of desmopressin. Among 1234 patients enrolled in the Italian Registry, 66 (35 unrelated families) were diagnosed with VWD2B because of RIPA < 0.7 mg mL−1 (normal range 0.7–1.2 mg mL−1). DNA sequence analysis was performed using oligonucleotides specifically designed to amplify selectively the VWF gene. The 5′ portion of exon 28 encoding for the VWF A1 domain was sequenced in all VWD2B cases with a normal VWF multimeric pattern.
Four unrelated families (13 individuals) had a normal multimeric pattern in plasma and no thrombocytopenia before and after stress situations. Mutation 3923G>T (R1308L) was responsible for this phenotype in one family (five patients), and has been documented in a previous study [15]. In the remaining three families (seven patients) more than one mutation was found, but all patients shared a substitution of proline 1266. The following nucleotide changes were identified: 3686T>G (V1229G), 3692A>C (N1231T), 3735G>A, 3789G>A and 3797C>T (P1266L) in family A (three patients), 3789G>A and 3797C>T (P1266L) in family B (two patients) and 3692A>C (N1231T), 3789G>A and 3797C>A (P1266Q) in family C (two patients) (Fig. 1). Nucleotide numbers are reported according to the VWF cDNA sequence [16].
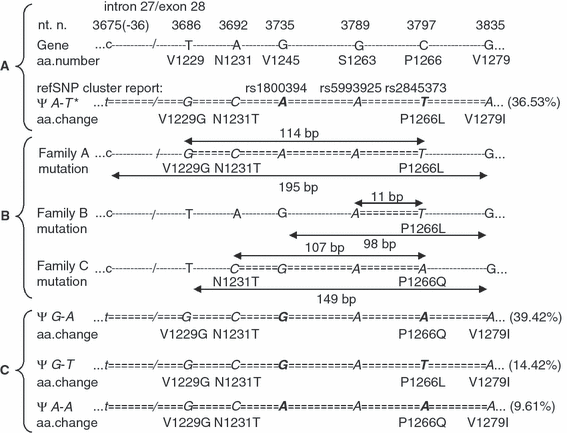
(A) Schematic representation of intron 27/exon 28 boundary of the VWF gene and pseudogene (Ψ). Nucleotide and amino acid numbers refer to the VWF cDNA sequence [6,16]. Only amino acid changes are reported in the pseudogene sequence. *Pseudogene sequence reported by Mancuso et al. [11]. The rs1800394, rs5993925 and rs2845373 identified the three reported SNP in the homonymous data base. (B) Nucleotide and amino acid changes found in family A, B and C. The minimal and maximal extent of gene conversion, in each family, is shown by the double-heads arrows. (C) The three additional pseudogene haplotypes identified in this study. Nucleotide changes are reported in bold. In brackets are reported the percentage of the different pseudogene haplotypes in the normal Italian population. Due to the technique used, the haplotype frequencies reported only regard nucleotides at position 3735 and 3797. The remaining nucleotides are ‘assumed’ to correspond to the sequence described by Mancuso et al. [11], confirmed in eight cases by direct DNA sequences.
The P1266L/Q genotypes correlate with the patients’ bleeding history and laboratory tests. Patients’ bleeding severity score was high (mean 2.85, range 1–6), although less than in other VWD2B variants (8, range 4–14; normal range 0 to −1)[14]. These lower bleeding severity scores are consistent with the mildly reduced or normal VWF levels and activities, reported herewith as mean values (ranges): VWF: Ag 56.4 (30–73) IU dL−1, and VWF:RCo 47 (21–66) IU dL−1, increased RIPA 0.53 (0.40–0.70) mg mL−1, presence of the HMWM and normal platelet count. We have further investigated additional causes for the bleeding tendency of these patients. However, only in family A did two patients show mildly reduced platelet secretion after stimuli with ADP, as reported by others [17]. VWF:CB (mean values 56.8, 48–64 IU dL−1) could be tested only in patients with mutation P1266L, and were similar to those of VWF:RCo (56.8, 45–66 IU dL−1), at variance with the discrepant values reported in VWD2B patients carrying the R1308L mutation [15].
The finding of several nucleotide substitutions in all the patients from each family suggested that all defects were on a single allele. As previously reported in several diseases [18], these multiple nucleotide substitutions were probably due to gene conversion mechanisms. In VWD the conversion between VWF gene (chromosome 12) and its pseudogene (chromosome 22) is interchromosomal and appears to be caused by a pair of chi-like sequences found in intron 27 and exon 28 [7]. In families A and B, from northern Italy, all substitutions indeed corresponded to the pseudogene sequence [11], whereas in family C, from Vietnam, nucleotides 3735G and 3797A did not match the pseudogene sequence (Fig. 1). Mutation 3797C>A (P1266Q) was previously reported in the ISTH-SSC on the VWF data base (http://www.vwf.group.shef.ac.uk/; accessed 14 February 2008) in a VWD2B patient who had a second mutation (3835G>A, V1279I) in the same allele. Due to the fact that mutation V1279I corresponds to the pseudogene sequence, we assumed that these defects were due to a gene conversion but with a pseudogene presenting a different sequence. To confirm our hypothesis, we amplified and sequenced, using oligonucleotides selective for the VWF pseudogene (5′-tgtagggctcagaagtgtccat, 5′-gccacgcggacccacttcta), eight normal individuals from Italy, Russia, Iran and India. Unexpectedly, three of them were homozygous for nucleotides 3735G and 3797A, four were double heterozygous for both 3735G/A and 3797A/T, and one was heterozygous for 3735G/A and homozygous for 3797T. Frequency of the different pseudogene sequences in the Italian population was evaluated using four primers to selectively amplify nucleotides 3735A (5′-gccgggaggcctggtggta), 3735G (5′-gccgggaggcctggtggtg), 3797T (5′-gctgcagtagaaatcgtgcaact) and 3797A (5′-gctgcagtagaa atcgtgcaaca). Four PCR were performed for each DNA sample using two primers at the time of the four possible combinations. PCRs showing an amplification product revealed, on the base of primers used, the nucleotide present at positions 3735 and 3797 on the same allele. Four different haplotypes were identified out of 104 chromosomes investigated, 39.42% with the sequence 3735G-3797A, 36.53% with the sequence 3735A-3797T (reported by Mancuso et al.) [11], 14.42% with the sequence 3735G-3797T and 9.61% with the sequence 3735A-3797A (Fig. 1). In addition, as patients with mutation P1266Q were originally from Vietnam, and no DNA samples were available to us from that population, we investigated 20 individuals from Iran and 20 from India to have some information regarding other populations. Sequence 3735G-3797A was the most common (52.5% in Iran and 62.5% in India), followed by sequence 3735A-3797T (30% and 27.5%), 3735G-3797T (15% and 7.5%) and 3735A-3797A (2.5% and 2.5%). At the end of this study we found out that nucleotide substitution at position 3735, 3789 and 3797 of the pseudogene sequence had already been reported in the single nucleotide polymorphism (SNP) data base (http://www.ensembl.org/index.html; accessed 14 February 2008) as rs1800394, rs5993925 and rs2845373, respectively (Fig. 1). Allele frequencies studies of these polymorphisms were available only for the SNP rs2845373 (3797 T/A). In the European population the allele frequencies reported (A 58.3%, T 41.7%) were different from those documented by us in the Italian population (A 49.05%, T 50.95%).
Gene conversion can cause different phenotypic expressions depending on its length or whether it is homozygous, heterozygous or compound heterozygous with a second defect. It can be associated with different types of VWD: type 2B (Type 1 Malmö and New York) [5,7–10], type 1 [19] and type 3 [8,20]. A gene conversion comparable with that found in family A, with the same minimum and maximum length of gene conversion (Fig. 1), was previously reported in a Russian patient [8], whereas the gene conversion found in family B was found in four of the five original cases identified with type 2B Malmö/New York [5]. Conversely, the set of substitutions found in family C is unique and, at first, appeared not to be related to gene conversion. However, our finding of nucleotides 3735G and 3797A in the VWF pseudogene of normal individuals from different ethnic groups confirmed the role of gene conversion also in this family. It was unexpected that an allele so common in the general population is so seldom involved with gene conversion events. Perhaps the frequency of these pseudogene sequences differs in different ethnic groups, and perhaps in those populations in whom the VWD2B Malmö/New York patients were identified the pseudogene sequence 3735G-3797A is rarer. However, this is apparently not the case in the Italian population. It is also possible that mutation P1266Q might cause a less severe VWD2B, so that the affected individuals come to attention less frequently. Also, the gene conversion event itself could be negatively influenced by the different pseudogene sequence 3735G-3797A (nucleotide 3735G is one base pair from the chi-like sequence) or by other possible nucleotide substitutions linked with this haplotype.
In conclusion, we evaluated the prevalence of VWD2B Malmö/New York in the Italian VWD2B population (35 families) and found it to be approximately 9% (three families). We showed that the mutation P1266Q is responsible, similarly to P1266L, for the VWD2B Malmö/New York phenotype. We have identified three different haplotypes in the VWF pseudogene, one of which (3735G-3797A) is quite common in the general population but rarely reported to be involved with gene conversions in patients with VWD.
Disclosure of Conflict of Interests
The authors state that they have no conflict of interest.




