Combined morphological and molecular analysis of individual nematodes through short-term preservation in formalin
Abstract
Small metazoans such as marine nematodes are increasingly identified using both molecular and morphological techniques. Formalin is the preferred fixative for morphological analysis but specimens become unsuitable for molecular study due to formalin-induced modification of DNA. Nematodes fixed in ethanol work well for molecular studies but become unsuitable for taxonomy due to shrinkage. Here we show for the first time that formalin can be used as a short-term fixative (≤ 7 days) for marine nematodes, allowing both morphological and molecular work to be conducted on the same individual. No sequence ambiguities were detected in polymerase chain reaction (PCR) amplifications of 18S ribosomal DNA (rDNA) following short-term formalin preservation.
Molecular techniques are increasingly being applied to address the phylogenetic relationships and rapid classification of many meiofaunal groups, including free-living marine nematode worms, which are often the dominant and ecologically most important component of the meiofauna (Austen 2004; Lambshead 2004). Because of the immature nature of the nematode taxonomy, we need to compare specimens according to traditionally classified morphological features observed through traditional microscopy with specimens subjected to molecular analyses. Preferably both techniques should be applied to the same specimen.
Traditionally, formalin and its derivatives have been the preferred choice of solvent for preserving or fixing marine specimens for long-term studies because these are inexpensive, effective and low maintenance preservatives, which maintain the morphological integrity of specimens. The application of molecular technologies has revolutionized research in marine biology in the last few decades (Rawson et al. 2000; Cook et al. 2005, etc.), but specimens fixed in formalin for long periods often cannot be used for molecular studies. This is because formalin preservation makes DNA poorly available for amplification and subject to amplification errors, besides other direct and indirect effects such as irreversible denaturation, modification and formations of covalent bonds between nucleotides (Chang & Loew 1994; Thomas 1994; Schander & Halanych 2003, etc.). As a result of these factors, it is becoming increasingly common to fix specimens in organic solvents such as absolute alcohol for molecular studies, despite the drawbacks these methods may have in terms of specimen integrity. Alcohol preservation of small, soft-bodied metazoans such as nematodes and polychaete worms results in shrinkage of specimens, often making accurate morphological identification impossible. Some authors (e.g. Dessauer et al. 1996) have suggested cryopreservation of specimens for molecular work, despite this having drawbacks such as high cost.
In a previous study, Thomas et al. (1997) fixed nematodes in formalin for 2 days and successfully carried out subsequent molecular analysis.
The aims of this study were to investigate whether nematodes preserved in formalin for relatively short periods of time could provide DNA suitable for molecular analyses, and, if so, to determine the duration of this time window. If DNA suitable for polymerase chain reaction (PCR) amplification could be obtained from nematode specimens subjected to short-term preservation in formalin, this would allow both morphological and molecular studies of the same specimen with relative ease. If successful, such an approach should be applicable to a range of similar small, soft-bodied metazoans.
For the purposes of this study, we examined individuals of the cosmopolitan free-living marine/estuarine nematode Terschellingia longicaudata (De Man 1907). This species is reported from most of the world's oceans and estuaries and is typically one of the dominant species in soft sediments in inshore water. It is 1.5–1.7 mm in length, a size typical of a wide range of related species.
Twenty-five grams of intertidal mud from the Tamar Estuary, Cornwall, UK were fixed in 200 mL of 5% unbuffered formalin and stored overnight at room temperature (20 °C). The formalin fixed sediment was subjected to meiofaunal extraction the next day following Somerfield & Warwick's (1996) flotation method. Extracted fauna was then stored in a 50 mL falcon tube containing 4% unbuffered formalin. Terschellingia longicaudata specimens (n = 6) were picked from the falcon tube after selected time intervals. Specimens were picked after 2, 3, 4, 5, 6, 7, 9, 11, 13 and 15 days and after 1 month. Each specimen was placed in 0.5 mL PCR tube containing 20 µL of 0.25 m NaOH for DNA extraction.
DNA was extracted using a modification of the method of Floyd et al. (2002). All 0.5 mL PCR tubes were frozen overnight (8–9 h) at −20 °C, then incubated overnight at 60 °C. The tubes were then heated for 3 min at 99 °C on a heating block and allowed to cool to room temperature before centrifugation (RCF = 16 000; 30 s). Four microlitres of 1 m HCl, 10 µL of 0.5 m Tris-HCl (pH 8.0) and 5 µL of 2% Triton X-100 was added to each tube. The contents of each tube were mixed briefly and centrifuged for 30 s in a bench top microcentrifuge (RCF = 16 000). The tubes were reheated for 3 min at 99 °C and allowed to cool to room temperature. The extract was then used for PCR amplification.
Two primers, namely G18F forward (5′-GCTTGTCTCAAAGATTAAGCC-3′) and 22R reverse (5′-GCCTGCTGCCTTCCTTGGA-3′), were used for PCR amplification of the partial nuclear small subunit ribosomal DNA (18S rDNA) (Blaxter et al. 1998), producing an amplification product of approximately 400 bp. Reactions were performed on 5 µL aliquots of the extracted DNA by adding 5 µL 10× MgCl2 buffer, 5 µL 2 mm dNTPs, 2 µL of each of the primers (10 pmol/µL), 0.5 µL of Taq DNA polymerase (Promega) and MilliQ water to a final volume of 50 µL. The PCR cycles were: 5 min at 94 °C, followed by 40 cycles for 1 min at 51 °C, 2 min at 72 °C and 1 min at 94 °C.
Pfx DNA polymerase (Invitrogen) is a proofreading enzyme with 3′ to 5′ exonuclease activity that provides higher fidelity than Pfu DNA polymerase (Cline et al. 1996). This was also used in addition to Taq polymerase to amplify individuals picked on the 11th, 13th, 15th and 30th days. In this case, PCRs were set up according to the manufacturer's instructions.
Five microlitres of each of the PCR products were analysed in 1% (w/v) agarose gels using the Mini-Sub Cell GT system (Bio-Rad Laboratories). A 100 bp ladder was used as a size marker.
In order to check the identity and integrity of the amplified region, PCR products (using Pfx DNA polymerase) from the 11th day were directly treated with ExoSAP-IT according to manufacturer's instructions (USB Corporation). A total of 2.5 µL of the ExoSAP-IT-treated PCR product was cycle sequenced using a BigDye Terminator Kit (Applied Biosystems) and cleaned using the Wizard Magnesil system (Promega). Sequencing was performed in both directions using an additional internal primer Mn18F (5′-CGCGAATRGCTCATTACAACAGC-3′) and 22R reverse primer in an ABI PRISM 3100 Genetic Analyser (Applied Biosystems). Generated sequence was then compared with that from alcohol-preserved T. longicaudata specimens to check for possible ambiguity.
The primary objective of this work was to investigate whether unbuffered formalin could be used as a fixative agent for short periods of time, without inhibiting or otherwise compromising subsequent molecular analyses, i.e. DNA extraction and PCR amplification. Nematode worms stored in formalin for between 2 and 15 days were subjected to DNA extraction and amplification, together with individuals that had been fixed in formalin for 1 month. The electrophoresis images clearly show that there is no apparent shearing of DNA or inhibition of PCR amplification in the case of individuals fixed in formalin for up to 9 days (Fig. 1). Visible changes in both DNA quality and PCR amplification were observed, however, in individuals stored in formalin for longer periods. Here the DNA was apparently highly sheared and gave little or no PCR amplification (Fig. 1).

Gel showing 18S rDNA amplification products of Terschellingia longicaudata specimens extracted from formalin after 2, 3, 4, 5, 6, 7, 9, 11, 13, 15 and 30 days.
A comparative test was carried out to check the specificity of the DNA polymerase used for amplification. AccuPrime Pfx DNA polymerase enzyme was used to amplify individuals stored in formalin for 11–15 days or worms that had been fixed for a month. No bands were detected from such worms when Taq polymerase enzyme was used (Fig. 1). There was partial amplification success for individuals fixed for 11 days, which produced some amplicons with Pfx (Fig. 2). However, there were no visible PCR products in the case of 13- and 15-day individuals or those fixed for 1 month. Sequences from 11-day worms showed no apparent increase in ambiguity or error compared to those obtained from alcohol-preserved material (Fig. 3).
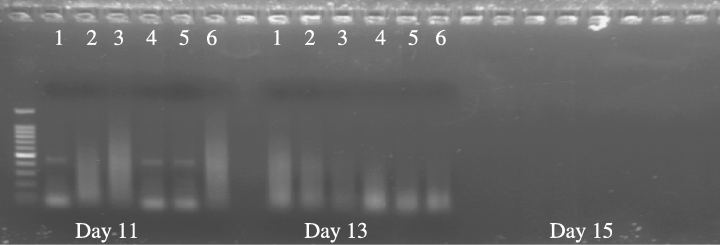
Gel showing results of PCR amplification of 18S rDNA after 11, 13 and 15 days using Pfx DNA polymerase. Limited amplification is visible in some of the day 11 samples, but no amplification can be detected after longer time periods. No amplification was observed from specimens fixed for 1 month.
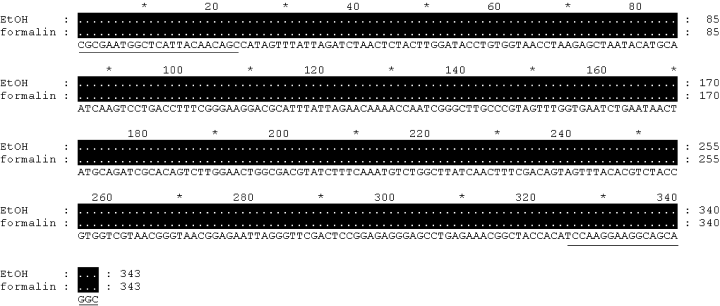
Comparison of DNA sequences from alcohol and formalin fixed Terschellingia longicaudata specimens. Lines indicate the position of forward and reverse primers.
This study shows that formalin can be used to fix small invertebrates such as marine nematodes for molecular biological work for a short period. This would allow both morphological and molecular work to be conducted on the same individual, provided that it is done rapidly after collection. The application of this study will be immensely important in terms of sediment samples taken offshore from places geographically separated from analytical laboratories. It is useful in many ways as it gives a chance to bring samples back to laboratory for further analysis. We have also shown that the average PCR yield after up to 9 days in formalin was relatively high, with a rapid drop-out subsequently. In some instances, it is apparently possible to get PCR products from specimens fixed in formalin for slightly longer using proofreading DNA polymerases.
Our expectation is that in future, small soft-bodied invertebrates such as nematodes could be stored in formalin for short periods (ideally < 9 days), allowing taxonomists to perform morphological analyses followed by the application of molecular investigations on the same individual. Such coupling of morphological and molecular analyses is critical in many groups where identification requires close examination, particularly if we are to avoid assigning sequence data to the wrong organism. The approach described here should enable taxonomists and molecular biologists to collaborate more effectively, allowing a better understanding of the biodiversity and evolutionary relationships of small metazoans.
Acknowledgements
This is a contribution to the PML Functional Biodiversity Project and to the Marine Genomics Network of Excellence funded by EU Framework 6 programme.




