COMMON EVOLUTIONARY ORIGIN OF STARCH BIOSYNTHETIC ENZYMES IN GREEN AND RED ALGAE1
1Received 10 March 2005. Accepted 17 August 2005.
Abstract
Plastidic starch synthesis in green algae and plants occurs via ADP-glucose in likeness to prokaryotes from which plastids have evolved. In contrast, floridean starch synthesis in red algae proceeds via uridine diphosphate-glucose in semblance to eukaryotic glycogen synthesis and occurs in the cytosol rather than the plastid. Given the monophyletic origin of all plastids, we investigated the origin of the enzymes of the plastid and cytosolic starch synthetic pathways to determine whether their location reflects their origin—either from the cyanobacterial endosymbiont or from the eukaryotic host. We report that, despite the compartmentalization of starch synthesis differing in green and red lineages, all but one of the enzymes of the synthetic pathways shares a common origin. Overall, the pathway of starch synthesis in both lineages represents a chimera of the host and endosymbiont glycogen synthesis pathways. Moreover, host-derived proteins function in the plastid in green algae, whereas endosymbiont-derived proteins function in the cytosol in red algae. This complexity demonstrates the impacts of integrating pathways of host with those of both primary and secondary endosymbionts during plastid evolution.
Abbreviations:
-
- AGPase,
-
- ADP-glucose pyrophosphorylase;
-
- BE,
-
- branching enzyme;
-
- GBSS,
-
- granule bound starch synthase;
-
- Glc-1-P,
-
- glucose-1-phosphate;
-
- Glc-6-P,
-
- glucose-6-phosphate;
-
- G(S)S,
-
- glycogen (starch) synthase;
-
- LSU,
-
- large subunit;
-
- PGM,
-
- phosphoglucomutase;
-
- SS,
-
- soluble starch synthase;
-
- SSU,
-
- small subunit;
-
- UDP,
-
- uridine diphosphate;
-
- UGPase,
-
- UDP-glucose pyrophosphorylase
Most living organisms accumulate either glycogen or starch as energy storage polymers. Glycogen, amylose, and amylopectin, the latter two of which comprise green algal and plant starch, are molecules of α-1,4-linked glucose units with α-1,6-branch points, the length and number of branches varying between organisms. The α-1,4-glucan chains are synthesized by glycosyltransferases, which use uridine diphosphate (UDP)-glucose or ADP-glucose as the sugar donor and a preexisting α-1,4-glucan chain as the acceptor. Glycogen is localized in the cytoplasm of bacteria, fungi, and animal cells. Both eukaryotic and prokaryotic glycogens are always amorphous and never form the crystalline granules characteristic of starch. Red algal starch, thought to be comprised purely of amylopectin chains (Marszalec et al. 2001, Yu et al. 2002), is cytosolic and is known as floridean starch, whereas in green algae and plants, starch accumulates within the plastid.
The eukaryotic ancestor of all plants, algae, and organisms derived from such acquired a plastid by the engulfment and subsequent retention of a cyanobacterium (Gray and Spencer 1996). Descendants of this ancestor have given rise to a diversity of plastid-containing eukaryotes. Many genes have been transferred from the endosymbiont to the nucleus: It is estimated that as many as 18% of the genes in Arabidopsis are of plastid origin (Martin et al. 2002). It was long assumed that the protein products of all of these genes would be targeted back to the organelle (Weeden 1981), but more recent work is calling the contribution of the endosymbiont to the eukaryotic cell as a whole into question (Abdallah et al. 2000, Martin et al. 2002, Leister 2003). Those that are redirected to the plastid are targeted using a transit peptide allowing them to fulfill their original role in the plastid (Peltier et al. 2000). Several plastidial reactions, however, have been replaced by enzymes derived from the host (Fast et al. 2001); similarly, some endosymbiont-derived proteins have been adopted by pathways in the cytoplasm (Brinkmann and Martin 1996).
Despite differences in structure between bacterial glycogen and plant starch, the plastidial location of plant starch and the use of ADP-glucose (as in bacteria), rather than UDP-glucose, as a substrate (as in fungi and animals) suggest that the plant pathway was inherited from its prokaryotic endosymbiont. On the other hand, the use of ADP-glucose and the cytosolic location of starch in red algae have led to the suggestion that these algae and their derivatives use a novel pathway likely derived from the host (Nyvall et al. 1999, Viola et al. 2001). There is little direct evidence for the nature of the starch biosynthesis pathway in red algae and until recently virtually no molecular data. However, the recent sequencing of two red algal genomes (Matsuzaki et al. 2004, Michigan State University Galdieria Database http://genomics.msu.edu/galdieria_public) and the accumulation of Expressed Sequence Tag (EST) and genomic data from several algal groups with plastids derived from red algae (Armbrust et al. 2004, Patron et al. 2004b) now allow us to investigate the evolutionary origins of their starch biosynthetic pathway. Here, we investigate the origins of these enzymes by examining their molecular phylogeny to determine whether the plant and algal proteins were inherited from the cyanobacterial endosymbiont or from the eukaryotic host. Particular attention is paid to the red algae, because little is known about starch synthesis in any rhodophyte, and the origin of the enzymes may resolve whether their pathway is similar to that found in the green plastid lineage or whether it evolved independently and is novel.
MATERIALS AND METHODS
Protein sequences were obtained from GenBank and from the Cyanidioschyzon merolae (http://merolae.biol.s.u-tokyo.ac.jp/) and Galdieria sulphuraria (http://genomics.msu.edu/galdieria/funding.html) genome projects and from an in-house Heterocapsa triquetra EST project (http://amoebidia.bcm.umontreal.ca/public/pepdb/agrm.php).
Protein alignments were made using Clustal X and manually edited. All ambiguous sites of the alignments were removed from the data set for phylogenetic analyses. The alignment data are available on request. Protein maximum likelihood analyses used PhyML (Guindon and Gascuel 2003) with input trees generated by BIONJ, the JTT model of amino acids substitution, proportion of variable rates estimated from the data, and nine categories of substitution rates (eight variable and one invariable). One hundred bootstrap trees were calculated with PhyML with four gamma correction categories. For distance analyses, gamma-corrected distances were calculated by TREE-PUZZLE 5.0 using the WAG substitution matrix with eight variable rate categories and invariable sites. Trees were inferred by weighted neighbor joining using WEIGHBOR 1.0.1a. Bootstrap resampling was performed using PUZZLEBOOT with rates and frequencies estimated using TREE-PUZZLE 5.0.
RESULTS
Phosphoglucomutase. Phosphoglucomutase (PGM; EC 2.7.5.1) catalyzes the interconversion of glucose-1-phosphate (Glc-1-P) and glucose-6-phosphate (Glc-6-P) at a pivotal metabolic trafficking point that controls the synthesis and degradation of carbohydrates. The direction of metabolic flow through PGM depends on the available carbon source. Glycogen and galactose degradation require PGM activity to convert Glc-1-P to Glc-6-P. The Glc-6-P produced is then used in glycolysis and the pentose-phosphate pathway. In the reverse reaction, PGM converts Glc-6-P to Glc-1-P for the synthesis of sugar nucleotides and ultimately glycogen, starch, trehalose, and lipopolysaccharides.
The PGM superfamily consists of true PGMs and other enzymes of closely related function, some of which have been shown to have residual PGM activity in addition to their primary functions. In humans, three loci for PGM were found and named PGM1, PGM2, and PGM3. Phosphoglucomutase 1 is a true PGM, whereas PGM2 is a phosphoribomutase (Ninfali et al. 1984) and PGM3 is an N-acetylglucosamine-phosphate mutase (Pang et al. 2002). Two isoforms of PGM are present in plants, one located in the cytosol and the other in the chloroplast stroma (Mühlbach and Schnarrenberger 1978). The plastid PGM is required for starch synthesis to store net photosynthate during the day (Dietz 1987, Periappuram et al. 2000). Deficiency in the plastid PGM activity resulted in a “starchless” phenotype in Arabidopsis (Kofler et al. 2000).
Phosphoglucomutases of all eukaryotes and prokaryotes are relatively well conserved at the primary amino acid level, and homologues from all sampled taxa were readily alignable. Prokaryotes form a moderately supported clade (82 Maximum Likelihood [ML] and 54 Weighbor [WBR] bootstrap support), Metazoan, fungal, chlorophyte-plastid, and chlorophyte-cytosolic forms also separate into well-supported clades, but there is no support for their position or order within the tree (Fig. 1). It is not possible to say conclusively if the proteins found in red and green algae and plants share a common ancestry or whether they are of cyanobacterial or eukaryotic origin, although the support for the prokaryotic branch suggests they are more likely eukaryotic. The red alga Cyandioschyzon merolae encodes three PGMs, one of which is most similar to a chloroplast isoform from spinach (data not shown). Both the C. merolae and spinach isoforms are highly divergent, and a reduced character set had to be used to analyze the data with these sequences (data not shown), which gave a tree with even less resolution and no conclusion to the origin of these isoforms. The amino-terminus of this C. merolae protein is extended by comparison to prokaryotic enzymes, but the extension does not have any of the characteristics of a transit peptide, such as frequent hydroxylated residues. The C. merolae sequence that branches between the prokaryotes and the eukaryotes has a similar amino-terminal extension, but there is no support for the positions of either this or the other rhodophyte sequences within the tree. Given that floridean starch is cytosolic, there is no expectation for plastid isoforms of PGM for the purpose of starch production. However, other plastid pathways, such as fatty acid and amino acid biosynthesis, also require Glc-6-P (Neuhaus and Emes 2000), and so a plastid isoform in rhodophytes would not be surprising, although it would not be evidence of a plastid carbon storage pathway.
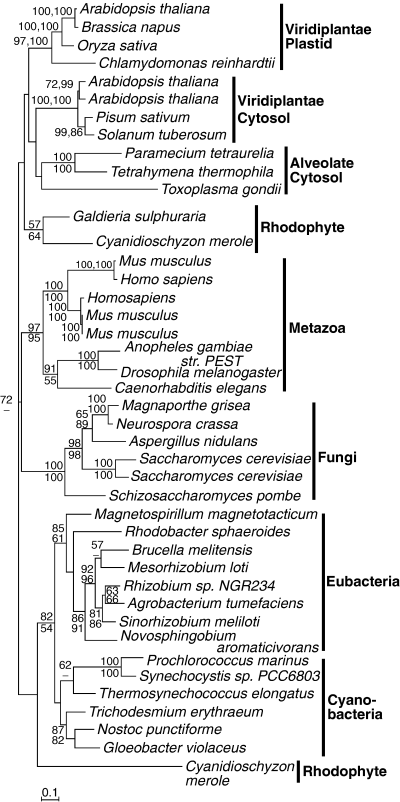
Protein maximum likelihood phylogeny of phosphoglucomutase. Numbers at nodes indicate bootstrap support from (top to bottom or left to right) maximum likelihood, as determined by PhyML and weighted neighbor joining. Analysis used 400 characters, the alpha parameter=0.083 and proportion of invariable sites=0.024.
Nucleotide diphosphate pyrophosphorylases. The first committed step in the biosynthesis of glycogen and starch is the synthesis of the nucleotide sugar, ADP- or UDP-glucose, from ATP or UTP and Glc-1-P (Preiss 1988, Smith 1999, Ball and Morell 2003). In bacteria, plants, and green algae, ADP-glucose is produced by ADP-glucose pyrophosphorylase (AGPase). Eukaryotes without plastids do not, as far as is known, encode AGPase. The α-glucan synthases of these organisms use UDP-glucose, synthesized by UDP-glucose pyrophosphorylase (UGPase), as a glucosyl donor. Bacteria and plants also encode UGPase, but in bacteria UGPase is not directly involved in glycogen synthesis but is, instead, essential for the metabolism of galactose and the synthesis of capsular polysaccharides. In plants and green algae, UGPase is only known to be present in the cytosol where it produces UDP-glucose for the synthesis of cell wall polysaccharides, the production of the carbohydrate moiety of glycolipids, glycoproteins and proteoglycans, and as a precursor to UDP-glucuronic acid (Kleczkowski et al. 2004). In nonphotosynthetic tissues, UGPase is linked to sucrose degradation by the use of UDP-glucose, produced by sucrose synthases, to provide Glc-1-P for the production of energy and other cellular components (Martin et al. 1993, Kleczkowski et al. 2004). The UGPases of eukaryotes and prokaryotes only share similarity within their “UTP-glucose-1-phosphate uridylyltransferase” domain (pfam 01704) and are otherwise too dissimilar to be aligned; thus, the prokaryotic UGPases, not being involved in glycogen synthesis, have been excluded from these analyses. There is no evidence in any species from a red- or green-plastid lineage for a UGPase of bacterial origin. All sampled red and green algae and plants contained one or more isoforms most similar to those found in other eukaryotes.
The AGPases contain a nucleotide-transferase domain (pfam 0483) that is shared by a range of nucleotide transferases such as GDP-mannose pyrophosphorylases and Glc-1-P thymidylyltransferases. Outside of this domain there is no similarity at the amino acid level between the enzyme groups. The AGPases are tetrameric enzymes (Haugen and Preiss 1979). The bacterial enzyme is composed of four identical subunits, whereas AGPase from higher plants is a heterotetramer composed of two closely related but different subunits. Both the small subunit (SSU) and large subunit (LSU) are responsible for catalysis and allosteric regulation (Cross et al. 2004). Both subunits are related to the prokaryotic AGPase.
The plant LSU and SSU separate into two distinct and well-supported clades (all supported at 100%) that represent an early duplication during the evolution of plants (Fig. 2). Multiple isoforms of AGPase LSU have been found in plants (e.g. three in potato, four in Arabidopsis, and four in rice). These form three well-supported clades indicating duplications in the ancestor of Viridiplantae and subsequent duplications in some lineages.
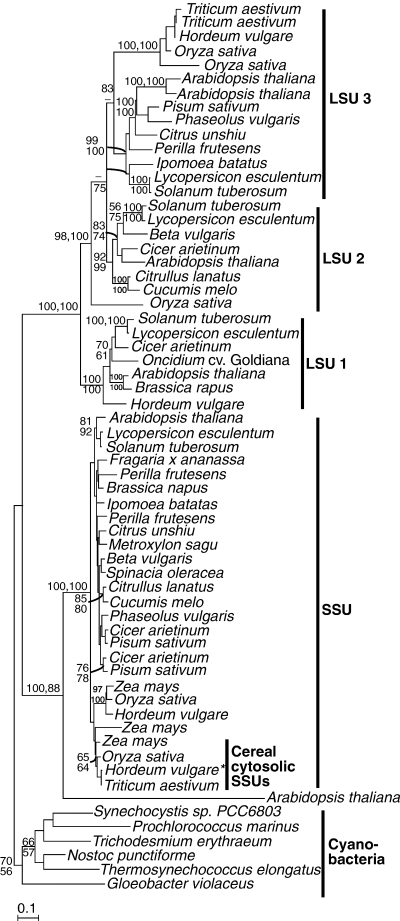
Protein maximum likelihood phylogeny of ADP-glucose pyrophosphorylase. Numbers at nodes indicate bootstrap support from (top to bottom or left to right) maximum likelihood, as determined by PhyML and weighted neighbor joining. Analysis used 424 characters, the alpha parameter=1.002 and proportion of invariable sites=0.014.*a plastidial protein is encoded by the same gene (see text for details). SSU, Small Subunit; LSU, Large Subunit.
In most plant species, the synthesis of ADP-glucose occurs entirely within the plastids in all tissues so far examined (Beckles et al. 2001). However, in the endosperm of grasses, a second form of AGPase synthesizes ADP-glucose outside the plastid and accounts for most (85%–95%) of the total AGPase activity in the endosperm (Denyer et al. 1996, Thorbjørnsen et al. 1996a, Beckles et al. 2001, Sikka et al. 2001). ADP-glucose is then transported into the plastid on a specific transporter, encoded at the Brittle1 locus (Shannon et al. 1998, Patron et al. 2004a). In barley, cytosolic and plastidial SSU mRNAs are produced from a single gene by the use of two alternate first exons (Thorbjørnsen et al. 1996b), although a separate SSU plastidial gene has been shown to be responsible for AGPase activity in the plastid (Johnson et al. 2003). Which of the LSUs is responsible for activity in the plastids of cereals is currently unknown. The cytosolic cereal SSUs form a sister clade to the cereal plastidial isoforms, implying a gene duplication after the divergence of cereals from other plants.
It is not completely clear whether starch in red algae is made using ADP-glucose or UDP-glucose. Although α-glucan synthases and branching enzymes (BE) isolated from floridean starch and the cytosol of red algae have shown a preference for UDP-glucose (Lluisma and Ragan 1998, Nyvall et al. 1999), ADP-glucose was shown to be a possible substrate for glucan synthases in Porphyridium purpureum (Sheath et al. 1979, Sesma and Iglesias 1998) and AGPase activity was characterized and an enzyme purified from Gracilaria gracilis (Sesma and Iglesias 1998). Molecular data are lacking from both of these species, but neither the genomes of C. merolae (Matsuzaki et al. 2004) nor Galdieria sulphuaria (Michigan State University Galdieria Database http://genomics.msu.edu/galdieria_public) encode an AGPase. Both contain a single isoform of UGPase that is weakly related to plant cytosolic homologues (Fig. 3). These red algal proteins do not have any N-terminal extension and are therefore likely to be located in the cytosol, as are green algal and plant homologues.
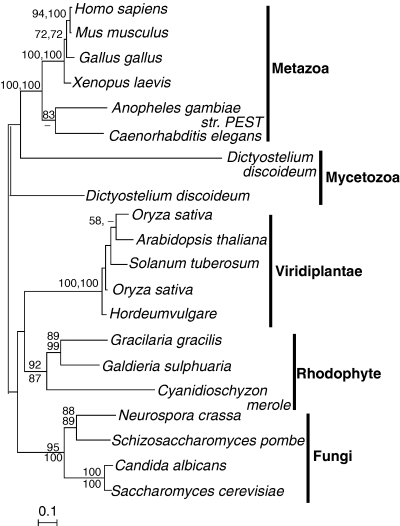
Protein maximum likelihood phylogeny of eukaryotic UDP-glucose pyrophosphorylase. Numbers at nodes indicate bootstrap support from (top to bottom or left to right) maximum likelihood, as determined by PhyML and weighted neighbor joining. Analysis used 420 characters, the alpha parameter=1.077 and proportion of invariable sites=0.098.
Glucan synthases. Based on sequence similarity, α-1,4-glucan synthases have been grouped in two families, glycogen synthases and glycogen (starch) synthases (G(S)S), which roughly correspond to the type of polysaccharide synthesized (Cida et al. 2002). There is no significant overall sequence similarity between the two families. These glycosyltransferases are partly responsible for the differences in structure of glycogen and starch because they direct the elongation of the linear glucan chains, although branching and debranching enzymes also contribute to the final structure of the molecule (Smith 1999, Ball and Morell 2003). Glycogen synthetases from yeasts and mammals use UDP-glucose as sugar donor, and their activities are regulated at two levels: inactivation by phosphorylation at serine/threonine sites and allosteric activation by Glc-6-P (Villar-Palasi and Guinovart 1997). Eubacteria and plant plastids use ADP-glucose as sugar donor, and neither inactivation by phosphorylation nor allosteric activation by Glc-6-P have been reported in this family.
Plants have several isoforms of G(S)S that have specific roles in determining granule structure (Edwards et al. 2002, Fulton et al. 2002). All are related to the G(S)S found in prokaryotes. Granule bound starch synthases (GBSS) are bound exclusively to the starch granule, whereas soluble starch synthases (SS) are localized predominantly in the plastid stroma.
Phylogenetic analyses split the eukaryotic isoforms into two major groups. The first is well supported (91% bootstrap support from both methods) and consists of SSI, SSII, and GBSS isoforms (plus isoforms from red, green, and dinoflagellate algae), whereas the second consists of SSIII and SSIV isoforms, which also group together with good support (99% and 100% bootstrap) with prokaryotic G(S)S. The two groups represent an ancient duplication, but whether this happened in cyanobacteria or in plastids is not certain. Some cyanobacteria have two isoforms, glg1 and glg2, but glg2 is closely allied to plant and green algal SSIII and SSIV isoforms and in Figure 4 is actually branched with relatively good support within the plant and green algal clade. At face value, this would suggest the cyanobacteria acquired this gene relatively recently from a plant or alga. Alternatively, the gene may have duplicated within cyanobacteria and plastids may have inherited both isoforms, with red plastids subsequently loosing SS3/SS4 isoforms.

Protein maximum likelihood phylogeny of glycogen (starch) synthases. Numbers at nodes indicate bootstrap support from (top to bottom or left to right) maximum likelihood, as determined by PhyML and weighted neighbor joining. Analysis used 346 characters, the alpha parameter=1.551 and proportion of invariable sites=0.037. GBSS, Granule-Bound Starch Synthase; SS, Soluble Starch Synthase.
Red algae have been reported as having starch synthase activity that preferentially uses UDP-glucose in the cytosol (Nyvall et al. 1999), which is the site of floridean starch. The use of UDP-glucose rather than ADP-glucose has led to suggestions that rhodophytes, unlike plants and green algae, have an α-glucan metabolic pathway of eukaryotic origin (Viola et al. 2001). However, the identity of the glucan donor is only part of the story. An interrogation of the genomes of G. sulphuraria and C. merolae reveals genes that encode proteins with high similarity to G(S)S but not glycogen synthase, that is, they were readily alignable with both bacterial and plant isoforms but not those from other eukaryotes. In the phylogeny, these rhodophyte proteins group within the well-supported SSI/SSII/GBSS clade and, more specifically, with GBSS isoforms on a long branch, which may reflect divergence for the use of UDP-glucose as a glucosyl donor. Physicochemically, floridean starch has properties more similar to those of amylopectin than amylose (Yu et al. 2002) and is generally described as being amylose free. However, amylose has been purified from Porphyra purpureum (McCracken and Cain 1981). The red algal proteins from C. merolae and G. sulphuraria do not appear to have N-terminal extensions (relative to the prokaryotic sequences) and are therefore likely to be cytosolic and to account for the production of floridean starch. Intriguingly, the marine dinoflagellate H. triquetra, which has a secondary plastid of red algal origin (Fast et al. 2001, Archibald and Keeling 2002, Patron et al. 2004b), also encodes a G(S)S, and this gene shows a specific relationship to GBSSI of plants and green algae. Very little is known about the properties or synthesis of starch in dinoflagellates, except that it is cytosolic and granular, or in other starch-storing organisms with secondary plastids. Whether or not the dinoflagellate and red algal isoforms shown in Figure 4 are responsible for the production of the cytosolic starch found in these species, they appear to represent a case of cytosolic gene replacement by a gene of plastid origin.
Branching enzymes. The branches of mature polysaccharides are produced by α-1,4-glucan-6-glucosyltransferases, known as BE, which cleave the α-1,4 glucosidic linkage, yielding a nonreducing end oligosaccharide chain, and attach the oligosaccharide to the α1,6 position (Smith 1999, Ball and Morell 2003). Branching increases the number of nonreducing ends, making the polymers more available for synthesis and digestion. Branching enzymes belong to the α-amylase family of enzymes, which includes α-amylases, pullulanase/isoamylase, cyclodextrin glucanotransferases, and BE. Branching enzymes are divided into three domains based on secondary structure prediction and comparison with other enzymes in the family: an amino-terminal domain, a carboxyl-terminal domain, and a central β-barrel catalytic domain. Multiple isoforms of BE have been reported in higher plants. These isoforms are classified as starch branching enzyme I or B (SBEI) and SBEII (or A) and have been reported to differ in substrate specificity and expression patterns (Rahman et al. 2001, Rydberg et al. 2001, Mutisya et al. 2003, Yao et al. 2004). In several cereal crops, it has been shown that the SBEII family can be further divided into IIa and IIb isoforms that are encoded by different genes (Boyer and Preiss 1978, Gao et al. 1996, 1997).
Prokaryotic and eukaryotic BE (from all species) are similar enough to align in the central β-barrel catalytic domain and the carboxyl terminal domain; however, the amino-terminal domain of plants is comparatively long and poorly conserved, even within the plant kingdom, and so most characters from this domain were excluded from analysis.
The prokaryotic and eukaryotic BEs form two discrete and well-separated clades (Fig. 5). Both plastid and nonplastid eukaryotic enzymes group together, but none is specifically related to the cyanobacteria. Moreover, all the eukaryote enzymes share three conserved insertions compared with the prokaryotes, altogether indicating that the original plastid isoform was lost and replaced by a host enzyme. This is in agreement with the phylogeny reported by Lluisma and Ragan (1998). Only one isoform was found in each of the rhodophyte genomes. Within the eukaryotic clade, fungi, rhodophytes, and each isoform from Viridaeplantae all form separate well-supported subclades. The metazoans also form a subclade but with no support. The Viridiplantae subclades are sisters, implying a gene duplication in that lineage.
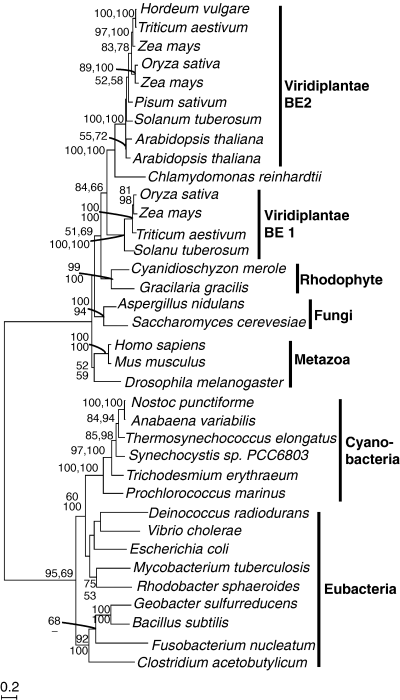
Protein maximum likelihood phylogeny of branching enzymes. Numbers at nodes indicate bootstrap support from (top to bottom or left to right) maximum likelihood, as determined by PhyML and weighted neighbor joining. Analysis used 546 characters, the alpha parameter=1.310 and proportion of invariable sites=0.043. BE, branching enzyme.
Isoamylases. Starch-debranching enzymes in plants are of two types. The limit dextrinases (also referred to as R-enzymes or pullulanases) hydrolyze α-1,6 linkages in amylopectin and α-limit dextrin but do not readily hydrolyze glycogen. The isoamylase-type debranching enzymes are most active on amylopectin, but they also are active on glycogen and α-limit dextrin substrates. Isoamylases are inactive on pullulan. It is known that isoamylases play an important role in plant starch synthesis (Mouille et al. 1996, Zeeman et al. 1998, Dauvillée et al. 2000) and granule initiation (Burton et al. 2002, Hussain et al. 2003, Bustos et al. 2004).
Although the two types of debranching enzyme have related primary amino acid sequences within the conserved α-amylase domains (James et al. 1995, Beatty et al. 1999, Hussein et al. 2003), which shows that all α-amylases probably had a common ancestor, the two types are sufficiently divergent that inclusion of all enzymes reduced the inclusion set to an uninformative number of characters. Consequently, limit dextrinases and all eukaryotic α-amylases of nonplastid lineages have been excluded from these analyses. Isoamylases from plastid-containing organisms readily align to bacterial isoamylases, from which they likely arose.
Plant isoamylase isoforms I, II, and III, green and red algal isoforms, form five well-supported clades, although there is no support for relationships between them (Fig. 6). Gene duplications have seemingly taken place independently in the lineages of vascular plants, green and red algae, although cyanobacteria also encode multiple isoforms. Whether more than one isoform was inherited by the plastid cannot be resolved from these data. Altogether, the plant and green algal plastid enzymes and the red algal cytosolic enzyme all form a weakly supported group that is specifically related to cyanobacteria, indicating that both the plastid and cytosolic enzymes from these organisms are all likely plastid derived.
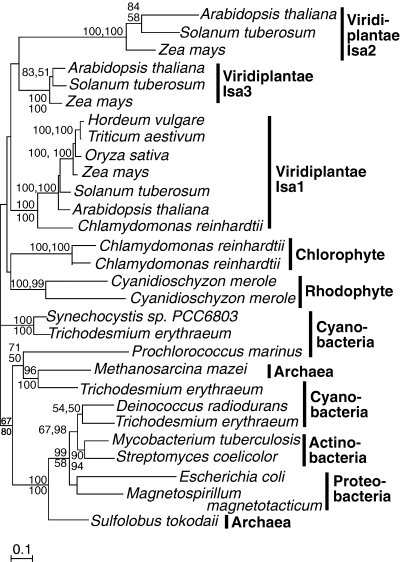
Protein maximum likelihood phylogeny of isoamylase-type de-branching enzymes. Numbers at nodes indicate bootstrap support from (top to bottom or left to right) maximum likelihood, as determined by PhyML and weighted neighbor joining. Analysis used 427 characters, the alpha parameter=1.176 and proportion of invariable sites=0.032.
DISCUSSION
It is recognized that plant (and green algal) starch, although different in structure, is related to bacterial glycogen. Its plastidial location and the use of AGP-glucose as a glucan donor have made this easy to infer. However, even the green-plastid starch synthetic pathway contains at least one enzyme (BE) of eukaryotic origin, which has replaced the bacterial enzyme in the plastid (Fig. 7) (Lluisma and Ragan 1998). Branching enzymes are important for the synthesis of amylopectin, which, in turn, is vital in ensuring the crystalline structure of the starch granule.
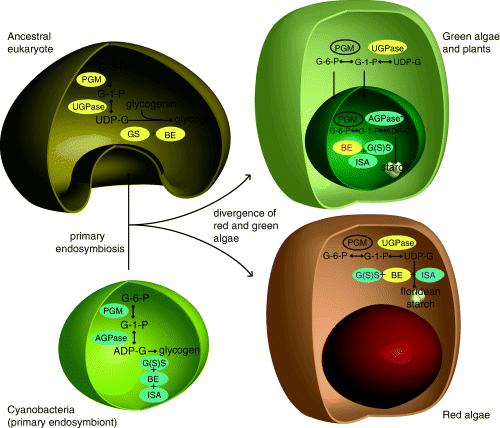
Schematic diagram showing the acquisition of a cyanobacterium by an ancestral eukaryote and subsequent integration of the eukaryotic and prokaryotic glycogen synthesis pathways to the pathways now observed in green algae (and plants) and red algae. Yellow ellipses indicate an enzyme of eukaryotic origin and blue ellipses indicate a prokaryotic (plastidial) origin. Red text indicates plastidial gene replacement. Purple text indicates cytosolic gene replacement. PGM, phosphoglucomutase; UGPase, UDP-glucose-pyrophosphorylase; AGPase, ADP-glucose pyrophosphoryase; GS, glycogen synthase; G(S)S, glycogen starch synthase; BE, branching enzyme; ISA, isoamylase *cereals also encode a cytosolic AGPase. This is a duplication of the plastidial gene. The Brittle–1 transporter imports ADPG from this reaction into the plastid.
Red algal starch accumulates in the cytosol, and this, along with biochemical evidence for the use of UDP-glucose in its synthesis (Nyvall et al. 1999, Viola et al. 2001), has led to speculation that the entire pathway was inherited from the host (Viola et al. 2001) and might therefore be quite distinct from the pathway in green plastids. This study, however, shows that this is not the case: Most of the enzymes of the red algal cytosolic starch synthetic pathway share a common origin with the plastid-targeted enzymes of green algae and plants, the exception being the absence of AGPase in red algae (whether their PGMs are closely related is unresolved). At the same time, in neither group are all the enzymes functioning in the compartment where they originated. Green algae, plants, and red algae all use glycogen (starch) synthases and isoamylases of bacterial (endosymbiont) origin and BE of eukaryotic (host) origin for the final steps of starch synthesis, giving them all the crystalline structure different from that seen in either eukaryotic or prokaryotic glycogen. It appears therefore that different enzymes have been recompartmentalized during the evolution of these lineages. In red algae, isoamylases and starch synthases are plastid-derived enzymes operating in the cytosol where they use UDP-glucose as a glucan donor, resulting in cytosolic starch synthesis. In the green lineage, on the other hand, a cytosolic BE, and probably the PGM, has relocated to the plastid so starch synthesis is completely plastidic. Based on the endosymbiont-derived nature of most of these enzymes, it is likely that the ancestor of green and red algae synthesized starch in the plastid and that the red algae subsequently relocated the pathway to the cytosol. Moreover, the use of ADP-glucose as a glucan donor in green G(S)S is likely ancestral (it is shared with prokaryotes), and so it is also likely that the use of UDP-glucose in red algal enzymes is recently derived.
The endosymbiont origin for most of the cytosolic red algal starch synthesis enzymes raises a number of very interesting questions about how this pathway fits into the larger picture of metabolism in red algae and also in algae with secondary plastids of red algal origin. Floridoside, α-D-galactopyranosyl-(1-2)-glycerol, is a major photosynthetic reserve in red algae, and carbon is presumably shunted between the floridoside and starch pools (Ekman et al. 1991). This is similar to the situation in plants where carbon moves between the starch and sucrose pools. Multiple pathways, such as that of agar synthesis, a sulfurated galactose polymer, compete for carbon reserves in the cytosol, and investigation of how these pathways interrelate will shed light on the interesting process of integration of endosymbiont enzymes into the algal cytosol. Further biochemical and molecular investigation into enzymes of starch synthesis from a diversity of red algae is also needed to complement the data from the genomes currently available. There is biochemical evidence for glucans in the plastids of some red algae (Yu and Pedersen 1993) and evidence for amylose in the cytosolic starch of some red algal lineages (McCracken and Cain 1981). This suggests that perhaps both cytosolic and plastidial pathways exist in some red algal lineages. Indeed, the genomic data come from some of the simpler, microscopic, and reduced red algae (Matsuzaki et al. 2004), so a great deal of variability in other taxa is expected.
Also to be investigated is the crystalline starch found in eukaryotes with secondary plastids. Although no starch-accumulating organisms with secondary green plastids are known (both known groups store β-1,3-glucans [Barras and Stone 1969, McFadden et al. 1997]), two large and diverse groups of protists with secondary plastids, likely to be of the same red algal origin, store granular starch. Dinoflagellates have relocated the starch synthetic pathway a second time from the cytosol of the red alga to the cytosol of the secondary host, where crystalline starch has been reported to accumulate in some species (Dodge and Lee 2000). We have now characterized the first starch synthetic enzyme from a dinoflagellate, and despite its cytosolic location, it is clearly derived from a bacterium (most likely the plastid endosymbiont), although its close relationship to green plastid GBSSI is enigmatic. It will be interesting to see whether all the enzymes of this pathway have also relocated from the secondary endosymbiont or whether dinoflagellate host cytosolic proteins have contributed to a fresh rebuilding of the starch synthetic pathway. The second group of algae with secondary plastids and starch are the cryptomonads. These represent an intermediate in the processes of secondary endosymbiosis because they have retained the relict nucleus of the red algal endosymbiont (Ludwig and Gibbs 1985, Archibald and Keeling 2002, Cavalier-Smith 2003). In this case, starch granules accumulate in the cytosol of this reduced endosymbiont, known as the periplastidial space (Gillot and Gibbs 1980). However, the genome of the endosymbiont has been completely sequenced and does not encode any starch synthesis enzymes (Douglas et al. 2001), so these proteins are presumably encoded in the host nucleus and targeted to the periplastidial space through the two outer membranes of the four-membraned plastid.
The emerging picture of the evolutionary history of starch synthesis is complex. Enzymes have been derived from various sources through endosymbiotic mergers and are now located in different compartments, and these compartments frequently do not correspond to the original site of activity of the enzyme. Moreover, this complex picture only includes two major lineages of starch producers, red and green algae. The initial data from dinoflagellates suggest they will add further wrinkles to the story. Altogether, such mixing and matching of enzymes and compartments between host and endosymbiont is characteristic of organelle evolution whose significance is becoming increasingly apparent and has affected other pathways in addition to starch biosynthesis (Ralph et al. 2004). The impact of the host and symbiont on one another is even deeper than was initially imagined, and their integration is an ongoing process.
Acknowledgments
This work was supported by the Protists EST Program (PEP) of Genome Canada/Genome Atlantic. We thank Drs. Kay Denyer and Ross Waller for critical reading of this manuscript.




