Identification of new sensitive biomarkers for the in vivo response to interferon-β treatment in multiple sclerosis using DNA-array evaluation
Abstract
Objective: Neutralizing antibodies (NAbs) occur in a proportion of multiple sclerosis (MS) patients treated with interferon (IFN)-β. NAbs impair the effect of treatment. The biological effect of IFN-β can be measured as the induction of the myxovirus resistance protein A (MxA) molecule. However, other markers could be more sensitive for evaluating the response to IFN-β. We used DNA array analysis to identify genes that are strongly induced in blood cells by IFN-β, and measured their expression in MS patients with different NAb levels.
Methods: Gene expression was studied on DNA arrays in untreated patients, in NAb negative patients, and in MS patients with varying NAb levels 9–12 h and 36–48 h after IFN-β administration. The expression of selected genes was measured by real-time PCR. NAb levels were assessed by a cytopathic effect assay.
Results: Several hundred genes were induced 9–12 h after an injection of IFN-β. The molecules CXCL10, CCL2 and IFI27 were among the most strongly induced. Gene induction was generally much less pronounced after 36–48 h, but IFI27 remained strongly induced. The strong induction of these molecules and MxA was confirmed by real-time PCR. Induction of MxA, CCL2, CXCL10 and IFI27 was reduced in patients with low NAb levels and lost in patients with intermediate/high NAb levels.
Conclusion: We identify IFI27, CCL2 and CXCL10 as sensitive biomarkers for the response to IFN-β. The expression of these markers adequately reflects bioactivity of IFN-ß as documented by the decreased induction in low NAb-positive patients and the lost induction in patients with moderate/high NAb levels.
Introduction
Interferon (IFN)-β treatment reduces the number of relapses and delays the development of irreversible neurological impairment in multiple sclerosis (MS) [1]. Unfortunately, many patients develop neutralizing antibodies (NAbs) to IFN-β within the first 1–2 years of treatment, and in patients with moderate or high NAb titres the effects of treatment are abolished [2–4]. However, in patients with low NAb titres treatment may still be partly or even fully efficacious, and patients who go on to develop NAbs may initially respond better to therapy than patients who remain NAb negative [5]. NAb titres may decrease upon prolonged treatment, but often persist at high levels after cessation of therapy [6–10]. However, NAb-positive patients who revert to the NAb-negative state regain full therapeutic efficacy of IFN-β [11].
The lowest NAb titre that leads to a loss of biological activity of IFN-β is not known. Traditionally, a blood sample with a titre of 20 measured with the Kawade technique has been defined as positive [12,13], although it has been shown that even lower titres reduce the biological response to IFN-β [14,15]. Biomarkers, in the form of molecules whose expression is induced by IFN-β, may aid in discriminating NAb positive MS patients with a full response from NAb positive patients with an impaired response to IFN-β therapy. A biological response to treatment demonstrated by increased serum concentrations of neopterin or β2-microglobulin was, indeed, seen in some patients with low NAb levels, whereas the induction of these molecules was abolished in patients with high NAb levels [16]. A loss of biological response was associated with increased disease activity in gadolinium-enhanced brain magnetic resonance imaging studies.
The IFN-β-induced molecule myxovirus resistance protein A (MxA) is among the most studied markers of treatment response to IFN-β. As observed with neopterin and β2-microglobulin, the induction of MxA is abolished in patients with moderate/high NAb titres [15,17–21]. However, in patients with lower NAb titres, the induction of MxA is more variable [22].
It has been demonstrated that some other interferon-induced molecules have expression patterns comparable to MxA in IFN-treated MS patients [23,24]. We hypothesized that the use of biomarkers more responsive than MxA might provide a better discrimination between patients with a full in vivo response to IFN-β therapy and patients with antibody-mediated decreased bioactivity [19]. We addressed these issues by a series of gene expression studies on DNA arrays and real-time reverse transcriptase polymerase chain reaction (RT-PCR) analysis of gene expression in patients with different levels of NAbs during IFN-β-therapy.
Methods
Blood samples were obtained from 25 patients with relapsing-remitting MS according to the McDonald criteria before and 3 months after beginning treatment with IFN-β. In addition, we studied 175 consecutive NAb positive patients treated with IFN-β for varying periods. Fifty healthy volunteers served as controls. The study was approved by the regional scientific ethics committee and the Danish National Board of Health, and informed consent was obtained from all participants.
Blood samples were routinely obtained 9–12 h after an injection of IFN-β. In 12 of the patients additional blood samples were obtained 9–12 and 36–48 h after an injection of the IFN-β after 6 months of treatment. Blood for Affymetrix GeneChip® (Affymetrix, Santa Clara, CA, USA) Human Genome Focus Array analysis of gene expression was drawn in EDTA tubes. Total RNA was extracted from mononuclear cells, and cRNA and hybridization was performed according to protocols from Affymetrix (Affymetrix): http://www.affymetrix.com) as previously described [25]. The Affymetrix Microarray Suite mas 5.0 software (Affymetrix) was used to process images, evaluate data quality and normalize, transform and filter data from the DNA arrays. Genes calling as present in at least one sample according to Affymetrix algorithms were considered for statistical analysis.
Blood for real-time RT-PCR studies of RNA expression was drawn in PAXgene™ tubes (Becton Dickinson, Brøndby, Denmark). The tubes were left at room temperature for 2–24 h, and stored at –20°C. Total RNA was extracted using the PAXgene blood RNA kit (Qiagen, Hilden, Germany FRG), and cDNA was prepared using the High Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA, USA). Real-time PCR was performed on an ABI PRISM 7500 Real Time PCR System (Applied Biosystems) using a commercially available TaqMan Universal PCR Master Mix and pre-designed, validated primer/probe kits (Applied Biosystems: MX1: Hs00182073; CXCL10: Hs00171042_m1; CCL2: Hs00234140_m1; IFI27: Hs00271467_m1). cDNA from healthy donors was used as reference sample for the gene expression studies, and an expression index was calculated by the 2-ΔΔCt method with GAPDH as reference gene. The mean inter-assay coefficient of variation for the gene expression indices was 13% for duplicate PAXgene tubes analysed by two different technicians on two separate days.
NAbs were measured by a viral cytopathic effect assay as previously described in detail [26]. Results are given as neutralizing capacities, which is the amount of IFN-β added to the assay that has been neutralized by NAbs in the patient sample in a medium-sensitive assay. In this assay neutralizing capacities below 20%, corresponding to a Kawade titre below 20, are considered negative [12,13]. Neutralizing capacities ranging from 20–80%, corresponding to Kawade titres in the range 20 to 160–200, are considered low, and neutralizing capacities in the 80–100% range are considered medium/high. Kawade titres were measured in a subgroup of the NAb positive samples [12,13].
Data analysis was carried out using Excel and spss software (SPSS Inc., Chicago, IL, USA). For DNA array studies paired t-tests with correction for number of expected false positive tests (false discovery rate, FDR) were carried out using a 1% FDR rate [27]. In addition, conventional t-tests, Mann–Whitney and Spearman-rank correlation coefficient (SRCC) analysis were used for data analysis (5% significance level). The boxes in the box and whisker plots denote the inter-quartile range, with the line denoting the median value. The whiskers denote the range of observations, excluding outliers and extremes, which are indicated by circles and asterisks.
Results
Gene expression studies on Affymetrix Gene Chips, with more than 8500 verified human transcripts, in 12 MS patients at four different time-points, showed 5996 probe sets that called as present according to the Affymetrix algorithm in at least one sample. When gene expression before treatment was compared with gene expression in the same patients 9–12 h after a scheduled intramuscular injection of IFN-β1a after 3 months of treatment, 1606 genes had lower expression levels (mean decrease 37%), and 315 had higher expression levels (mean increase 185%) than at baseline at a 1% FDR. After 6 months of treatment 1190 genes had lower expression levels (mean decrease 39%), and 288 had higher expression levels (mean increase 175%) 9–12 h after an injection of IFN-β than at baseline. The twenty most induced genes at these time points are listed in Table 1. The genes encoding the chemokines CCL2, CCL8, and CXCL10 and IFI27 (interferon, alpha-induced protein 27) were the four most induced genes at these time points, but MX1 (encoding MxA protein) was also strongly induced 9–12 h after an injection of IFN-β (Table 1).
| Gene symbol | Gene title | Representative public ID | Month 3 early FDR | Month 3 early-baseline ratio | Month 6 early FDR | Month 6 early-baseline ratio | Month 6 late FDR | Month 6 late-baseline ratio |
|---|---|---|---|---|---|---|---|---|
| FFAR2 | Free fatty acid receptor 2 | NM_005306 | 0.00068 | 6.305 | 0.00838 | 7.588 | 0.02259 | 2.218 |
| IFI44L | Interferon-induced protein 44-like | NM_006820 | 0.00026 | 6.563 | 0.00056 | 6.307 | 0.01077 | 3.887 |
| MX1 | Myxovirus (influenza virus) resistance 1 | NM_002462 | <0.00001 | 6.699 | 0.00005 | 7.257 | 0.03254 | 2.628 |
| IFI6 | Interferon, alpha-inducible protein 6 | NM_022873 | 0.00007 | 6.823 | 0.00036 | 7.518 | 0.00689 | 3.245 |
| JUP | Junction plakoglobin | NM_021991 | 0.00021 | 6.959 | 0.00050 | 7.974 | 0.00524 | 2.735 |
| LY6E | Lymphocyte antigen 6 complex, locus E | NM_002346 | 0.00015 | 7.035 | 0.00070 | 7.557 | 0.00462 | 3.567 |
| HERC5 | Hect domain and RLD 5 | NM_016323 | 0.00001 | 7.953 | 0.00009 | 8.590 | 0.05280 | 2.205 |
| RGS1 | Regulator of G-protein signalling 1 | S59049 | 0.01267 | 8.058 | 0.00182 | 5.084 | 0.54248 | 1.251 |
| USP18 | Ubiquitin specific peptidase 18 | NM_017414 | 0.00008 | 8.248 | 0.00005 | 8.727 | 0.01564 | 3.216 |
| IFIT3 | Interferon-induced protein with tetratricopeptide repeats 3 | NM_001549 | 0.00003 | 9.061 | 0.00011 | 10.401 | 0.00895 | 3.025 |
| IL1RN | Interleukin 1 receptor antagonist | U65590 | 0.00006 | 9.858 | 0.00004 | 10.430 | 0.01503 | 1.657 |
| ISG15 | ISG15 ubiquitin-like modifier | NM_005101 | <0.00001 | 10.488 | 0.00003 | 11.191 | 0.01295 | 3.811 |
| SERPING1 | Serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 | NM_000062 | 0.00008 | 10.526 | 0.00027 | 11.914 | 0.08963 | 2.086 |
| IFIT1 | Interferon-induced protein with tetratricopeptide repeats 1 | NM_001548 | <0.00001 | 11.652 | 0.00003 | 13.081 | 0.00979 | 4.872 |
| LAMP3 | Lysosomal-associated membrane protein 3 | NM_014398 | 0.00005 | 12.958 | 0.00020 | 12.267 | 0.67920 | 1.303 |
| CXCL11 | Chemokine (C-X-C motif) ligand 11 | AF030514 | 0.00221 | 16.648 | 0.00247 | 18.587 | 0.51033 | 1.284 |
| IFI27 | Interferon, alpha-inducible protein 27 | NM_005532 | 0.00009 | 17.083 | 0.00030 | 20.098 | 0.00111 | 20.370 |
| CXCL10 | Chemokine (C-X-C motif) ligand 10 | NM_001565 | 0.00008 | 29.210 | 0.00005 | 34.226 | 0.21192 | 2.008 |
| CCL8 | Chemokine (C-C motif) ligand 8 | AI984980 | 0.00003 | 37.373 | 0.00005 | 41.168 | 0.14819 | 2.229 |
| CCL2 | Chemokine (C-C motif) ligand 2 | S69738 | 0.00018 | 58.725 | 0.00005 | 59.759 | 0.05854 | 7.409 |
- This table lists the 20 genes that are most strongly induced early (9–12 h) after an injection of interferon (IFN)-β. Gene expression was analysed serially in 12 patients before treatment start, early (9–12 h) after an injection of IFN-β after 3 and 6 months of treatment and late (36–48 h) after an injection of IFN-β after at least 6 months of treatment. The false discovery rate (FDR) for each gene and the ratio between gene expression at the given time point and baseline were calculated.
Gene expression studies performed 36–48 h after an injection of IFN-β1a showed that gene expression changes at this time were now much less prominent than after 9–12 h, with only 64 genes having lower expression levels (mean decrease 38%) and 39 genes having higher expression levels (mean increase 159%) than at baseline. The twenty most induced genes at this time point are listed in Table 2. IFI27 was still strongly induced, whereas CCL2, CCL8 and CXCL10 were much less induced than 9–12 h after an injection of IFN-β (Tables 1 and 2).
| Gene symbol | Gene title | Representative public ID | Month 3 early FDR | Month 3 early-baseline ratio | Month 6 early FDR | Month 6 early-baseline ratio | Month 6 late FDR | Month 6 late-baseline ratio |
|---|---|---|---|---|---|---|---|---|
| S100A11 | S100 calcium binding protein A11 | NM_005620 | 0.00076 | 2.395 | 0.00013 | 2.875 | 0.00824 | 1.792 |
| IFITM2 | Interferon induced transmembrane protein 2 (1-8D) | NM_006435 | 0.00011 | 2.120 | 0.00019 | 2.401 | 0.00582 | 2.024 |
| --- | --- | M10098 | 0.08240 | 1.469 | 0.35318 | 1.226 | 0.00544 | 2.198 |
| MARCKS | Myristoylated alanine-rich protein kinase C substrate | NM_002356 | 0.00007 | 3.777 | 0.00003 | 4.053 | 0.00832 | 2.201 |
| IFITM3 | Interferon induced transmembrane protein 3 (1-8U) | BF338947 | 0.00007 | 2.957 | 0.00010 | 3.198 | 0.00717 | 2.208 |
| BCL2A1 | BCL2-related protein A1 | NM_004049 | 0.00017 | 2.169 | 0.00037 | 1.983 | 0.00355 | 2.211 |
| CTSL | Cathepsin L | NM_001912 | 0.00065 | 1.796 | 0.00144 | 1.881 | 0.00548 | 2.246 |
| OASL | 2′-5′-Oligoadenylate synthetase-like | AF063612 | <0.00001 | 4.143 | 0.00007 | 4.383 | 0.00718 | 2.267 |
| RHOB | Ras homolog gene family, member B | AI263909 | 0.00006 | 3.174 | 0.00068 | 3.581 | 0.00956 | 2.572 |
| LGALS3BP | Lectin, galactoside-binding, soluble, 3 binding protein | NM_005567 | 0.00030 | 3.764 | 0.00050 | 4.088 | 0.00552 | 2.580 |
| CDKN1C | Cyclin-dependent kinase inhibitor 1C (p57, Kip2) | N33167 | 0.00537 | 1.631 | 0.00800 | 2.0030 | 0.00497 | 2.625 |
| JUP | junction plakoglobin | NM_021991 | 0.00021 | 6.959 | 0.00050 | 7.974 | 0.00524 | 2.735 |
| --- | --- | M10098 | 0.04277 | 1.803 | 0.27731 | 1.443 | 0.00829 | 3.018 |
| IFIT3 | Interferon-induced protein with tetratricopeptide repeats 3 | NM_001549 | 0.00003 | 9.061 | 0.00011 | 10.401 | 0.00895 | 3.025 |
| OAS1 | 2′,5′-Oligoadenylate synthetase 1, 40/46 kDa | NM_002534 | <0.00001 | 5.574 | 0.00004 | 6.362 | 0.00695 | 3.102 |
| IFI6 | Interferon, alpha-inducible protein 6 | NM_022873 | 0.00007 | 6.823 | 0.00036 | 7.518 | 0.00689 | 3.245 |
| LY6E | Lymphocyte antigen 6 complex, locus E | NM_002346 | 0.00015 | 7.035 | 0.00070 | 7.557 | 0.00462 | 3.567 |
| MS4A4A | Membrane-spanning 4-domains, subfamily A, member 4 | NM_024021 | 0.00082 | 4.560 | 0.00063 | 5.382 | 0.00830 | 4.144 |
| IFIT1 | Interferon-induced protein with tetratricopeptide repeats 1 | NM_001548 | <0.00001 | 11.652 | 0.00003 | 13.081 | 0.00979 | 4.872 |
| IFI27 | Interferon, alpha-inducible protein 27 | NM_005532 | 0.00009 | 17.083 | 0.0003 | 20.098 | 0.00111 | 20.370 |
- This table lists the 20 genes that are most strongly induced late (36–48 h) after an injection of interferon (IFN)-β. Gene expression was analysed in 12 untreated multiple sclerosis patients and in the same patients early (9–12 h) after an injection of IFN-β after 3 and 6 months of treatment and late (36–48 h) after an injection of IFN-β after at least 6 months of treatment. The false discovery rate (FDR) for each gene and the ratio between gene expression at the given time point and baseline were calculated.
We selected CCL2, CXCL10 and IFI27 for further study by real-time RT-PCR analysis, and compared the expression of these molecules with that of MxA. In the 50 healthy control subjects the MxA index was log-normal distributed, with an upper reference level that was calculated as log(MxA index) + 3SD to a value of 5.0. However, the distribution of the CCL2-index, CXCL10-index and IFI27-index was skewed, and could not be transformed to a normal distribution. Hence, the 95 percentiles (3.8 for the MxA-index, 12.3 for the IFI27-index, 3.0 for the CCL2-index and 2.5 for the CXCL10-index) were used as upper normal limits.
Figure 1 shows the MxA-index, IFI27-index, CCL2-index and CXCL10-index in untreated MS patients, NAb negative MS patients treated with IFN-β and healthy controls. All biomarker indices differed significantly in these groups. Interestingly, untreated MS patients had a higher MxA-index (P = 0.002), CXCL10-index (P < 0.001) and IFI27-index (P < 0.001) than the healthy controls (Fig. 1). IFN-β-treated MS patients without NAbs had higher values of all indices than untreated MS patients and healthy controls (Fig. 1: all P < 0.001). The upper 95 percentiles for the gene expression indices in untreated MS patients were 25.9 for MxA, 35.5 for IFI27, 20.2 for CCL2, and 30.0 for CXCL10.
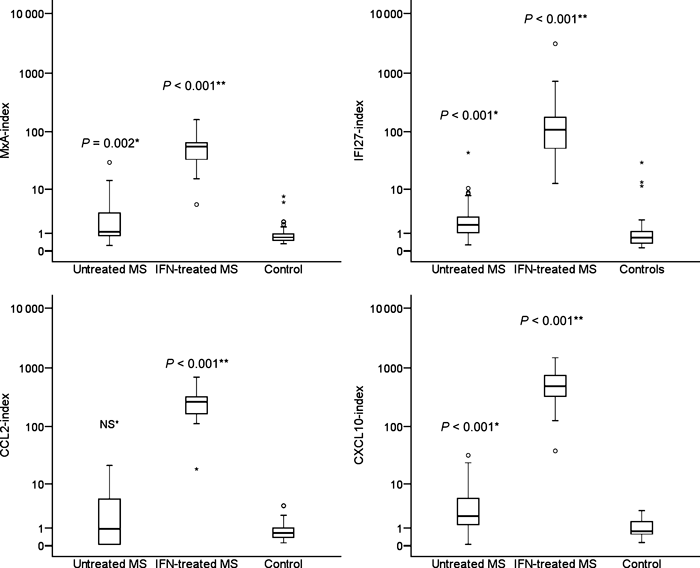
Expression of MxA, CCL2, CXCL10 and IFI27 measured by real-time PCR in 25 MS patients before and 9–12 h after an injection of interferon-β after 3 months of treatment, and in 50 healthy control subjects. Data are presented as index-values reflecting fold induction compared with a pool of healthy controls. Statistical analysis was by the Mann–Whitney U-test. *Compared with healthy controls. **Compared with untreated multiple sclerosis patients.
Comparing the expression of the four markers evaluated by PCR revealed a high degree of correlation. MxA-expression correlated with the expression of IFI27 (Spearman’s rank correlation coefficient, SRCC = 0.76), CCL2 (SRCC = 0.78) and CXCL10 (SRCC = 0.71). IFI27-expression correlated with the expression of CCL2 (SRCC = 0.81) and CXCL10 (SRCC = 0.72), and the expression of CCL2 and CXCL10 (SRCC = 0.86) also correlated (all P < 0.001 by Spearman rank correlation analysis).
Finally, we analysed the relationship between NAb levels and expression of each of the four IFN-β response biomarkers (Fig. 2). These plots show that all markers were significantly higher expressed in NAb negative IFN-β-treated patients compared with untreated patients. In patients with moderate/high NAb levels the MxA-index was comparable with that seen in untreated MS patients, but, interestingly, the CCL2-index, CXCL10-index and IFI27-index was significantly lower than in untreated patients. In patients with low NAb levels the MxA-index and the CCL2-index were only marginally increased. The expression of CXCL10 and IFI27 did not differ significantly between patients with low NAb levels and untreated MS patients.

The MxA-index, CCL2-index, CXCL10-index and IFI27-index were measured by real-time PCR analysis in 25 untreated MS patients and in 43 patients negative for anti-interferon (IFN)-β neutralizing antibodies (NAbs), 28 patients with low NAb levels and 95 patients with moderate/high NAb levels. Blood samples for gene expression analysis were obtained 9–12 h after an injection of the patient’s usual IFN-β preparation. Statistical analysis was by the Mann–Whitney U-test for comparisons with untreated patients.
The relationship between the Kawade titre and the four IFN-β response biomarkers was analysed in 25 Nab-negative patients and 28 NAb positive patients with Kawade titres ranging from 32 to 24,107 (median 599). As shown in Fig. 3 the Kawade titre showed a strong negative correlation with the expression of RNA encoding MxA (SRCC = 0.85, P < 0.001), IFI27 (SRCC = 0.85, P < 0.001), CCL2 (SRCC = 0.86, P < 0.001) and CXCL10 (SRCC = 0.84, P < 0.001).
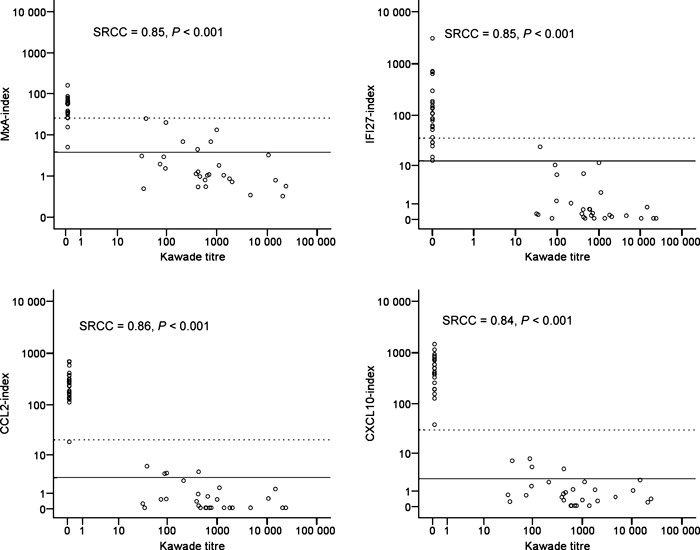
The MxA-index, CCL2-index, CXCL10-index and IFI27-index were measured by real-time PCR analysis in 25 untreated MS patients and in 28 Nab positive patients for whom Kawade titres were available. Statistical analysis was by calculation of Spearman rank correlation coefficients (SRCC). Full lines denote upper 95 percentiles in healthy control subjects and broken lines denote upper 95 percentiles in untreated MS patients.
Discussion
We used DNA array analysis to identify biomarkers that reflect the in vivo activity of IFN-β in patients with MS. The study confirms the strong induction of MX1 mRNA (encoding MxA) by IFN-β known from numerous previous studies [15,17–21], and identifies the even more pronounced induction of IFI27 and three chemokines (CCL2, CCL8 and CXCL10).
We recently completed a series of DNA array studies of gene expression in MS patients beginning therapy with IFN-β. These studies showed marked changes in gene expression induced by IFN-β within the first 24 h, but none of these effects persisted for 1 week [25]. We also confirmed that MxA is easily detected and strongly induced after the administration of IFN-β. In these studies we did, however, only analyse genes that were expressed in untreated MS patients at levels that are sufficiently high to allow their identification on DNA arrays, and the time points chosen for gene expression studies in that study (14–20 h and 7 days after an injection of IFN-β may not have been optimal for the detection of all IFN-β-induced genes.
Gene expression induced by IFN-β is highly dependent on the time from the administration of IFN-β [25,28,29], and the majority of IFN-induced genes are induced for <24 h. The induction of IFI27 was more long-lasting than the expression of the other markers studied, and did not differ in samples obtained 9–12 and 36–48 h after an injection of IFN-β. This is consistent with the results of a recent study, which also found long-lasting induction of IFI27 by IFN-β [29]. This makes IFI27 an attractive candidate as a biomarker for the effect of IFN-β, as the practical problems imposed by the strict time-dependency of MxA-induction assays should not apply to this molecule.
Increased expression of MxA in untreated MS patients was previously reported [30]. In the present study, we found that in addition to the expression of MxA, IFI27, and CXCL10 expression were higher in untreated MS patients than in healthy controls. Interestingly, we found that CXCL10 and IFI27 mRNA expression was comparable in patients with low NAb titers and untreated MS patients; in patients with moderate/high NAb levels the expression was even lower than in untreated MS patients. We hypothesize that this may reflect a role of endogenous IFN-β in basal expression of these molecules in MS patients. Although the MxA- and CCL2 mRNA expression was somewhat higher in patients with low NAb titers than in untreated MS patients, expression of these molecules was not increased in patients with moderate/high Nab levels.
Interferon-alpha-inducible protein 27 (IFI27) is a gene which encodes an IFN-inducible, putative highly hydrophobic protein of 122 amino acids with a 33% overall sequence similarity to the product of the IFI6 gene [31]. The biological function of IFI27 is unknown, but the related IFI6 gene encodes a protein that interferes with apoptosis induced by the TRAIL molecule [32].
CCL2 and CXCL10 were found to be induced by treatment with IFN-β in other studies [33,34]. A possible advantage of these molecules is that they are released in the circulation and, hence, can be measured by ELISA assays, which are less technically demanding than real-time PCR methods for the analysis of gene expression [33,34]. A correlation between mRNA expression and plasma concentrations of CXCL10 has been established [34], but it remains to be shown that the assessment of plasma chemokine concentrations can replace mRNA expression studies for the assessment of the in vivo response to treatment with IFN-β.
The finding of a grey zone in terms of MxA-index values that might either reflect endogenous MxA induction or MxA induction because of treatment with IFN-β is important for the interpretation of the results of in vivo MxA induction-tests in patients with NAbs. A recent study found that the magnitude of the MxA induction in response to IFN-β therapy correlates well with the clinical effect of treatment with IFN-β, but the optimal MxA induction level in IFN-β remains to be established [35]. An advantage of the other markers was that, compared with MxA, the induction of these molecules was much stronger, but a grey zone also existed for IFI27, CCL2, and CXCL10, although the overlap between untreated and IFN-β-treated patients was only marginal for CXCL10. Hence, these molecules could be of particular interest as biomarkers of IFN-β bioactivity.
Our findings indicate that the induction of even the most strongly IFN-β-induced molecules is lost in patients with moderate/high NAb levels, supporting the notion that all biological effects of IFN-β are lost in such patients. This supports the guidelines of the European Federation of Neurological Sciences, which recommends treatment with IFN-β to be discontinued in MS patients with persisting, high NAb levels [3].
In conclusion the present study identifies IFI27 and CXCL10 as candidate biomarkers for assessing the response to IFN-β in MS. Compared with MxA, IFI27 appears superior in terms of degree and duration of induction. CXCL10 has the advantage of distinguishing untreated from IFN-β-treated patients very clearly, and can be measured by ELISA assays. Finally, the induction of both CXCL10 and IFI27 is lost in patients with high levels of IFN-β NAbs. We therefore suggest that the biomarkers IFI27 and CXCL10 should be included in future studies of the biological effect of treatment with IFN-β in MS.
Acknowledgements
The study was performed as a part of the Neutralizing Antibodies in Multiple Sclerosis (NABINMS) Project under the European Commission 6th Framework Programme. Supported by the Danish Multiple Sclerosis Society, the Warwara Larsen Foundation, the Danish Medical Research Council, Brdr. Rønje Holding and unrestricted research grants from Biogen Idec, Merck-Serono and Sanofi-Aventis.




