The TCF7L2 Diabetes Risk Variant is Associated with HbA1C Levels: a Genome-Wide Association Meta-Analysis
Summary
Genome-wide association (GWA) studies have identified around 20 common genetic variants influencing the risk of type 2 diabetes (T2D). Likewise, a number of variants have been associated with diabetes-related quantitative glycaemic traits, but to date the overlap between these genes and variants has been low. The majority of genetic studies have focused on fasting plasma glucose levels; however, this measure is highly variable. We have conducted a GWA meta-analysis of glycated haemoglobin (HbA1C) levels within three healthy nondiabetic populations. This phenotype provides an estimate of mean glucose levels over 2–3 months and is a more stable predictor of future diabetes risk. Participants were from three isolated populations: the Orkney Isles in the north of Scotland, the Dalmatian islands of Vis, and Korčula in Croatia (total of 1782 nondiabetic subjects). Association was tested in each population and results combined by meta-analysis. The strongest association was with the TCF7L2 gene (rs7903146, P= 1.48 × 10−7). This is also the strongest common genetic risk factor for T2D but it has not been identified in previous genome-wide studies of glycated haemoglobin.
Introduction
Glycated haemoglobin (HbA1C) is generated from the normal exposure of haemoglobin in red blood cells to plasma glucose. HbA1C levels are therefore proportional to average blood glucose levels over the past 60–90 days. As such, HbA1C is a good measure of long-term plasma glucose concentrations, which are important in the monitoring of glycaemic control in people with diabetes. HbA1C provides a more accurate long-term measure of glucose homeostasis than fasting glucose (FG) measures and does not require a fasting sample to be collected.
Large genome-wide association (GWA) studies have identified and replicated 18 loci associated with the risk of type 2 diabetes (T2D) (Zeggini et al., 2008) and 5 with FG levels (Bouatia-Naji et al., 2009; Prokopenko et al., 2009), but few loci have been associated with both T2D and FG or any other disease-related intermediate phenotype. The melatonin receptor 1B (MTNR1B) locus has shown association with T2D and FG (Bouatia-Naji et al., 2009; Prokopenko et al., 2009) as has the glucokinase regulatory protein (GCKR) (Sparso et al., 2008). Solute carrier family 30 (zinc transporter) member 8 (SLC30A8) has previously been identified by association with T2D (Sladek et al., 2007) and more recently with HbA1C (Pare et al., 2008). This limited overlap between T2D and either FG or HbA1C loci may reflect the diversity of mechanisms involved in the genetic control of diabetes risk, and the fact that a genetic variant which directly affected FG or HbA1C levels would have a weaker association with T2D due to age of onset and confounding environmental influences.
Previous GWA studies of HbA1C have confirmed association with two known FG loci, glucokinase (GCK) and islet-specific glucose-6-phosphatase-related (G6PC2), and with one T2D locus (SLC30A8), as well as showing a novel association with the hexokinase type 1 (HK1) locus (Pare et al., 2008). However, so far no study has demonstrated association between transcription factor 7-like 2 (TCF7L2), which imparts the largest risk of any known T2D susceptibility locus (Grant et al., 2006), and HbA1C.
We have conducted a high density GWA meta-analysis of HbA1C levels in three independent, genetically isolated, populations. This approach has greater power to detect common variants than studying the populations individually and provides internal replication of results.
Materials and Methods
Populations
The Scottish samples are from the Orkney Complex Disease Study (ORCADES), an ongoing family based, cross-sectional study in the isolated Scottish archipelago of Orkney. Genetic diversity in this population is decreased compared to Mainland Scotland, consistent with the high levels of historical endogamy (Vitart et al., 2005; McQuillan et al., 2008). Data for healthy volunteers from a subgroup of 10 islands, aged 17–97 years (mean age 53 years; 54% female) with known Orcadian ancestry, were used for this analysis. Fasting blood samples were collected during 2005–2007 and over 200 health-related phenotypes and environmental exposures were measured in each individual. All participants gave informed consent and the study was approved by Research Ethics Committees in Orkney and Aberdeen.
The Vis study consists of unselected individuals aged 18–93 years (mean age 56 years, 57% female) from the villages of Vis and Komiža on the Dalmatian island of Vis in Croatia, who were phenotyped for over 200 disease-related quantitative traits in 2003–2004. There is marked genetic isolation of the villages from the Croatian mainland and from surrounding islands (Vitart et al., 2006) and this population has already yielded novel and replicated quantitative trait loci (Vitart et al., 2008). All participants gave informed consent and the study was approved by Research Ethics Committees in Scotland and Croatia.
The Korčula study was carried out in the isolated population of the Dalmatian island of Korčula, Croatia, with volunteers from the villages of Lumbarda, Žrnovo, and Račišće, as well as Korčula town (Polašek et al., 2009). This population is also isolated genetically from the Croatian Mainland (Vitart et al., 2006) and there was considerable background inbreeding historically (Rudan et al., 2003). Examinees were aged between 21 and 98 years and are a subgroup from a larger genetic epidemiological study carried out in 2007. Over 100 disease-related quantitative traits were measured. The mean age of participants was 57 years and 66% of subjects were female. All participants gave informed consent and the study was approved by the Research Ethics Committee at the University of Zagreb.
Phenotype
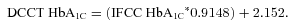
As part of the extensive phenotyping in each cohort, participants reported their medical history and any medication and had their height and weight measured by research nurses.
Genotype
All genotyping was performed using Illumina Beadchips, the Hap300 for ORCADES and Vis and CNV370 for Korčula. Genotypes were called using the BeadStudio V3 software (Illumina, San Diego, CA) according to the recommended parameters for the Illumina Infinium assay and using genotype cluster files provided by Illumina.
Individuals with low call rates (<98%) and one of each set of identical twins were excluded from analysis. Within each population multi-dimensional scaling was used to identify genetic outliers. The first three principal components were estimated and each individual's distance from the population mean was calculated. Individuals who were significantly distant (by χ2-test) from the main population cluster were excluded from analysis. Visual inspection of principal component analysis plots was used to rule out the possibility of population admixture.
Two nonindependent groups were analysed. Initially individuals who were taking oral glucose-lowering medications or insulin injections were excluded from analysis as these interventions would be expected to significantly lower HbA1C levels. The proportion of individuals excluded from the studies was 2.2% (15/679) for ORCADES, 8.5% (66/773) for Vis, and 6.2% (27/438) for Korčula. The association analysis was repeated on a second group that excluded both the medicated individuals and those with an FG level of 7 mmol/l or more (i.e. hyperglycaemics) to ensure that the effect seen is not due to untreated and undiagnosed diabetics.
SNP markers with less than 98% call rate, less than 1% minor allele frequency, or which showed significant deviation from Hardy-Weinberg equilibrium (p < 0.001) were excluded from the analysis. After these quality control requirements 256,587 markers were available in all three cohorts. The Bonferroni-corrected significance threshold for this number of tests is P= 1.95 × 10−7, for a genome-wide threshold of 0.05.
Analysis
Statistical analyses were carried out using the R-packages GenABEL (Aulchenko et al., 2007) and MetABEL. Age and body mass index (BMI) were both significant covariates for HbA1C. The HbA1C measures were corrected for age, BMI and sex effects. After exclusion of individuals receiving treatment for diabetes and correction for covariates the HbA1C distribution continued to show a positive skew and was significantly different from normal. To control for this the distribution of residual values was rank-transformed to normality with a mean of 0 and a standard deviation of 1.
As the study participants were from small isolated populations there is considerable relatedness between study subjects. Genomic kinship matrices were therefore calculated for participants within each study using identity-by-state sharing, weighted by allele frequency. The kinship matrix was used in a linear polygenic mixed model to account for nonindependence of data points due to relatedness.
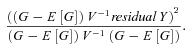
Meta-analysis was performed in MetABEL using a weighted inverse variance method. Briefly, the weight for each study is equal to one over the square of the standard error of the estimate for that study. The pooled effect estimate is then calculated by multiplying the study effect estimate by the study weight, summing over all studies and then dividing by the sum of all study weights.
Results
There was a large difference in the proportion of people receiving diabetes medication between Orkney (2.2%) and the two Croatian sites (6.2–8.5%). Considering only the untreated population, this difference was also reflected in the number of prevalent cases of diet-controlled and previously undiagnosed diabetes (defined by FG ≥7 mmol/l) between the Scottish (1.8%) and Croatian (6.8%) sites (Table 1). Distributions of age and BMI are comparable between study populations. HbA1C values in the untreated populations were somewhat lower in Orkney and Vis than in Korčula (Table 1).
| Population | Orkney | Vis | Korčula |
|---|---|---|---|
| Untreated N | 664 | 707 | 411 |
| Untreated, but FPG ≥7 mmol/l% (N) | 1.8% (12/664) | 6.8% (48/707) | 6.8% (28/411) |
| Mean (SD) Age (years), among untreated | 53.5 (15.3) | 55.6 (15.4) | 56.6 (13.9) |
| % Female, among untreated | 53.8 | 57.3 | 65.9 |
| Mean (SD) BMI, among untreated | 27.7 (4.8) | 27.2 (4.3) | 27.7 (4.0) |
| %HbA1C DCCT, among untreated | 5.4 (0.57) | 5.3 (0.56) | 5.7 (0.63) |
It is not surprising, given the relatively small numbers in each individual cohort that no SNPs reached genome-wide significance in the single population analyses. The results of the GWA meta-analysis are shown in Figure 1. In the analysis excluding only treated diabetics, using the highly conservative Bonferroni significance threshold of 1.95 × 10−7, a single marker shows genome-wide significant association with HbA1C: rs7903146 on chromosome 10 (P= 1.48 × 10−7). After further exclusion of hyperglycaemic individuals, this association is slightly diminished (P= 7.08 × 10−7).
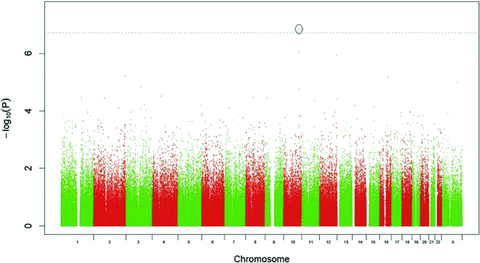
GWA meta-analysis results for rank-transformed HbA1C levels fitting effects for age, sex, and BMI.
As shown in Table 2 the marker showed consistent size and direction of effects across the three populations, but was not highly associated in any of the individual populations, due to lack of power. In terms of ranking within each study the rs7903146 SNP was 37th in Orkney, 1363rd in Korčula, and 3912th in Vis. The effect of rs7903146 is estimated to be −0.054% HbA1C per C allele (Fig. 2) in the meta-analysis.
| Population | N | Freq | Effect | SE | P-value |
|---|---|---|---|---|---|
| Meta-analysis | 1782 | 0.72 | −0.054 | 0.014 | 1.48 × 10−7 |
| Orkney | 664 | 0.74 | −0.067 | 0.024 | 1.41 × 10−4 |
| Vis | 707 | 0.71 | −0.044 | 0.021 | 1.54 × 10−2 |
| Korčula | 411 | 0.71 | −0.056 | 0.033 | 4.21 × 10−3 |
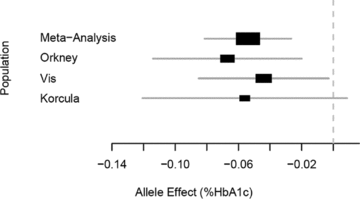
Forest plot of rs7903146 effect estimates using untransformed HbA1C in individual populations and the combined meta-analysis. The X-axis represents the change in %HbA1C per C allele.
The second most strongly associated SNP in the meta-analysis is rs12255372, located 50 kb away from, and in strong linkage disequilibrium with, rs7903146. Both SNPs are located in the TCF7L2 gene on chromosome 10, rs7903146 having the largest effect of any known common variant on T2D risk (Tong et al., 2009).
Discussion
We have described a genome-wide significant genetic association between HbA1C levels and the T2D susceptibility locus TCF7L2 among people not receiving insulin or oral anti-diabetic agents. Using a meta-analysis of data from three isolated and independent populations we have shown a replicable lowering effect of the rs7903146 C allele on HbA1C levels, consistent with the protective effect of this allele in T2D. To date TCF7L2 is the common genetic variant with the largest effect size for T2D risk (odds ratio of ∼1.4 for each copy of the minor allele) (Tong et al., 2009).
HbA1C levels have been shown to be a strong predictor of future T2D (Pradhan et al., 2007). Since all diabetic individuals receiving insulin or oral hypoglycaemic agents were excluded from our analysis, the results demonstrate that variation in the TCF7L2 gene influences HbA1C levels within a nontreated population that includes people with diet-controlled diabetes and previously undiagnosed diabetes. After subsequent exclusion of hyperglycaemic individuals with FG levels of 7 mmol/l or more, the association is diminished with a P-value of 7.08 × 10−7. The exclusion of hyperglycaemic individuals has no significant effect on the distribution of HbA1C in our populations, but does reduce the sample size by approximately 5%.
TCF7L2 is a widely expressed transcription factor and is known to regulate expression of the glucagon gene (GCG) (Yi et al., 2005). The mechanism by which polymorphisms in this gene contribute to T2D risk has not yet been fully determined though current evidence suggests a role in beta-cell proliferation and insulin secretion (Pearson, 2009). Knockdown of TCF7L2 expression in human pancreatic islets results in reduced beta cell proliferation and knockdown of TCF7L2 expression in mice has shown altered expression of SLC30A8 which is the only gene previously associated with both T2D and HbA1C (da Silva Xavier et al., 2009). Since the association between HbA1C and TCF7L2 reported here is found in nondiabetic individuals, this would support the proposition that the T2D risk contributed by this gene acts through a reduction in insulin secretion from pancreatic beta cells and hence higher blood glucose levels. Indeed TCF7L2 variants have recently been associated with FG and 2 h glucose in very large meta-analyses (Dupuis et al., 2010; Saxena et al., 2010).
This is the only the fifth time one of the 18 well-replicated T2D loci has been identified by GWA to be associated with a related quantitative phenotype, the previous examples being the association of MTNR1B, GCKR, and now TCF7L2 with T2D risk and FG (Sparso et al., 2008; Bouatia-Naji et al., 2009; Dupuis et al., 2010) and SLC30A8 with T2D and HbA1C (Sladek et al., 2007; Pare et al., 2008). The fact that so many of the known T2D risk loci do not appear to show an effect on the FG or HbA1C phenotypes may be due to the high variability of single measures of FG, which can vary due to exercise or the previous day's diet. HbA1C provides a much more stable measure of long-term glucose levels and so HbA1C variants may be more likely to overlap with T2D variants, in as much as mechanisms of glucose regulation in the normoglycaemic range overlap with those of disease progression. However, since far fewer GWA studies have been carried out on HbA1C than on FG, this has yet to be tested.
The prior evidence for association between the TCF7L2 locus and T2D and FG, along with the strong relationship between T2D, FG, and HbA1C makes it more likely that the association reported here is true. However, since the individual sample sizes reported here are small, replication of this result in an independent sample would be beneficial.
One previous high-density genome scan for HbA1C using data from the Women's Genome Health Study (WGHS) did not identify TCF7L2 at a genome-wide significant level (P value was 3.5 × 10−3) despite having a sample size of over 14,000 (Pare et al., 2008). This may indicate an increased relative effect of this locus in our populations due to the reduced environmental and genetic heterogeneity found in these isolated islands (Rudan et al., 2008) compared to the urban samples used in the WGHS. Although the WGHS cohort has a similar mean age to our study, the range is far smaller since all participants were of working age at recruitment. The HK1 variant associated with HbA1C in the WGHS did not show association here, perhaps due to the lower power of our study.
In summary, this work suggests that the TCF7L2 T2D risk polymorphism is associated with the quantitative phenotype HbA1C. We have shown that each copy of the rs7903146 C allele decreases an individual's HbA1C level by 0.054% which is in the same direction as the ∼1.4-fold decrease in T2D risk (Tong et al., 2009).
Acknowledgements
ORCADES was supported by the Chief Scientist Office of the Scottish Government, the Royal Society and the European Union framework program 6 EUROSPAN project (contract no. LSHG-CT-2006-018947). DNA extractions were performed at the Wellcome Trust Clinical Research Facility in Edinburgh. We would like to acknowledge the invaluable contributions of Lorraine Anderson and the research nurses in Orkney, the administrative team in Edinburgh and the people of Orkney.
The VIS and KORČULA studies in the Croatian islands of Vis and Korčula were supported through the grants from the Medical Research Council UK to H.C., A.F.W. and I.R.; and Ministry of Science, Education and Sport of the Republic of Croatia to I.R. (number 216-1080315-0302). The authors collectively thank a large number of individuals for their help in organising, planning, and carrying out the field work related to the project and data management: Professor Pavao Rudan and the staff of the Institute for Anthropological Research in Zagreb, Croatia (organisation of the field work, anthropometric and physiological measurements, and DNA extraction in Vis); Professor Stipan Jankovic and the staff of the University of Split Medical School (organization of the field work, anthropometric and physiological measurements, and DNA extraction in Korčula); Professor Ariana Vorko-Jovic and the staff and medical students of the Andrija Stampar School of Public Health of the Faculty of Medicine, University of Zagreb, Croatia (questionnaires, genealogical reconstruction and data entry in Vis and Korčula); Dr Branka Salzer from the biochemistry lab “Salzer,” Croatia (measurements of biochemical traits in Vis and Korčula); local general practitioners and nurses (recruitment and communication with the study population); and the employees of several other Croatian institutions who participated in the field work, including but not limited to the University of Rijeka, Croatia; Croatian Institute of Public Health; Institutes of Public Health in Split and Dubrovnik, Croatia. SNP Genotyping of the Vis samples was carried out by the Genetics Core Laboratory at the Wellcome Trust Clinical Research Facility, WGH, Edinburgh, and for Korčula by the German Research Centre for Environmental Health in Neuherberg, Munich, Germany.