The T-box genes H15 and optomotor-blind in the spiders Cupiennius salei, Tegenaria atrica and Achaearanea tepidariorum and the dorsoventral axis of arthropod appendages
Abstract
SUMMARY Dorsoventral axis formation in the legs of the fly Drosophila melanogaster requires the T-box genes optomotor-blind (omb) and H15. Evolutionary conservation of the patterning functions of these genes is unclear, because data on H15 expression in the spider Cupiennius salei did not support a general role of H15 in ventral fate specification. However, H15 has a paralogous gene, midline (mid) in Drosophila and H15 duplicates are also present in Cupiennius and the millipede Glomeris marginata. H15 therefore seems to have been subject to gene duplication opening the possibility that the previous account on Cupiennius has overlooked one or several paralogs. We have studied omb- and H15-related genes in two additional spider species, Tegenaria atrica and Achearanea tepidariorum and show that in both species one of the H15 genes belongs to a third group of spider H15 genes that has an expression pattern very similar to the H15 pattern in Drosophila. The expression patterns of all omb-related genes are also very similar to the omb expression pattern in Drosophila. These data suggest that the dorsoventral patterning functions of omb and H15 are conserved in the arthropods and that the previous conclusions were based on an incomplete data set in Cupiennius. Our results emphasize the importance of a broad taxon sampling in comparative studies.
INTRODUCTION
Arthropod appendages develop along three different axes. The molecular mechanisms of appendage axis formation have been studied best in the fly Drosophila melanogaster. The anterior–posterior axis builds on the mechanisms that also establish the anterior–posterior polarity of the body segments (Cohen 1990). The second axis, the proximal–distal axis, is governed by a hierarchic gene cascade comprising three principal levels (e.g., Lecuit and Cohen 1997; Rauskolb and Irvine 1999; Rauskolb 2001): first, the morphogens Wingless (Wg) and Decapentaplegic (Dpp) erect a system of protein concentration gradients. These gradients are read out by the next level, the leg gap genes, that include genes like dachshund and Distal-less. The ensuing subdivision into leg segments (podomeres) is the responsibility of the lower level of the cascade that includes mostly members of the Notch signaling pathway or its target genes.
The developmental mechanism governing the formation of the third appendage axis, the dorsal–ventral axis, is poorly known even in Drosophila. The two morphogens Dpp and Wg that organize the proximal-distal leg axis also organize the formation of the dorsal–ventral leg axis. The dpp gene has been shown to activate the T-box gene optomotor-blind (omb) along the dorsal side and misexpression of omb has been shown to lead to dorsalizing effects (Brook and Cohen 1996; Maves and Schubiger 1998). The wg gene activates another T-box gene, H15, in a large ventral domain (Wilder and Perrimon 1995; Brook and Cohen 1996; Abu-Shaar and Mann 1998). This ventral expression domain suggests a ventralizing role of H15, but experimental evidence for this is still lacking. Apart from these two T-box containing factors no other genes have been identified yet that are involved in dorsal–ventral leg axis formation in Drosophila.
The mechanisms of anterior–posterior and proximal–distal axis specification have been studied in other arthropods as well and seem to be conserved to a certain extent among arthropods (e.g., Abzhanov and Kaufman 2000; Prpic et al. 2003). The patterning of the dorsoventral leg axis, however, is not well studied in other arthropods. As a first step toward a better understanding of dorsal–ventral patterning in arthropod appendages, we have previously studied the expression of omb and H15-related genes in the American wandering spider Cupiennius salei and the pill millipede Glomeris marginata (Prpic et al. 2003, 2005). In both species, the omb gene showed a conserved expression pattern along the dorsal side of all appendages, compatible with a role in dorsal specification. With regard to the H15 genes, we recovered two different genes in each species. Also in Drosophila two H15-type genes are present, H15 and midline (mid), but they have identical expression patterns in the legs (Prpic et al. 2003) and probably identical functions in development (e.g., Buescher et al. 2004, 2006; Miskolczi-McCallum et al. 2005; Qian et al. 2005; Reim et al. 2005). Confusingly, however, the gene pairs in Cupiennius and Glomeris could neither be directly homologized with the Drosophila H15-related genes in a phylogenetic analysis, nor did they show expression patterns readily comparable with the Drosophila genes or with each other. The expression of the H15-related genes in Cupiennius was not compatible with a conserved function in ventral fate determination under the control of Wg signaling.
Here we present a study of omb and H15 type genes in three spider species representing three different superfamilies of entelegyne spiders (see Fig. 1D): Cupiennius salei (Fig. 1A), Tegenaria atrica (Fig. 1B), and Achaearanea tepidariorum (Fig. 1C). The most important finding is the discovery of a third H15-type gene in Tegenaria and its ortholog in Achaearanea that show an expression pattern very similar to the Drosophila H15-type genes. Our new data show that our previous conclusions with respect to the H15-type genes were based on an incomplete data set. All available data now suggest that the dorsalizing and ventralizing roles of omb and H15, respectively, are conserved in the arthropods. Further studies are now needed in order to confirm the dorsalizing and ventralizing functions of these genes, to establish their regulation by upstream factors (e.g., dpp, wg), and to identify further genes involved in dorsal–ventral axis formation in the arthropods.
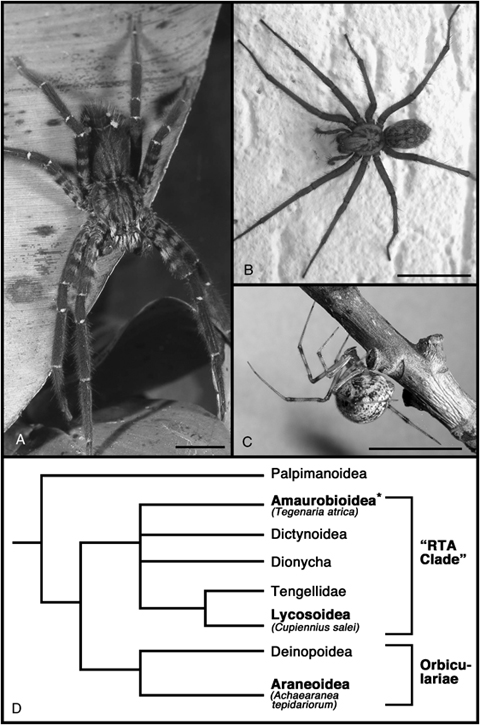
Spider species used in this study. (A) Cupiennius salei, the Central-American wandering spider, belongs to the family Ctenidae (Superfamily Lycosoidea). (B) Tegenaria atrica, a member of the Agelenidae (Superfamily Amaurobioidea). (C) Achaearanea tepidariorum belongs to the Theridiidae (Superfamily Araneoidea). (D) Phylogeny of the higher entelegyne spiders (simplified after Coddington and Levi 1991) illustrating the interrelationships of the spider species in this study. Superfamilies containing a species used in this study are in bold type. The asterisk indicates that the monophyly of the Amaurobioidea is not firmly established. Images taken by and courtesy of Andreas Hartl (A), Nikola-Michael Prpic (B), Matthias Pechmann (C). Scale bar in A, B, C is 10 mm.
MATERIALS AND METHODS
Embryo fixation and staging
Embryos of Cupiennius were obtained and treated as described (Damen and Tautz 1998; Damen et al. 1998). Female and male Tegenaria were collected at several sites in Cologne (Germany). Embryos of Tegenaria were obtained and treated as has been described for Cupiennius embryos. Embryos of Achaearanea were obtained from a laboratory culture in Cologne and fixed as described by Akiyama-Oda and Oda (2003). Spider embryos develop first by germ band extension, but then undergo a process called inversion, in which the germ band splits ventrally and which precedes dorsal closure (see Prpic and Damen, 2005a, for details). Embryos of all three spider species were staged roughly into stages before inversion (germ band elongation stages) and stages during inversion. For further details about spider embryology see Foelix (1996).
Cloning of T-box genes and sequence analysis
RNA from each species was isolated from a mixture of embryos of different developmental stages using TRIZOL (Invitrogen, Carlsbad, CA, USA). Poly-A RNA (PolyATtract mRNA isolation system III, Promega, Madison, WI, USA) was used to produce cDNA (SuperscriptIII first strand synthesis system, Invitrogen) for reverse-transcriptase polymerase chain reaction (RT-PCR). Two independent duplicates were performed for each gene cloning assay. The primers used to obtain fragments of omb and H15/mid type genes have been published previously (Prpic et al. 2003, 2005). In addition, we used the following primers: Cs-omb-2 was recovered with the primers CAN AAY GAR ATG ATH GTN CA (forward primer) and AA NGG RTT RTA YTT DAT YTT (reverse primer) in the initial PCR and the primers CAN AAY GAR ATG ATH GTN CA (forward primer) and TC RTT YTG RTA NGC NGT NAC (reverse primer) in the nested PCR. The sequences of At-H15-1, At-H15-2, and At-omb were available from GenBank. We used gene specific primers to amplify fragments of these genes by PCR (At-H15-1: nt 15–610; At-H15-2: nt 30–619; At-omb: 1071–1513). Sequences were determined from both strands on an ABI-3100 automated sequencer (Applied Biosystems, Foster City, CA, USA), using Big Dye dye-terminators (Perkin Elmer, Waltham, MA, USA). The sequences are available from GenBank with the following accession numbers: Cs-omb: AJ518937; Cs-omb-2: AM774407; Cs-H15-1: AJ518938; Cs-H15-2: AJ518939; At-H15-1: AB177874; At-H15-2: AB177875; At-omb: AB177876; Ta-H15-1: AM404110; Ta-H15-2: AM404111; Ta-H15-3: AM404111; Ta-omb-1: AM404113; Ta-omb-2: AM404114. Sequence analysis was performed as described previously (Prpic et al. 2005).
In situ hybridization and preparation
Whole-mount in situ hybridization was essentially performed as described for Drosophila (Tautz and Pfeifle 1989) with modifications for spider embryos (Damen and Tautz 1998). We used DIG-labeled RNA probes. Embryos were analyzed as whole mounts in PBST under a Leica dissection microscope equipped with an Axiocam (Zeiss, Jena, Germany). Appendages were dissected with fine tungsten needles and photographed under a Zeiss Axioplan compound stereomicroscope. Brightness, contrast and color values were corrected in all images using the image processing software Adobe Photoshop (Version 7.0 for Apple Macintosh).
RESULTS
T-box genes from the spider species Cupiennius salei, Tegenaria atrica, and Achaearanea tepidariorum
From the spider Tegenaria we have isolated in a PCR screen two different gene fragments with high similarity to omb from Drosophila, and three different gene fragments with high similarity to H15 and mid from Drosophila. New PCR screening in the spider Cupiennius resulted in the identification of a second omb-type gene fragment, in addition to the previously reported omb-type T-box sequence and the two H15-related T-box sequences in this spider species (Prpic et al. 2003). In addition the sequence of one omb-type gene and two H15-type genes of the common house spider Achaearanea are available in GenBank (accession numbers: see methods).
In order to establish the orthology of the spider omb and H15-type genes we performed a phylogenetic analysis using all T-box genes present in the Drosophila genome and all omb and H15-types genes identified in the three spider species, supplemented with sequences from the pill millipede Glomeris marginata (Prpic et al. 2005). The phylogenetic tree (Fig. 2) shows that all omb related sequences form a cluster of closely related sequences. The omb gene of Drosophila forms the sister group of all other omb genes. It is not possible to designate one of the two omb-like genes from either Cupiennius or Tegenaria as being more closely related to Drosophila omb and we designate them therefore simply as Ta-omb-1 and Ta-omb-2 (in the case of Tegenaria) and as Cs-omb (this name has been given previously; Prpic et al. 2003) and Cs-omb-2 (in the case of Cupiennius).

Phylogenetic analysis of arthropod T-box gene fragments. Shown is the unrooted Puzzle tree computed from 1000 intermediate trees produced with the Quartet Puzzling method (Strimmer and von Haeseler 1996). The numbers at the tree edges are the reliability values. For reasons of clarity the branch with the omb genes has been magnified (see inset). Dm, Drosophila melanogaster; At, Achaearanea tepidariorum; Cs, Cupiennius salei; Ta, Tegenaria atrica; Gm, Glomeris marginata.
The phylogenetic analysis also shows that all H15-like sequences form a well-supported cluster of closely related sequences, but also shows that they cluster together according to arthropod class (chelicerate/myriapod/insect) (Fig. 2). Furthermore, the phylogenetic analysis reveals that, within the spiders, the H15 sequences form three groups, one group consisting of At-H15-1, Cs-H15-1 and Ta-H15-1 (group-1 H15 genes), a second group consisting of Cs-H15-2 and Ta-H15-2 (group-2 H15 genes), and a third group consisting of At-H15-2 and Ta-H15-3 (group-3 H15 genes). These three groups are also supported by the expression pattern analysis (see below).
Please note that the Achaearanea genes have been named previously (Akiyama-Oda and Oda 2004); both sequence and expression analysis suggest that At-H15-2 despite its name is a group-3 H15 gene. Nevertheless we decided not to rename the gene as this could lead to new confusion, because it would suggest that three H15-type genes have already been isolated in this species.
Embryonic expression of omb homologs
We next examined the expression of the omb-related genes in all three spider species. First we describe the general expression pattern of the genes; the expression in the prosomal appendages is reported in the following section. The body of spiders is divided into two tagmata, the anterior prosoma and the posterior opisthosoma. The prosoma bears six pairs of appendages (from anterior to posterior): the chelicerae (venom fangs), the pedipalps (mainly sensory appendages) and four pairs of walking legs. The opisthosoma bears the breathing organs (book lungs and tubular tracheae) and the spinnerets.
The expression of Cs-omb has been described before (Prpic et al. 2003). Here we focus on the newly isolated Cs-omb-2 gene. The expression of Cs-omb-2 is virtually identical to the expression of Cs-omb. Like Cs-omb, Cs-omb-2 is expressed in the brain centers of the lateral eyes (Fig. 3A) and in a segmental pattern in the opisthosoma (Fig. 3B). When at later stages the four opisthosomal limb buds develop, Cs-omb-2 is expressed in the dorsal portion of all of them. However, Cs-omb-2 is also expressed in more posterior segments that do not develop appendages or appendage-derived organs (Fig. 3B).
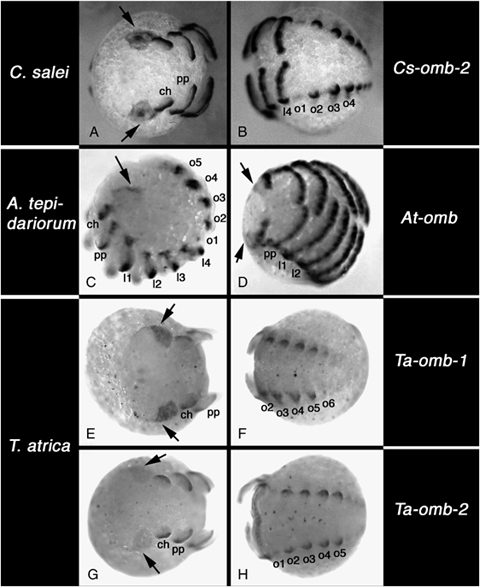
Embryonic expression of omb-type T-box genes. (A, B) Expression of Cs-omb-2. (A) Frontal view showing expression in the optic lobes (arrows). (B) View of the opisthosoma. (C, D) Expression of At-omb. (C) Lateral view, (D) ventral view; arrows denote expression in the head lobes. (E, F) Expression of Ta-omb-1. (E) Frontal view showing expression in the optic lobes (arrows). (F) View of the opisthosoma. (G, H) expression of Ta-omb-2. (G) Frontal view showing expression in the optic lobes (arrows). (H) View of the opisthosoma. Note that the irregular black dots in E-H are unspecific precipitation of NBT/BCIP caused by the in situ procedure in Tegenaria. All panels: anterior to the left. A, B, and H are mid inversion stages. C, E, F, and G are early inversion stages. D is a late inversion stage. Abbreviations: ch, chelicera; pp, pedipalp; l1–l4, walking legs 1 to 4; o1–o6, opisthosomal segment 2–6.
Like Cupiennius, Tegenaria also has two different omb-related genes. Both genes are expressed in the head lobes in a large region that also encompasses the primordia of the lateral eyes and corresponding brain regions (Fig. 3, E and G). The expression of Ta-omb-2 in this area is significantly weaker than the expression of Ta-omb-1. Ta-omb-1 is also expressed in the labrum (not shown). Both genes are expressed in the opisthosoma in virtually identical patterns. Expression is detected in the primordia of the book lungs, tubular tracheae and spinnerets, but only in their dorsal part (Fig. 3, F and H). At younger stages there is also very weak expression in opisthosomal segment 6, at a position obviously serially homologus to the expression in the breathing organs and spinnerets. However, this segment does not develop appendages or appendage derived organs.
At-omb, the only omb-type gene known from Achaearanea, is expressed similar to the omb-related genes from the other two spider species (see also Akiyama-Oda and Oda 2006). However, the expression in the optic lobes is very weak and restricted to a thin stripe in early stages (Fig. 3, C and D). Like in the other two spider species, At-omb is expressed in a segmental pattern in the opisthosoma (Fig. 3C).
Expression of omb homologs during leg development
In the chelicerae of Cupiennius both omb-related genes are expressed along the dorsal side in early stages (Fig. 4, A and G). Later, Cs-omb fades in the chelicerae (Fig. 4B), but expression of Cs-omb-2 remains strong (Fig. 4H). In both Achaearanea (Fig. 4, M and N) and Tegenaria (Fig. 4, S,T, Y, and Z) all omb-related genes are expressed along the dorsal side of the chelicerae at all examined stages of development.
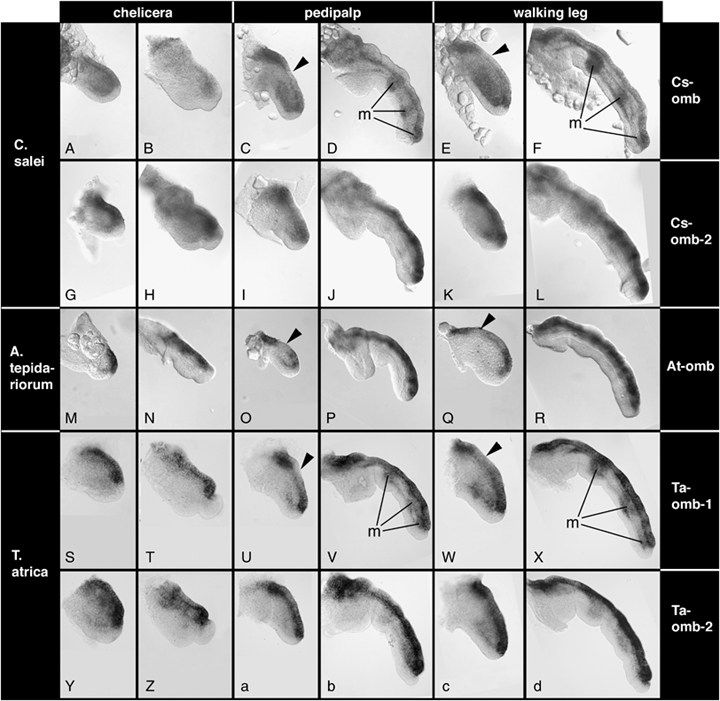
Expression of omb-type genes in the developing appendages. Genes are arranged in rows and appendage types are arranged in columns and these are given to the right and on the top, respectively. Species are arranged in rows and the species names are given to the left. For each appendage type, the first column shows young appendages at early stages of germ band elongation and the second column shows old appendages at late stages of inversion. The arrowhead in some panels denotes discontinous expression of omb homologs along the dorsal side. “m” denotes mesodermal expression in some appendage types. For details see text.
The pedipalps and walking legs of all three species also show expression of all omb-related genes along the dorsal side (Fig. 4, C–F, I–L, O–R, U–X, a–d). However, some genes show additional expression features: Cs-omb and Ta-omb-1 are expressed in the mesoderm in a repeated fashion at later stages (Fig. 4, D,F, V, and X). In addition, the dorsal expression of Cs-omb is weaker in late stages in both the pedipalps and the walking legs (Fig. 4, D and F). In earlier stages, Cs-omb (Fig. 4, C and E), At-omb (Fig. 4, O and Q) and Ta-omb-1 (Fig. 4, U and W) display a discontinuous dorsal expression. Expression at the proximal and distal ends of the appendages is strong, but these expression domains are connected by weaker expression that in some cases appears to form a distal-to-proximal gradient.
Embryonic expression of H15 homologs
The general expression of both H15-related genes from Cupiennius has been reported before (Prpic et al. 2003).
Apart from expression in the appendages that is described in the next section, both H15-related genes from Achaearanea are expressed in spots in the central nervous system (CNS) (Fig. 5, A and C) and in the developing dorsal vessel (heart) of the embryos. However, the expression of At-H15-2 in the heart is visibly weaker than the expression of At-H15-1 (Fig. 5, A,B, and D).
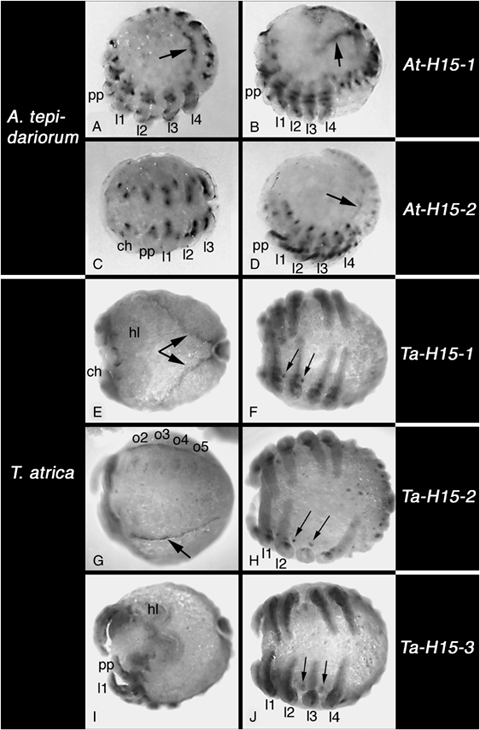
Embryonic expression of H15-type T-box genes. All embryos are late inversion stage, except for A, C, D and G, which are mid inversion stage. (A, B) Expression of At-H15-1, lateral view, arrow denotes expression in the heart. (C, D) Expression of At-H15-2. (A) Ventral view of the prosoma. (D) Lateral view, arrow denotes expression in the heart. (E, F) Expression of Ta-H15-1. (E) Dorsal view, arrows point to expression in the heart. F, ventral view, arrows denote expression in the neuroectoderm. (G, H) Expression of Ta-H15-2. (G) Ventral view of the opisthosoma, arrow denotes expression in the heart. (H) Ventral view, arrowheads denote expression in the neuroectoderm. Note that the third right walking leg has been removed. (I, J) Expression of Ta-H15-3. (I) Dorsal view. (J) Ventral view, arrows denote expression in the neuroectoderm. All panels: anterior to the left. Abbreviations: see Fig. 2.
All three H15-related genes of Tegenaria have similar but clearly different patterns of expression. Ta-H15-1 is expressed in the labrum and the developing heart (Fig. 5E). The gene is also expressed in the CNS: there is a small group of cells in the cheliceral neuromere (not shown) and similar spot-like expression domains are seen in all remaining prosomal neuromeres (Fig. 5F). Expression in the opisthosoma includes the primordia of the spinnerets and a segmental row of spots in the opisthosomal neuroectoderm extending into the posterior end of the germband (not shown). Ta-H15-2 is also expressed in the labrum (not shown) and the developing heart (Fig. 5G). Also the expression in the prosomal neuroectoderm is similar to Ta-H15-1 (Fig. 5H), but we did not detect expression of Ta-H15-2 in the cheliceral neuromere. In the opisthosoma there is expression in the spinnerets, but expression in the opisthosomal neuroectoderm was not detected (not shown). In contrast to the other two H15-related genes, Ta-H15-3 is not expressed in the labrum or the developing heart (Fig. 5I). It is expressed in the neuroectoderm of the prosoma including the cheliceral neuromere (Fig. 5J and data not shown), but in a pattern different from the point-like expression of the other two genes. The expression domain is larger and is more posterior in each hemisegmental neuromere. In the opisthosoma there is a faint segmental expression in the neuroectoderm, but this is so weak that it is hardly above the level of artificial background staining of our in situ hybridization technique; the only stronger opisthosomal expression of Ta-H15-3 is in the primordia of the breathing organs (not shown).
Expression of H15 homologs during leg development
As previously reported (Prpic et al. 2003), the two H15-related genes in Cupiennius are expressed in a restricted area along the proximal ventral side of the appendages. The expression domain of Cs-H15-1 (Fig. 6, A–F) is larger and it is expressed earlier in development than Cs-H15-2 (Fig. 6, G–I). Cs-H15-1 also has additional expression domains in the proximal dorsal area and in the ventral tip of pedipalp (Fig. 6D) and leg (Fig. 6F).
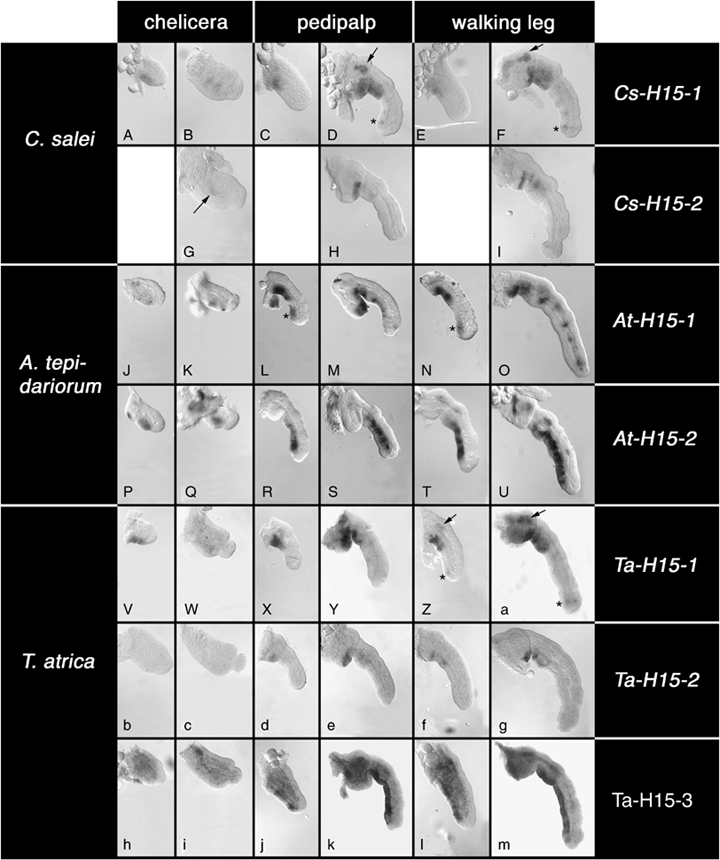
Expression of H15-type genes in the developing appendages. Genes are arranged in rows and appendage types are arranged in columns and these are given to the right and on the top, respectively. Species are arranged in rows and the species names are given to the left. For each appendage type, the first column shows young appendages at early stages of germ band elongation and the second column shows old appendages at late stages of inversion. The arrow and/or asterisk in panels D, F, L, N, Z, and a denote smaller expression domains in addition to the main ventral expression. The arrow in G denotes very weak ventral expression in the chelicera. For details see text. Please note that the black staining at the basis of the leg in U is a staining artifact: in many animals at this stage cuticle formation starts at the leg base and attracts artificial color precipitate.
At-H15-1 has an expression pattern similar to Cs-H15-1. Ventral expression is restricted to the proximal portions (Fig. 6, J–O) and there is an additional spot of expression in the ventral distal tip of pedipalp and leg (Fig. 6, L and N). In the legs at late stages there is a repeated pattern of expression dots (Fig. 6O). By contrast, At-H15-2 is not expressed in the ventral proximal area, but is expressed along the entire distal ventral side (Fig. 6, P–U). This is similar to H15 and mid in Drosophila and fundamentally different from both Cupiennius genes. It is interesting to note that the ventral patterns of At-H15-1 and At-H15-2 in pedipalps and legs are complementing each other and together they extend along the entire ventral side of the appendages.
The expression of Ta-H15-1 (Fig. 6, V–Z and a) is similar to Cs-H15-1 and At-H15-1. Expression is restricted to the proximal ventral side and, in the legs, there are additional expression domains proximally on the dorsal side and in the ventral tip (Fig. 6, Z and a). The expression of Ta-H15-2 (Fig. 6, B–G) is similar to Cs-H15-2. It is expressed more weakly and more restricted than Ta-H15-1 and lacks the additional expression domains. In the chelicerae we could not detect expression of Ta-H15-2 (Fig. 6, b and c). Ta-H15-3 (Fig. 6, h–m), finally, is expressed similar to At-H15-2 and to H15 and mid in Drosophila. Ta-H15-3 is expressed in the ventral ectoderm along the entire ventral side. However, in the pedipalps and legs proximal expression is weaker than in medial and distal areas (Fig. 6, j–m). In addition, there is a relatively strong expression within the appendages, especially at earlier stages (Fig. 6, h, j, and l).
DISCUSSION
H15-related genes in three spider species from different superfamilies
We have isolated H15-type genes from three spider species that belong to three different superfamilies, the Araneoidea (Achaearanea tepidariorum), the Amaurobioidea (Tegenaria atrica) and the Lycosoidea (Cupiennius salei) (see Fig. 1 for an overview). The fact that we find different numbers of paralogs (two each in Achaearanea and Cupiennius, and three in Tegenaria) might be explained simply by a number of independent duplication events during spider evolution. However, there is a clear pattern in the data: the phylogenetic analysis (Fig. 2) groups together those spider H15-related genes that also show similar expression patterns. Together these data strongly suggest that in the spiders there are three groups of paralogus genes that where already present in their last common ancestor (Fig. 7): Group-1 H15 genes (At-H15-1, Ta-H15-1, and Cs-H15-1); group-2 H15 genes (Ta-H15-2 and Cs-H15-2) and group-3 H15 genes (At-H15-2 and Ta-H15-3). In group 2 and group 3 the respective genes from Achaearanea and Cupiennius seem to be missing (as indicated by question marks in Fig. 7). The alternative hypothesis of a duplication event in the spider ancestor plus a recent duplication in the lineage leading to Tegenaria cannot be ruled out formally, but is not supported by our phylogenetic analysis and expression data. Although we cannot exclude that the group-3 H15 gene in Cupiennius and the group-2 H15 gene in Achaearanea have been lost secondarily, a more likely explanation for these “missing genes” is that these genes are present in the genomes, but have not yet been isolated in the PCR screens. Thus, we assume that a set of three H15-related genes is present in all three spider species (Fig. 7).
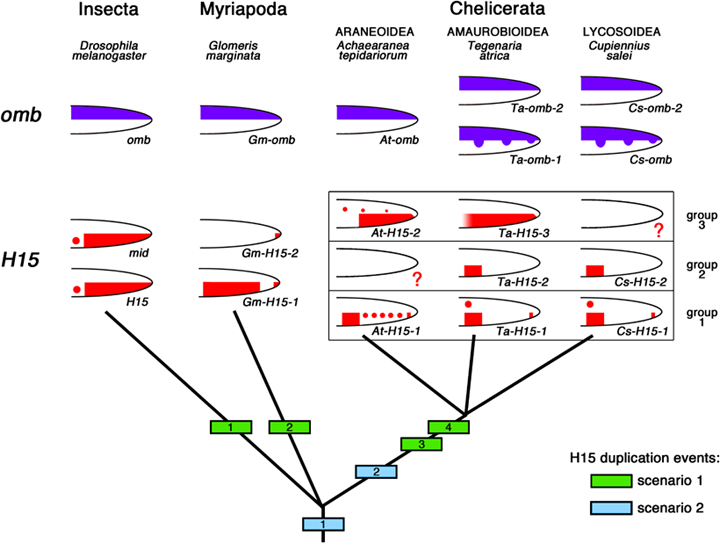
Alternative duplication histories of H15-type T-box genes in the Arthropoda. The green boxes 1–4 denote the four independent duplication events for H15 that have to be postulated if the ancestral state for the arthropods is a single H15-type gene (scenario 1; this is the hypothesis compatible with our phylogenetic analysis of the gene sequences, see Fig. 1). (Note that duplication event 4 affected only one of the two H15 genes produced in event 3.) If the sequence analysis, however, is subject to artefacts due to gene conversion, then the ancestral state in the arthropods could be already two H15-type genes (scenario 2; light blue box at the bottom of the tree denotes duplication of H15 before the split of extant arthropod lineages). A second duplication then occurred in the spider lineage before the split between the three superfamilies of Amaurobioidea, Araneoidea, and Lycosoidea. The top of the diagram depicts the simplified expression patterns of the genes in the different species. Please note that the schematic drawings summarize mainly those expression domains that are important for the discussion of DV patterning evolution. Please refer to the original data in this paper and the following references for details about the temporal and spatial expression profiles of the genes. The images are based on Abu-Shaar and Mann (1998) for H15 and omb from Drosophila; Prpic et al. (2003) for mid in Drosophila; and Prpic et al. (2005) for H15 and omb genes from Glomeris. The question marks in the Cupiennius and Achaearanea drawings indicate that these genes have not yet been isolated from these species, but are being postulated to exist, because of the corresponding data from Tegenaria. See text for further discussion. Note that the arrangement of the drawings is not intended to indicate orthology of genes, except for the H15 genes in the spiders that have been arranged in the three groups discussed in the text. The orthology of the spider omb genes is presently unclear; there might be a second omb gene in Achaearanea that is more similar to Ta-omb-1 and Cs-omb.
omb and H15 in dorsoventral appendage patterning
The data we present in this report suggests that the role of omb and H15 as instructors of dorsal and ventral fate in the appendages is conserved in all arthropods. Our new data on omb and H15 genes in the spiders Achaearanea and Tegenaria shows that the two T-box genes omb and H15 are acting in a dorsal and ventral domain, respectively, in the legs of all arthropod groups studied so far (Fig. 7).
In Drosophila omb is activated by decapentaplegic (dpp), which is expressed in a dorsal sector, and the H15 gene is activated by wingless (wg), which is expressed in a ventral sector (e.g., Brook and Cohen 1996). In other arthropod species investigated so far dpp is not expressed along the dorsal side of the appendages like it is in Drosophila. Rather it is restricted to the distal tip early and forms rings later on (Jockusch et al. 2000; Niwa et al. 2000; Prpic et al. 2003; Prpic 2004). Thus, it is difficult to imagine how Dpp could activate omb expression along the dorsal side of the legs in these species. It remains unclear, therefore, whether omb is activated directly by Dpp in these species. However, because Dpp protein is a morphogen, the Dpp protein spreading from the tip might be sufficient to trigger dorsal omb expression (see also Prpic et al. 2003). Alternatively, omb is activated via an as yet unknown mechanism. It is important to note that in more basal insects like the beetle Tribolium castaneum (Sanchez-Salazar et al. 1996) and the milkweed bug Oncopeltus fasciatus (Angelini and Kaufman 2005a) dpp is expressed similar to the millipede Glomeris and the spiders Cupiennius and Achaearanea (Akiyama-Oda and Oda 2003; Prpic et al. 2003; no dpp expression data is available yet for Tegenaria). The expression domain of dpp along the entire dorsal side is thus not characteristic for insects and could be an adaptation to leg development from imaginal discs in flies. Subsequently, this dorsal dpp expression could have been used for activating omb in the dorsal domain in flies, replacing a more ancestral system for dorsal omb activation.
In contrast to dpp expression, the expression of wg on the ventral side is highly conserved among all arthropods including the expression in the imaginal discs of Drosophila. The expression of wg in the legs of Tegenaria has not yet been studied, but if we assume it to be expressed like wg in the two other spider species, Cupiennius and Achaearanea (Prpic et al 2003, N. M. Feitosa unpublished data), then it is interesting to note that the ectodermal expression of the group 3 H15 genes (i.e., Ta-H15-3 and At-H15-2) is almost identical to the wg expression in the spider legs. We conclude from this that H15 may have a role in ventral fate determination in all arthropods and that H15 may be a regulatory target of wg in spiders as in Drosophila. This conclusion is consistent with the obtained expression patterns in this study. This possible involvement of wg (via H15) in spider DV leg axis development would also be consistent with the ventral leg expression of Notum (a modulator of the Wg protein gradient in Drosophila) in Cupiennius (Prpic and Damen 2005b). However, Angelini and Kaufman (2005a, b) have already argued that the involvement of Wg signalling in leg development may have evolved very late in arthropod evolution, probably in the lineage leading to the higher dipterans. Clearly, more work is needed in order to establish the role of Wg signalling in arthropod leg development.
Evolution of the H15-related genes: how many gene duplications?
In the phylogenetic analysis the H15 genes cluster together according to arthropod class. This indicates that the H15 genes in each class are more closely related among themselves than to any of the H15 genes in a different arthropod class. This suggests independent duplication events in each class. In this scenario (Fig. 7, green squares) the ancestral state is a single H15 like gene that gave rise to the multiple H15 genes in the fly, the millipede, and the spiders by independent duplication events. In this case an additional duplication of one of the genes must have taken place in the chelicerate lineage to arrive at the complement of three genes in spiders.
On the other hand, this scenario invokes a high number of independent gene duplication events and, therefore, is not very parsimonious. An alternative interpretation is that the sequence analysis is influenced by concerted evolution. Concerted evolution can be the result of gene conversion and has been documented for genes or portions of genes that share a high sequence similarity and that are located close to one another in the genome (e.g., Peel et al. 2006). Thus, although our phylogenetic analysis suggests independent duplications of H15 in all arthropod classes (with high reliability values at all three edges), the analysis might be suffering from artefacts caused by concerted evolution. In the latter scenario, also a lower number of gene duplication events has to be postulated (Fig. 7, blue squares): the ancestral state for arthropods then would be two H15-like genes and an additional duplication event has taken place in the lineage leading to the spider species. Further work is needed to investigate this in more detail.
CONCLUSIONS
Although comparative analyses of gene expression patterns can give important clues as to the evolutionary conservation or change of gene regulation, our data indicate that the presence of multiple paralogus genes can be a potential source of misinterpretation. Our previous analysis of T-box genes in Cupiennius demonstrated that omb was expressed along the dorsal side of the appendages, compatible with a role in dorsal fate determination like in Drosophila. However, the two H15-related genes recovered from Cupiennius are only expressed in a very small portion of the ventral side and thus only in a fraction of wg expressing cells. These data argued against a general role in ventral fate determination as well as against a general activation of H15 by Wg signaling. The discovery of a third H15-type gene in two other spider species, Tegenaria and Achaearanea, suggests that this conclusion presumably was premature. This third H15-type gene is expressed along most of the ventral side of all appendages in these two spider species much like the Drosophila genes and thus is fully compatible with a role in ventral fate determination downstream of Wg-signaling. It also suggests that our previous account on the H15-type genes in Cupiennius was incomplete and that the group-3 H15 gene has not yet been isolated from Cupiennius. Taken together all data now suggest that the role of omb and H15 in dorsal-ventral axis formation in the legs is conserved. In particular, all present data firmly establish the link between omb expression and the dorsal surface of the appendages. Thus, omb expression might be a useful marker for identifying the dorsal side in highly specialized appendages (e.g., the mandible in insects or the gnathochilarium in millipedes), where it has been difficult in the past to unambiguously identify homologies with other, more normal appendages.
Acknowledgments
We thank Diethard Tautz for support and advice. Many thanks to Andreas Hartl for the Cupiennius image, and to Matthias Pechmann for the Achaearanea image. This work has been supported in part by the DFG via SFB 572 of the University of Cologne and the European Union via the Marie Curie Research and Training Network ZOONET (MRTN-CT-2004-005624) (grants holder: W. G. M. D.) and by the DFG Emmy-Noether-Programme (ENP grant PR1109/1-1) (grant holder: N. M. P.).




