THE MOLECULAR PHYLOGENETIC AND GEOMETRIC MORPHOMETRIC EVALUATION OF MICRASTERIAS CRUX-MELITENSIS/M. RADIANS SPECIES COMPLEX1
Received 28 July 2009. Accepted 28 January 2010.
Abstract
We investigated nine strains of the Micrasterias crux-melitensis (Ehrenb.) Hassall ex Ralfs and M. radians W. B. Turner species complex. A combination of molecular, morphological, and geometric morphometric data was used to reveal the patterns of their phenotypic and phylogenetic differentiation. The molecular data based on internal transcribed spacer (ITS) rDNA, glycine transfer RNA (trnGuuc) intron, and SSU rDNA sequences revealed three phylogenetic lineages. One of them comprised the six European and North American strains that were morphologically identified as M. crux-melitensis. Phenotypic data illustrated high morphological variability of strains within this genetically homogenous lineage that spanned several traditional infraspecific taxa, including strains corresponding to M. crux-melitensis var. janeira (Racib.) Grönblad and M. crux-melitensis var. superflua W. B. Turner, whose morphometric characteristics profoundly differed. Three strains of M. radians formed two separate phylogenetic lineages corresponding to traditional varieties M. radians var. evoluta (W. B. Turner) Willi Krieg. and M. radians var. bogoriensis (C. J. Bernard) G. S. West. The morphological types corresponding to the former variety have, so far, only been reported from Africa. Therefore, we cannot preclude that geographic isolation may play a role in species differentiation of relatively large freshwater protists, such as Micrasterias.
Abbreviations:
-
- AIC
-
- Akaike information criterion
-
- BI
-
- Bayesian inference
-
- CAUP
-
- Culture Collection of Algae of Charles University in Prague
-
- CVA
-
- canonical variates analysis
-
- GPA
-
- generalized Procrustes analysis
-
- ILD
-
- incongruence length difference test
-
- ITS
-
- internal transcribed spacer
-
- ML
-
- maximum likelihood
-
- PCA
-
- principal component analysis
-
- SVCK
-
- Sammlung von Conjugaten-Kulturen am Institut für Allgemeine Botanik der Universität Hamburg
-
- trnG
-
- glycine transfer RNA gene
-
- wMP
-
- weighted maximum parsimony
The members of the green algal genus Micrasterias are some of the most conspicuous protists. Similarly to other Desmidiaceae, their cells are composed of two semicells divided by a narrow isthmus that contains the centrally located nucleus. Semicells of Micrasterias are typically further divided into several lobes and lobules resulting in highly complex cells. Most Micrasterias species actually have the highest level of cell complexity, evaluated by their deviation from ideal globularity, among the Desmidiales (Neustupa et al. 2009). Since the 19th century, ∼110 species and subspecific taxa have been recognized. The extraordinarily rich taxonomic history of the genus Micrasterias has been summarized by Krieger (1939) and Prescott et al. (1977). The European taxa were compiled by Růžička (1981), who considered many subspecies and forms, originally described on the basis of slight morphological differences, as synonyms of broadly perceived plastic taxa. Consequently, species concept within this genus has not been stable, despite a relative abundance of morphological markers in comparison to most other protist groups. The species complex of M. crux-melitensis and M. radians represents a well-known example of indistinct species boundaries within a genus.
M. crux-melitensis is one of the most frequently occurring species of this genus. It prefers phytobenthos of mesotrophic, slightly acidic wetlands, and it has been reported worldwide, typically from temperate habitats or from high-altitude localities of tropical ecosystems (Vyverman 1992, Coesel 1996). Notable morphological variation in this species was reported and resulted in the description of several varieties and forms on the basis of different morphology of vegetative cells (Krieger 1939, Růžička 1981). However, most of these subspecific taxa were defined solely on the basis of quantitative differences in depth of the incisions between lobes and lobules, cell dimensions, or their degree of lobulation. Notably, M. crux-melitensis var. janeira was defined by its shallow incisions between lobes. On the other hand, M. crux-melitensis var. superflua was characterized by deep incisions and the lobules of third order on vegetative cells, whereas lobulation of up to second order was reported for the type variety (Krieger 1939). Růžička (1981) synonymized most of these varieties and forms of M. crux-melitensis and proposed the single, broadly perceived and morphologically variable M. crux-melitensis. The second species of this morphological complex, M. radians, differs primarily by slightly greater cell dimensions, by deeper incisions between lobes and lobules, as well as by a presumably tropical distribution (Krieger 1939, Scott and Prescott 1961). Vyverman and Viane (1995) investigated morphometric variation (based on conventional measurements of distances and angles) of different populations of traditional M. crux-melitensis and M. radians from Papua New Guinea. The main difference between the typical M. crux-melitensis and M. radians populations relied on the relative length of the incisions, differences in cell dimensions, and also in the shape of a cell’s apex. The apex was generally narrower and U-shaped in M. radians, but more opened and wider in populations of M. crux-melitensis. However, there was a rather gradual shift in morphology from typical M. radians in lowland localities to typical M. crux-melitensis in high-altitude Papua New Guinea samples. Therefore, Vyverman and Viane (1995) hypothesized that traditional M. crux-melitensis and M. radians may actually belong to a single species with temperature-related morphological variation reflecting the altitudinal gradient of the tropical ecosystems.
Taxonomy of desmids has recently undergone major changes based on molecular phylogenetic analyses (McCourt et al. 2000, Gontcharov 2008, Gontcharov and Melkonian 2008, Hall et al. 2008). Most major desmid genera were found to be polyphyletic (Gontcharov and Melkonian 2005, 2008). Thus far, there is limited taxon sampling available for species of the genus Micrasterias, but the present data suggest that the genus might be paraphyletic (Gontcharov et al. 2003, Hall et al. 2008). Interestingly, Triploceras gracile Bailey, which is morphologically quite distinct from Micrasterias and rather resembles some aradiate mutants of Micrasterias (i.e., cells without lateral semicell lobes, Pickett-Heaps 1975), was repeatedly nested within the genus Micrasterias in molecular phylogenetic studies (Gontcharov 2008, Hall et al. 2008).
Most of the molecular phylogenetic investigations of Desmidiales were concentrated on reconstruction of major lineages corresponding to families and orders. Few studies have specifically evaluated taxonomic concepts of traditional morphology-based desmid species. Denboh et al. (2001) demonstrated monophyly of several morphologically closely similar Closterium species groups. However, Denboh et al. (2003) illustrated higher phylogenetic diversity within the Closterium moniliferum/ehrenbergii species complex than was expected on the basis of traditional taxonomic identification. In addition, they demonstrated reproductive isolation between individual clades that indicated possible cryptic or pseudocryptic species diversity within the investigated complex. A similar study based on mating experiments between geographically distant, but morphologically very similar isolates of Micrasterias thomasiana W. Archer, revealed the existence of several reproductively isolated mating groups (Blackburn and Tyler 1987). Gontcharov and Melkonian (2008) illustrated a probable pseudocryptic or cryptic diversity in two strains of Cosmarium punctulatum Bréb. that were morphologically very similar but were positioned in distant parts of the phylogenetic tree of Desmidiaceae. Similarly, two strains of Staurodesmus extensus (Borge) Teiling were also only distantly related and apparently polyphyletic (Gontcharov and Melkonian 2008). These scarce data may indicate that traditional concepts of at least some desmid species have been too broad, and there may be conspicuous pseudocryptic or cryptic species diversity. Consequently, the estimates of total species richness of ∼3,200 species in Desmidiales (Gontcharov 2008) might be underestimated, and the traditional species concepts should be revised utilizing molecular and quantitative morphometric methods.
In the present study, we evaluated the phylogenetic position of nine strains of M. crux-melitensis and M. radians on the basis of molecular data in order to test for the monophyly and phylogenetic differentiation of this complex. In addition, morphology of these strains was analyzed using geometric morphometric methods and SEM of cells. The variation in shape among strains, and eventual phylogenetic groups, was revealed. We asked whether the investigated strains, originating from geographically distant locations, would be homogenous in highly variable nuclear and chloroplast sequences, thus indicating probable cosmopolitanism of broadly perceived species comprising the traditional M. crux-melitensis and M. radians. Alternatively, we attempted to identify morphological and possible distributional differences among phylogenetic lineages within the investigated complex.
Materials and methods
Origin and cultivation of strains, LM and SEM observations. The investigated strains were acquired from algal culture collections at the Hamburg (SVCK) and Prague (CAUP) universities (Table 1). Cultures were initiated with an inoculum of 10–15 cells and grown for 6 weeks in 100 mL Erlenmeyer flasks containing liquid oligotrophic medium developed by the Culture Collection of Algae of Charles University of Prague (CAUP) (http://botany.natur.cuni.cz/algo/caup.html). Strains were maintained at temperatures of 20°C and illuminated at 40 μmol photons · m−2 · s−1 from 18 W cool fluorescent tubes (Philips TLD 18W/33, Royal Philips Electronics, Amsterdam, the Netherlands), at a light:dark (L:D) regime of 12:12. Microphotographs were taken on an Olympus BX51 light microscope with Olympus Z5060 digital microphotographic equipment (Olympus Corporation, Tokyo, Japan). Projection of light microscopic images was made using the Deep Focus 3.0 module (Promicra s.r.o, Prague, Czech Republic) implemented in the QuickPHOTO CAMERA 2.3 software (Promicra s.r.o). For SEM, the acetone-washed glass coverslips (10 mm in diameter) were placed on a heating block and coated three times with a poly-L-lysine solution (1:10 in distilled water) to ensure better adhesion of cells. After cooling, a drop of the formaldehyde-fixed cell suspension was placed on the glass, and when almost dry, it was transferred into 30% acetone and dehydrated by an acetone series (10 min successively in 30, 50, 70, 90, 95, 99% and 2x in 100%). Finally, cells were dried to a critical point with liquid CO2, subsequently sputter-coated with gold (Bal-Tec Sputter Coater SCD 050, Capovani Brothers Inc., Sconia, NY, USA), and examined using the JEOL 6380 LV scanning electron microscope (JEOL Ltd., Tokyo, Japan).
| Strain no. | Taxon name | Origin | Accession nos. | ||
|---|---|---|---|---|---|
| ITS | trnG | SSU | |||
| SVCK 266 | M. crux-melitensis Ralfs var. superflua W. B. Turner | Kiebitzmoor peat bog near Hamburg, Germany | FN424415 | FN424424 | – |
| SVCK 431 | M. crux-melitensis Ralfs var. crux-melitensis | N. Deming Pond, Itasca State Park in Minnesotta, USA | FN424416 | FN424425 | – |
| SVCK 128 | M. crux-melitensis Ralfs var. superflua W. B. Turner | A pond in Rheinland, Germany | FN424417 | FN424426 | – |
| SVCK 98 | M. crux-melitensis Ralfs var. janeira (Raciborski) Grönblad | A bog near Korvanen, Finland | FN424418 | FN424427 | – |
| CAUP K602 | M. crux-melitensis Ralfs var. crux-melitensis | Borkovicka Blata peat bog in South Bohemia, Czech Republic | FN424419 | FN424428 | – |
| CAUP K607 | M. crux-melitensis Ralfs var. crux-melitensis | Brezina bog in North Bohemia, Czech Republic | FN424420 | FN424429 | – |
| SVCK 518 | M. radians W. B. Turner var. evoluta (W. B. Turner) Willi Krieg. | Lake Ol Bolossat, Kenia | FN424421 | FN424430 | FN424433 |
| SVCK 519 | M. radians W. B. Turner var. evoluta (W. B. Turner) Willi Krieg. | Lake Ol Bolossat, Kenia | FN424422 | FN424431 | – |
| SVCK 389 | M. radians Ralfs var. bogoriensis (C. J. Bernard) Willi Krieg. | Kuching, Malaysia | FN424423 | FN424432 | FN424434 |
DNA isolation, amplification, and sequencing. DNA was extracted from the strains listed in Table 1. After centrifugation in an MPW 223 centrifuge (MPW Med. Instruments, Warsaw, Poland), algal cells were mechanically disrupted by shaking in the presence of glass beads (0.5 mm diameter, Sigma-Aldrich, St. Louis, MO, USA). Genomic DNA was extracted using the Invisorb Spin Plant Mini Kit (Invitek, Berlin, Germany) following the manufacturer’s instructions. The ITS rDNA regions were amplified by PCR using the algal-specific primer nr-SSU-1780-5′ (5′-CTGCGGAAGGATCATTGATTC-3′; Piercey-Normore and DePriest 2001) and a universal primer ITS4-3′ (5′-TCCTCCGCTTATTGATATGC-3′; White et al. 1990). Additionally, the trnGuuc intron was amplified using newly designed primers trnG-uuc-F-5′ (5′-AGCGGGTATAGTTTAGTGGT-3′) and trnG-uuc-R-3′ (5′-GGTAGCGGGAATCGAACCCGC-3′). The new primers were designed based on a published chloroplast genome of Staurastrum punctulatum (GenBank accession no. AY958085; Turmel et al. 2005). All PCRs were performed in 20 μL reaction volumes (15.6 μL sterile Milli-Q Water [Millipore Corp., Bedford, MA, USA], 2 μL 10′ PCR buffer [Sigma-Aldrich], 0.4 μL dNTP [10 μM], 0.25 μL of primers [25 pmol · mL−1], 0.5 μL Red Taq DNA Polymerase [Sigma-Aldrich] [1 U · mL−1], and 1 μL of DNA [not quantified]). PCR reactions were performed in either an XP thermal cycler (Bioer, Tokyo, Japan) or a Touchgene gradient cycler (Techne, Cambridge, UK). PCR amplification of the ITS rDNA began with an initial denaturation at 95°C for 5 min and was followed by 35 cycles of denaturing at 95°C for 1 min, annealing at 54°C for 1 min and elongation at 72°C for 1 min, with a final extension at 72°C for 7 min. Identical conditions were used for the amplification of the trnGuuc intron, except that an annealing temperature of 66°C was used. The PCR products were quantified on a 1% agarose gel stained with ethidium bromide and cleaned either with the JetQuick PCR Purification Kit (Genomed, Löhne, Germany) or with QIAquick Gel Extraction Kit (Qiagen Inc., Valencia, CA, USA) according to the manufacturer’s protocols. The purified amplification products were sequenced with the PCR primers with an Applied Biosystems (Seoul, Korea) automated sequencer (ABI 3730xl) at Macrogen Corp. in Seoul, Korea. Sequencing reads were assembled and edited using SeqAssem software (SequentiX, Klein Raden, Germany). After checking the diversity in ITS rDNA and trnGuuc intron sequences, SSU rDNA regions were amplified for selected strains. PCR amplification was performed as described in Neustupa and Škaloud (2007). In total, nine ITS rDNA, nine trnGuuc intron, and two SSU rDNA sequences were generated and submitted to GenBank (Table 1).
Sequence alignment. ITS2 rDNA and trnGuuc intron sequences were manually aligned in MEGA 4 (Kumar et al. 2008). The trnGuuc intron sequences were all of equal length (750 bases); their alignment was straightforward and unambiguous. However, ITS2 rDNA sequences showed considerable variability that made it difficult to create precise alignment. Thus, the common ITS2 rRNA secondary-structure transcript has been constructed using the mfold program (version 2.3; Walter et al. 1994, Zuker 2003) to align the sequences correctly. The final alignment (374 bases) was generated in 4SALE (Seibel et al. 2006, 2008), according to the secondary-structure information (Fig. S1 in the supplementary material). The final SSU rDNA alignment was generated as follows. First, we downloaded 280 aligned sequences of Desmidiales from the SILVA database, version 98 (Pruesse et al. 2007); then, the alignment was reduced to Desmidiaceae and analyzed by the neighbor-joining (NJ) method in PAUP*, version 4.0b10 (Nylander 2004). On the basis of an inferred phylogenetic tree and literary data (Gontcharov et al. 2003, Gontcharov and Melkonian 2008, Hall et al. 2008), the alignment was reduced to 63 sequences that were selected to encompass all Desmidiaceae lineages. In addition, the long-branched Cosmarium sequences AJ428113, AJ428114, AY964133, and AY964135 were removed to avoid the long-branch attraction (LBA) effect in phylogenetic analyses. Finally, two new SSU rDNA Micrasterias sequences, together with one closely related sequence revealed by BLAST searches, were added to the alignment and aligned with the help of the published SSU rRNA secondary-structure transcript of the genus Closterium (Denboh et al. 2001). The final alignment thus comprises 62 sequences (Table S1 in the supplementary material). Ambiguously aligned regions (loops at the end of stem 17 and between the stems 45 and 46) and positions with deletions in most sequences were removed from the alignment, resulting in an alignment comprising 1,725 base positions. All of the alignments were submitted to TreeBASE (ID: SN4729).
Phylogenetic analyses. The phylogenetic trees were inferred with Bayesian inference (BI) using MrBayes version 3.1 (Ronquist and Huelsenbeck 2003). The most appropriate substitution model was estimated for each data set using the Akaike information criterion (AIC) with PAUP/MrModeltest 1.0b (Nylander 2004). In the BI analysis, two parallel Markov chain Monte Carlo (MCMC) runs were carried out for three million generations, each with one cold and three heated chains. Bootstrap analyses were performed by maximum-likelihood (ML) and weighted parsimony (wMP) criteria using PAUP*, version 4.0b10. ML bootstrap analysis (100 replications) consisting of heuristic searches with 10 random sequence addition replicates, tree bisection-reconnection (TBR) swapping, and a rearrangement limit of 5,000 for each replicate. The wMP bootstrapping (1,000 replications) was performed using heuristic searches with 100 random sequence addition replicates, TBR swapping, random addition of sequences (the number limited to 10,000 for each replicate), and gap characters treated as a fifth character state. Congruence between ITS2 rDNA and trnGuuc intron data sets was tested using the incongruence length difference (ILD) test (Farris et al. 1995), as implemented by the partition homogeneity test in PAUP* (heuristic search, simple addition, TBR branching swapping, 100,000 replicates).
Morphometric analyses. For each strain, 50 adult semicells were randomly chosen and photographed. On each semicell, 31 structurally corresponding landmarks and semilandmarks were depicted (Fig. 1). Whereas the landmarks were placed in fixed positions, the semilandmarks (nos. 10, 11, 15, 17, 21, and 22) were allowed to slide along the abscissa connecting adjacent landmarks (Zelditch et al. 2004). For most of the geometric morphometric analyses, the TPS-series software (available at http://life.bio.sunysb.edu/morph/) was used. Positions of landmarks and length and width of the cells were digitized in TpsDig, ver. 2.12. The landmark configurations of the entire set of 450 objects were superimposed by generalized Procrustes analysis (GPA) in TpsRelw, ver. 1.42. This widely used method standardizes the size of the object and optimizes the rotation and translation so that the distances between corresponding landmarks of investigated objects are minimized (Bookstein 1991, Zelditch et al. 2004). Correlation between Procrustes and the Kendall tangent space distances was assessed using TpsSmall, ver. 1.20, to ensure that the variation in shape was small enough to allow subsequent statistical analyses (Zelditch et al. 2004). The correlation of Procrustes and Kendall shape spaces was very high (r = 0.999), so we proceeded with further analyses. The Micrasterias semicells are bilaterally symmetrical, and their anterior and posterior sides do not differ (Neustupa and Škaloud 2007). As the asymmetry of semicells was not of interest in this study, cells were symmetrized prior to analysis following the standard formula of Klingenberg et al. (2002). This involved reflecting the landmark configurations (by multiplying the x-coordinates in all landmarks by −1). Then, the paired landmarks in the reflected copy were relabeled, and the original and mirrored configurations were averaged in the general Procrustes superimposition (Klingenberg et al. 2002). A principal component analysis (PCA) of geometric morphometric data was conducted on the entire set of 450 semicells. Scores of the objects on the all nonzero 29 principal component (PC) axes were used for canonical variate analysis (CVA) in PAST, ver. 1.89 (Hammer et al. 2001), to test for the differences in shape of individual strains. In addition, scores on PC axes were used for the two-group linear discrimination analyses between all pairs of investigated strains. Significance of the difference between group mean shape configurations was assessed by the Hotelling’s T2 test. In parallel, the K-means clustering, a method based on the nonhierarchical clustering of multivariate data into a specified number of groups (Bishop 1995), was used to illustrate relative differences in shape of the individual strains. The K-means clustering into two groups was conducted in pairs of all investigated strains. Number of cells classified out of the original pattern of 50 versus 50 objects from two strains in each pair indicated their overall shape similarity. K-means clustering pattern identical with the underlying origin of cells from two strains indicated clear separation of groups, while a high number of cells placed into the wrong group illustrated the close relationship of two groups on the basis of their landmark-based shape data.
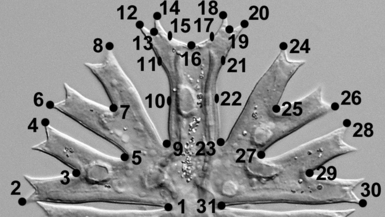
Position of structurally corresponding landmarks on Micrasterias cells.
Results
Phylogenetic analyses. The genetic diversity of nine Micrasterias strains was determined using the ITS2 of rDNA and the group II intron sequences of the plastid gene that encodes transfer RNA-Gly (trnGuuc). Results of genotype and haplotype groupings among the strains are illustrated in Figure 2. These two markers differed considerably in the amount of their variation. The ITS2 rDNA showed a higher average genetic distance (Kimura 2-parameter model) between the lineages (0.168) than the trnGuuc intron (0.032). The ITS2 rDNA sequences were so variable that the alignment could only be established with the aid of the common ITS2 rRNA secondary structure transcript (Fig. S1). On the other hand, the alignment of trnGuuc intron sequences was straightforward and unambiguous, allowing us to analyze the phylogeny of nine investigated Micrasterias strains, together with the three related strains (M. crux-melitensis NIES 152, GenBank accession no. FN562171; M. truncata CAUP K606, FN562173; and Staurodesmus dickiei ASW 07056, FN562172). In general, genotype groups observed in the ITS2 rDNA sequences (Fig. 2A) corroborated the haplotype groupings observed in the trnGuuc intron sequences (Fig. 2B). The congruence of both phylogenies was supported by the statistically significant partition homogeneity test (ILD test; P = 0.52). Generally, three lineages could be established from the phylogenetic analyses: (i) a large lineage comprising all the European and the single North American M. crux-melitensis isolates (CAUP K602, CAUP K607, SVCK 98, SVCK 128, SVCK 266, and SVCK 431, respectively); (ii) the single M. radians strain originating from Southeast Asia (SVCK 389); and (iii) two M. radians isolates of tropical African origin (SVCK 518 and SVCK 519). The single difference between the ITS2 and trnGuuc phylogenies pertained to the genetic identity of the European strains. The ITS2 analysis showed the divergence of SVCK 431, whose sequence differed by two substitutions and one insertion from that of the other strains in the European lineage. By contrast, the trnGuuc analysis revealed a single substitution in the SVCK 98 sequence that differentiated it from the rest of the strains. Moreover, the trnGuuc data revealed the monophyly of M. crux-melitensis/radians complex (BI/ML/MP support 0.98/93/99). However, the strains pertaining to the same traditional morphospecies did not cluster together (Fig. 2B). According to the trnGuuc sequences, European/North American M. crux-melitensis isolates formed a well-supported clade (1.00/98/100) with Asian M. radians SVCK 389. Similarly, the pair of African M. radians isolates significantly clustered with M. crux-melitensis NIES 152 (1.00/100/100).
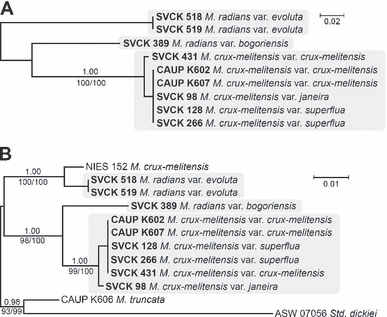
Unrooted Bayesian analyses based on ITS2 rDNA (A) and trnGuuc intron (B) sequences using a GTR model for ITS2, and a GTR + I model for trnGuuc data set. Values at the nodes indicate statistical support estimated by three methods: MrBayes posterior node probability (upper), maximum-likelihood bootstrap (bottom left), and maximum-parsimony bootstrap (bottom right). Three lineages recognized from the phylogenetic analyses are highlighted by gray boxes. Scale bars = expected number of substitutions per site. GTR, generalized time reversible; ITS, internal transcribed spacer; trnG, glycine transfer RNA gene.
Although both ITS2 rDNA and trnGuuc markers were very useful to reveal genetic relationships among nine investigated Micrasterias strains, they were too variable for determining phylogenetic position of the three recognized lineages within the genus Micrasterias and Desmidiaceae as a whole. Therefore, SSU rDNA sequences of the selected representative strains (CAUP K602, SVCK 389, and SVCK 518) were compared with the SSU rDNA data set of 59 desmid species (Fig. 3). All three lineages formed a well-supported independent clade, together with M. crux-melitensis NIES 152, M. truncata CAUP K606, and S. dickiei SVCK 38. The strain of M. crux-melitensis CAUP K602 belonging to the European/North American cluster formed a clade (74/95/0.99) with the Asian M. radians SVCK 389, which did not include the African M. radians SVCK 518 (Fig. 3). Although the exact position within Desmidiaceae remained ambiguous, BI, ML, and MP analyses (latter two not shown) consistently showed all other Micrasterias sequences as the next closest relatives to this clade. However, neither the relationship with any of these other Micrasterias species nor the lineage encompassing the genus as a whole received significant internal branch support.
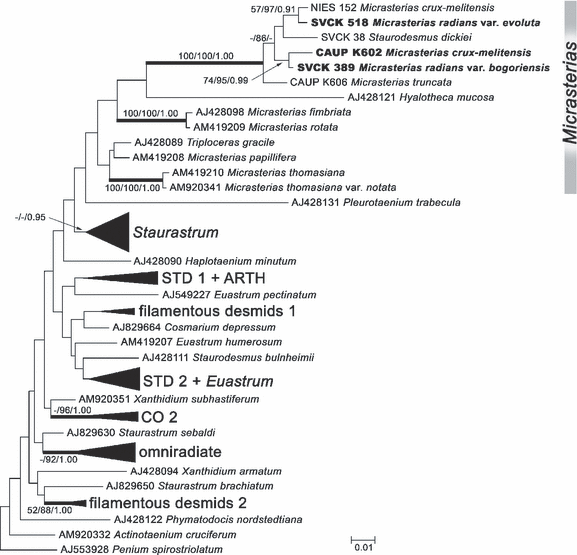
Bayesian analysis of SSU rDNA sequences of Micrasterias species and other representatives of Desmidiaceae using a GTR + I + Γ model. Values at the nodes indicate statistical support estimated by three methods: maximum-likelihood bootstrap (left), maximum-parsimony bootstrap (in the middle), and MrBayes posterior node probability (right). Thick branches represent nodes receiving the highest PP support (1.00). The sequences representing three lineages recognized from the phylogenetic analyses of ITS2 rDNA and trnGuuc intron sequences (Fig. 2) are shown in bold. Collapsed groups of sequences correspond to clades proposed by Gontcharov and Melkonian (2008). The tree is rooted with Penium spirostriolatumAJ553928. Scale bar = expected number of substitutions per site. GTR, generalized time reversible; ITS, internal transcribed spacer; PP, posterior probability; trnG, glycine transfer RNA gene.
Morphology of cells. The cells of all strains were readily identifiable into M. crux-melitensis/M. radians species complex (Fig. 4). However, there were apparent differences in morphology of individual strains. Cell dimensions profoundly differed (Fig. 5, A and B). The strains SVCK 266, SVCK 431, and SVCK 98 consistently had the smallest cells, while cells of the SVCK 128, CAUP K602, CAUP K607, and SVCK 518 strains were ∼35% to 45% larger. Moreover, the degree of lobulation differed among strains. Most strains had well-developed second-order lobules present in all semicells (Fig. 4, A, B, E–I), with the notable exception of the SVCK98 strain, whose second-order lobules were only weakly developed (Fig. 4D). At the same time, this strain had the relatively smallest cells (Fig. 5, A and B), and it corresponded well to the M. crux-melitensis var. janeira, according to the traditional taxonomy (Krieger 1939). On the other hand, the SVCK128 strain of M. crux-melitensis var. superflua had third-order lobules present on most semicells (Fig. 4C) and relatively large cells (Fig. 5, A and B), so that it fit well into the description of this variety. The second strain, originally assigned as M. crux-melitensis var. supeflua, SVCK 266, was different from SVCK 128 as it had considerably smaller cells and most semicells did not develop the third-order lobules. The SVCK 431, CAUP K602, and K607 fit into descriptions of the type variety M. crux-melitensis var. crux-melitensis, even if their cell dimensions differed among strains. In contrast to other strains, the SVCK 518 and SVCK 519 had a pair of spines on each side of their polar lobes (Fig. 6, A–C). However, these polar lobe spines were not always symmetrically developed on all semicells, and, sometimes, there was just a single asymmetrical spine present (Fig. 6B). This characteristic presence of polar lobe spines, as well as the long furcate lobules of their semicells, led us to identify these strains as M. radians var. evoluta (W. B. Turner) Willi Krieg. according to traditional criteria (Krieger 1939). The SVCK 389 strain of M. radians var. bogoriensis (C. J. Bernard) Willi Krieg. fit well into the taxonomic description of this variety, mainly by the presence of long, slightly curved spines on tips of the polar lobes and lateral lobules (Fig. 6F). Furthermore, the third-order lobules were sometimes present, in accordance with the taxonomic delimitation of this variety. The cell walls of all the investigated strains were consistently provided with pore-linked mucilage extrusions (Fig. 6, D–F), and no differences among strains were detected in this genus-wide feature.
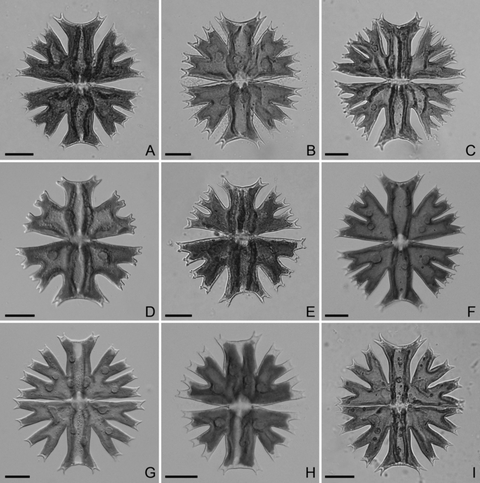
Morphology of Micrasterias strains. (A) M. crux-melitensis var. superflua, strain SVCK 266. (B) M. crux-melitensis var. crux-melitensis, strain SVCK 431. (C) M. crux-melitensis var. superflua, strain SVCK 128. (D) M. crux-melitensis var. janeira, strain SVCK 98. (E) M. crux-melitensis var. crux-melitensis, strain CAUP K602. (F) M. crux-melitensis var. crux-melitensis, strain K 607. (G) M. radians var. evoluta, strain SVCK 518. (H) M. radians var. evoluta, strain SVCK 519. (I) M. radians var. bogoriensis, strain SVCK 389. Scale bar = 20 μm.
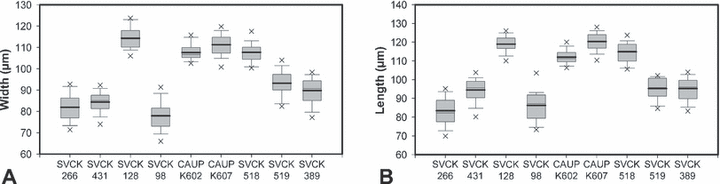
Size data of investigated strains: (A) width and (B) length of cells.
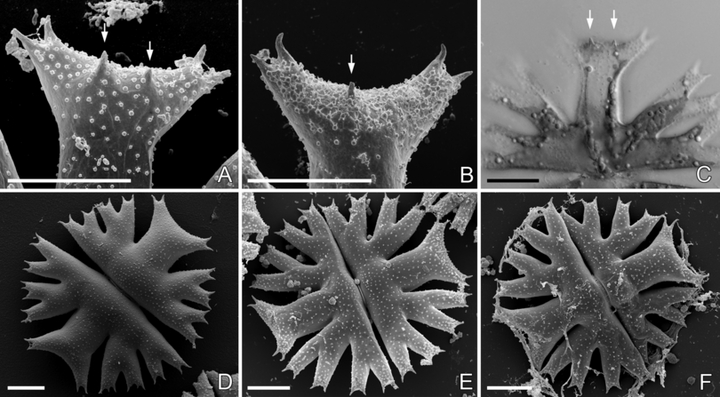
Morphology of Micrasterias strains. (A–B) Scanning electron microscopic images of the polar lobes with prominent spines in M. radians var. evoluta: (A) SVCK 518 and (B) SVCK 519. (C) Projection of two light microscopic images visualizing the spines in the polar lobe, SVCK 518. (D–F) SEM images of whole cells illustrating the consistent nodulation of the cell walls: (D) CAUP K607, (E) SVCK 518, and (F) SVCK 389. Spines present in the polar lobes are marked by arrows. Scale bar = 20 μm.
Geometric morphometrics. The PCA of the symmetrized Procrustes-superimposed landmark data yielded 29 nonzero PC axes. The first PC axis spanned 59.5% of the total variation. Semicells varied from being deeply lobed, with a narrow polar lobe in negative values, to compact shapes with shallow incisions and a wide polar lobe in positive marginal positions along the first PC axis (Fig. 7). Consequently, the strains SVCK 518, SVCK 519, and SVCK 389, which were characterized by deeply lobed semicells of the M. radians type were positioned close to the negative margin on PC1. Conversely, the SVCK 98 strain had the most positive PC1 scores corresponding to its compact semicells with shallow incisions (Fig. 8, A and B). The second PC axis, which spanned 10.44% of the total shape variation, was related to width of the upper lateral lobule. Semicells with negative scores on PC2 had narrow upper lateral lobules and open incisions, while semicells with positive PC2 scores were characterized by closed incisions and wide upper lateral lobules (7, 8). This axis primarily separated strains SVCK 518, SVCK 519, SVCK 389, and SVCK 98 positioned mostly in negative PC2 values from the rest of the strains.
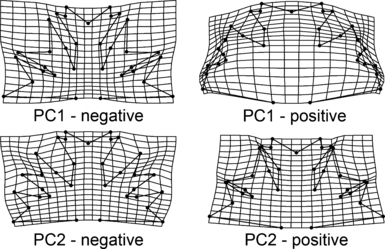
The deformation grids illustrating shapes reconstructed at the extreme positions of first and second axes of the principal component analysis of the entire investigated set.
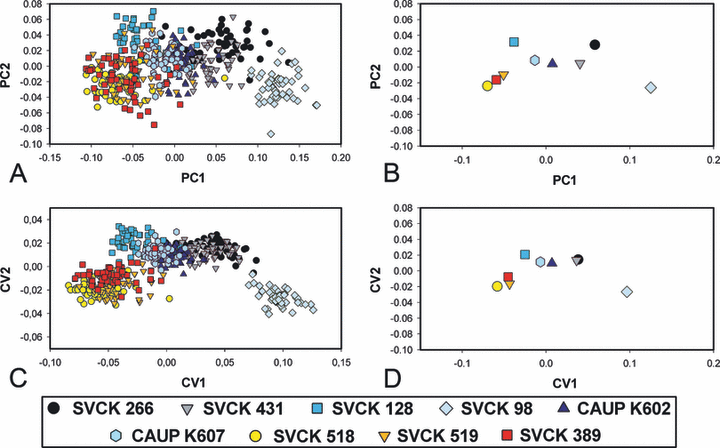
Multivariate analyses of geometric morphometric data. (A) The ordination plot of first and second axes of the principal component analysis (PCA) of all the 450 investigated objects. (B) Position of the group centroids in the ordination plot of first and second axes of the PCA of all the 450 objects. (C) The ordination plot of first and second axes of the canonical variates analysis (CVA) aimed at separation of individual groups. (D) Position of the group centroids in the ordination plot of first and second axes of the CVA aimed at separation of individual groups.
The CVA illustrated the separation of strains on the basis of their shape characteristics (Fig. 8, C and D). The overall separation among strains was highly significant (Wilk’s λ = 1.46e10−5, P = 0). The SVCK 98 strain was the most dissimilar from all other strains. In addition, the three strains SVCK 518, SVCK 519, and SVCK 389, assigned originally as M. radians, were grouped together and more or less separated from the remaining strains (Fig. 8, C and D). The pair comparisons revealed that all strains were mutually statistically significantly different in their mean shape characteristics (Table 2). The P-value of the Hotelling’s T2 tests was <10−5 in all pairs, and there were 100% correctly classified cells in most of the pair comparisons. The K-means clustering algorithm illustrated a higher number of objects classified across the original group assignment (Table 2). In this respect, the pairs of M. crux-melitensis strains from the Czech Republic (CAUP K602, K607) and the pair of African M. radians var. evoluta strains SVCK 518 and SVCK 519 had the most similarly shaped semicells.
| SVCK 266 | SVCK 431 | SVCK 128 | SVCK 98 | CAUP K602 | CAUP K607 | SVCK 518 | SVCK 519 | SVCK 389 | |
| SVCK 266 | – | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| SVCK 431 | 17 | – | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| SVCK 128 | 4 | 2 | – | 100 | 100 | 100 | 100 | 100 | 100 |
| SVCK 98 | 3 | 0 | 0 | – | 100 | 100 | 100 | 100 | 100 |
| CAUP K602 | 14 | 5 | 3 | 0 | – | 99 | 100 | 100 | 100 |
| CAUP K607 | 8 | 2 | 6 | 0 | 35 | – | 100 | 100 | 100 |
| SVCK 518 | 1 | 1 | 2 | 1 | 1 | 1 | – | 99 | 100 |
| SVCK 519 | 2 | 1 | 2 | 0 | 4 | 6 | 33 | – | 100 |
| SVCK 389 | 1 | 3 | 0 | 0 | 1 | 3 | 2 | 5 | – |
Discussion
Our molecular phylogenetic analyses clearly did not confirm taxonomic homogeneity of the M. crux-melitensis/M. radians species complex. The phylogenetic inferences based on the nuclear ITS2 rDNA and chloroplast trnGuuc intron sequences consistently demonstrated that the investigated nine strains clustered into three different lineages. The phylograms of both molecular markers were highly congruent, except for slight differences in the relationships of the M. crux-melitensis strains (Fig. 2). However, because the differences were based on just a few substitutional changes in the rapidly evolving molecular markers, they may correspond to an among-population structure of a single species. This slight incongruence in genetic variability between the two molecular markers within the European lineage clearly illustrates the importance of using several independent molecular markers for inferring phylogenies of closely related organisms.
The SSU rDNA phylogeny unambiguously grouped all the investigated strains into the highly supported clade within Desmidiaceae (Fig. 3). In addition to several Micrasterias sequences, this lineage also included the single sequence of the morphologically completely different S. dickiei (strain SVCK 38, accession no. AJ428101). Based on the substantial morphological dissimilarity between Micrasterias species and S. dickiei, we decided to resequence this species to disprove possible contamination or misidentification of strain SVCK 38. Since the Staurodesmus strain SVCK 38 is no longer available in the culture collections, we acquired the SSU rDNA and trnGuuc intron sequences from the S. dickiei strain ASW 07056 (GenBank accession no. FN562174). The obtained SSU rDNA sequence indeed differed from AJ428101 by just a single substitution change. Therefore, we confirmed the phylogenetic position of S. dickiei among the Micrasterias species, as previously reported by Gontcharov et al. (2003). Such close relationship between the members of the genus Micrasterias and other morphologically dissimilar desmid species was already reported by Hall et al. (2008), who illustrated the position of Triploceras gracile among the Micrasterias species. This phenomenon may indicate repeated reductive morphological evolution of morphologically different desmid species from Micrasterias-like ancestors. The SSU rDNA phylogeny did not reveal internal structure of the well-supported clade that encompassed the M. crux-melitensis/radians sequences, S. dickiei, and Micrasterias truncata. However, the phylogenetic inference based on the trnGuuc intron sequences clustered all the M. crux-melitensis/radians sequences into a well-supported monophyletic lineage (BI/ML/MP support 0.98/93/99), sister to the pair of M. truncata and S. dickiei (Fig. 2B).
In Desmidiaceae, the SSU rDNA phylogenies yielded rather weak resolution and support of internal branches (Besendahl and Bhattacharya 1999, Denboh et al. 2001, Gontcharov et al. 2003). However, several recent phylogenetic studies that combined various nuclear, chloroplast, and mitochondrial markers resulted in increased phylogenetic resolution and resolved inconsistencies among particular single-gene phylogenies (Gontcharov et al. 2004, Gontcharov and Melkonian 2008, Hall et al. 2008). Although we used the single-gene phylogeny, the Bayesian inference of SSU rDNA sequences was generally consistent with phylogenies of Desmidiaceae based on concatenated data sets (Gontcharov and Melkonian 2008, Hall et al. 2008). The close relationship of Micrasterias and Staurastrum lineages was demonstrated, although it was not supported by statistical tests (Fig. 3).
Comparison of phylogenetic results with morphological and morphometric data provides evidence for the probable unreliability of some traditionally delimited subspecific taxa of M. crux-melitensis. This species, represented in our study by strains from temperate Europe and North America, was genetically homogenous, despite its high morphological variability. In fact, SVCK 98, morphologically corresponding to M. crux-melitensis var. janeira, and SVCK 128, corresponding to M. crux-melitensis var. superflua, occupied opposite ends of the overall morphospace of the investigated complex. Despite this morphological dissimilarity, they were phylogenetically homogenous. Therefore, we can certainly not reject the hypothesis of Růžička (1981), who suggested that subspecific varieties of M. crux-melitensis bear no taxonomic value and should be synonymized into a single taxon. On the other hand, an evident pseudocryptic diversity was illustrated for M. radians. Therefore, we can reject the hypothesis of possible conspecificity of M. crux-melitensis and M. radians (Vyverman and Viane 1995). The pair of African strains (SVCK 518 and SVCK 519) formed a tight cluster separate from the Asian SVCK 389, which remained in an isolated phylogenetic position within the investigated complex. The overall cell-shape properties of these three strains were relatively similar as they occupied adjacent or overlying parts of the morphospace (Fig. 8). However, they clearly differed in the pattern of spine formation on cells. The African strains formed spines on central parts of semicell polar lobes that corresponded to traditionally delimited M. radians var. evoluta. On the other hand, the SVCK 389 strain had conspicuous spines, mainly on lateral tips of polar lobes, and occasionally formed the third-order semicell lobules corresponding to the taxonomic delimitation of M. radians var. bogoriensis. Based on the degree of their phylogenetic divergence and presence of the unambiguous discriminating morphological characters, we suppose that these two lineages are in fact separate species. However, we would be reluctant to describe these species formally on the basis of an investigation of only three strains. Evidently, further natural populations corresponding to M. radians var. evoluta and to M. radians var. bogoriensis should be sampled and sequenced before taxonomic conclusions can be made. Nevertheless, our study demonstrated that M. radians is heterogenous and clearly nonmonophyletic. This species has mainly been reported in floristic studies from tropical and subtropical habitats worldwide, and the morphology of these findings was typically documented by illustrations or microphotographs. M. radians var. evoluta, unambiguously illustrated with spines in the central parts of polar lobes, which morphologically corresponds to our SVCK 518/519 phylogenetic lineage, was reported, for example, from Congo (Van Oye 1957), Mali (Couté and Tell 1981), Uganda (Lind 1971), Mozambique (Rino 1972), South Africa (Williamson 1994), and Madagascar (Bourelly and Manguin 1949, Coesel 2003). Turner (1892) described this taxon from East India, but no illustration was provided. Therefore, we can speculate that occurrence of the SVCK 518/519 lineage, corresponding to the traditional M. radians var. evoluta, may be limited to tropical regions of Africa, with most reports stemming from the sub-Saharan region. M. radians var. bogoriensis was taxonomically characterized by the presence of third-order lobules and by conspicuous spines on polar lobe tips (Krieger 1939). This taxon has frequently been reported from tropical and subtropical Asia and Australia (e.g., North Australia, Scott and Prescott 1958, Indonesia, Scott and Prescott 1961). In addition, Franceschini (1992) illustrated the M. radians specimen from tropical South America, morphologically similar to M. radians var. bogoriensis. The variety has further been reported from this region by Heckman (1998). The single European report of M. radians var. bogoriensis from Aquitaine, France (Capdevielle 1982), should be considered as uncertain, because the illustrated specimens, characterized solely by the presence of the third-order lobules, may very well correspond to M. crux-melitensis var. superflua. This is known from several European localities and belongs to the temperate M. crux-melitensis lineage according to our molecular data.
In conclusion, we have established that morphology may not provide a reliable species identification, even in morphologically very complex protists such as Micrasterias. The strains of M. crux-melitensis, originating from European and North American localities, encompassed a large part of the observed shape variation within the investigated complex, but this phenotypic variability was not reflected in the molecular data. On the other hand, morphologically similar, subtropical and tropical M. radians strains from Africa and Asia, corresponding to traditional subspecific taxa, were revealed to be of separate lineages. These results suggest that there may be more Micrasterias species than indicated solely by morphological data. In addition, the actual species diversity of these relatively large protists may be related to the patterns of their geographic distribution, or to the climatic factors, rather than to purely morphological features, as indicated, for example, by the continent-wide European distribution of the morphologically diverse M. crux-melitensis lineage.
Acknowledgments
We are indebted to Monika Engels for kind provision of strains from the SVCK culture collection for our research. Frans Kouwets was kind enough to provide us some important literature on the topic of this study. We thank the anonymous reviewers for their recommendations that led us to improvements of the manuscript. The study has been supported by the grant no. 206/09/0906 of the Czech Science Foundation and by the research project no. 21620828 of Czech Ministry of Education. We thank Dr. Marina Julian from JustMeEditing, Manuscript Editor for the Biological Sciences, for the language and style corrections.