Horse mackerel egg identification using DNA methodology
Abstract
Egg identification in plankton samples is usually needed for purposes of stock assessment. Until recently, only morphological characters were used for identifying the eggs of different fish species. Late developmental stages are easily distinguishable due to pigmentation as well as egg and oil globule size range. However, for early stages, these characteristics are difficult to be discriminated and may overlap with other species. European horse mackerel species (Trachurus trachurus, T. mediterraneus, T. picturatus) overlap significantly in their spawning areas in some European waters. Because of the fact that their eggs are morphologically similar, genetic methodologies are needed to identify eggs and larvae to species correctly. In the present study, formalin- and ethanol-preserved eggs were tested to estimate the efficacy of genetic methodologies for species identification of eggs when different preservatives are used. A 370-bp fragment of cytochrome b was successfully amplified followed by restriction fragment analysis with two restriction enzymes aiming to identify the eggs to species. Horse mackerel egg identification was accomplished with the maximum success in ethanol-preserved eggs. However, it seems that various preservatives had different effects on the DNA quality of eggs as genetic identification was less successful in formalin-preserved eggs. Preserving in ethanol a part of the eggs obtained in plankton surveys is suggested for purposes of accurate genetic identification, even if their morphological features are distorted in time.
Problem
Fisheries represent a global resource that is increasingly threatened by overexploitation. There are over 3 million fishing vessels operating in the world, and 70% of fisheries stocks are either fully exploited or overfished (Pauly et al. 2002; http://www.fao.org). For this reason, an appropriate management of these valuable resources requires realistic estimation of stock sizes. However, until now, the stock assessment methods depend mostly on catch and effort data. Recently, considerable concern has arisen over the increasing unreliability of catch and effort data caused by mis-reporting (Watson & Pauly 2001).
An alternative way to estimate stock biomass sizes and forecast adult abundance of target fish species is to employ the egg production method. To estimate the egg abundance for both mackerel and horse mackerel species, ichthyoplankton surveys are carried out covering the whole area of spawning. These surveys are executed every 3 years in ICES waters and are designed within the ICES working Group of Makerel and Horse mackerel eggs surveys (WGMEGS). Egg identification in plankton samples is needed for purposes of stock assessment as the egg production method uses the earliest development stage eggs to estimate the adult biomass. The most useful morphological characters for the identification of the pelagic fish eggs are egg shape, egg size, character of the chorion and yolk, presence/absence of oil globule, and characters associated with embryo development (Ahlstrom & Moser 1980). However, as the chorion is smooth and most species have eggs with a single oil globule, in practice, egg identification depends mostly on egg size and oil globule diameter. Late development stages are easily distinguishable due to pigmentation, morphological characteristics of eggs and oil globule size range, while in the case of early developmental stages, the range of variation in these measurements is short and may overlap with other species (Moser et al. 1984). Therefore, species determination by visual inspection is inaccurate in many cases.
Molecular techniques applied with species-specific genetic markers may allow identification of all life-history stages from eggs through metamorphosis to adult. DNA-based methods are the most useful tools for identification of marine organisms (Feral 2002; Shao et al. 2002). One of the major concerns when developing species-specific markers is the type of marker to be employed. Mitochondrial genes are frequent target sequences (Medeiros-Bergen et al. 1995; Palumbi & Cipriano 1998; Taylor et al. 2002) because their large number of copies per cell enable PCR amplification even in early developmental stages, when the number of cells per egg is small. On the other hand, sequences must be conserved within species and variable between species. Highly conserved coding genes such as cytochrome b and 16S rRNA are thus the first choice (Carr & Marshall 1991; Rehbein et al. 1999;Stubbs et al. 2000).
Another important issue for genetic identification of eggs is the preservatives used for conservation of plankton samples. Three different preservatives normally employed are: DMSO, ethanol and formaldehyde. Eggs are generally preserved in formaldehyde to avoid shape and colour changes that DMSO and ethanol usually provoke and may complicate the visual inspection of eggs for determining their developmental stage. Employing formaldehyde as a preservative, visual identification of eggs could be possible even long time after sampling.
At present, very few studies have dealt with genetic identification of eggs to species. Aoyama et al. (2001) employed sequence analysis of the 16S rDNA to identify ethanol-preserved eggs and larvae of Japanese eel, while Perez et al. (2005) genetically identified formaldehyde-fixed hake and megrim eggs using fragment size analysis of the 16S rDNA gene. Garcia-Vazquez et al. (2006) also identified formalin-preserved eggs from different species based on PCR-SSCP analysis of this gene.
The purpose of the present work was to apply molecular methodologies to validate their use in genetically identifying eggs of Trachurus species (Trachurus trachurus, T. mediterraneus and T. picturatus) that are present in ichthyoplankton samples. These species are distributed in overlapping marine areas and are commercially exploited especially in the N.E. Atlantic waters (International Council for the Exploration of the Seas 2001). Observation of morphologic features provides a good means for adult Trachurus species identification. However, the three horse mackerel species share partially, spatial and temporal niches during the spawning, and have eggs with very similar morphological characteristics (chorion aspect, egg size, oil globule diameter). As visual methods cannot identify all Trachurus eggs at the species level, a reliable genetic approach is needed and was applied to identify accurately the horse mackerel species present in samples from ichthyoplankton survey. Additionally, formalin- and ethanol-preserved eggs were tested to estimate the efficacy of genetic methodologies for species identification of eggs when different preservatives are used.
Material and Methods
Horse mackerel (T. trachurus) eggs were obtained by artificial fertilization at sea, and preserved in 4% buffered formaldehyde or 95% ethanol. Additional ichthyoplankton samples were obtained off the Portuguese coast employing Bongo nets (60 cm diameter, 303-μm-mesh nets) towed to a nominal depth of 200 m, and retrieved obliquely. Fish eggs at each tow were visually identified and sorted before the ship departed from the sampling station, then fixed in 4% buffered formaldehyde solution (pH 7.5). We used the artificial fertilized eggs so as to verify the applicability of the genetic methodology in the already determined eggs to species, while the ichthyoplankton samples were used so as to determine the species present in the sampling in a real world survey. The horse mackerel ichthyoplankton samples were visually identified only as belonging to genus Trachurus because it is extremely difficult to discriminate morphologically the eggs of other Trachurus species.
DNA was extracted from individual eggs following the Chelex-based protocol described by Estoup et al. (1996). Each individual egg was placed in an Eppendorf tube and rinsed in distilled water for 5 min. Then water was removed, the egg was gently squashed with a pipette tip and it was embedded in 50 μl of Chelex (12%) solution heated to 60 °C and 2 μl of 20 mg μl−1 Proteinase K. The mixture was incubated at 55 °C for 1 h and 40 min, shaking every 15 min, and finally the mixture was heated at 100 °C for 20 min.
DNA was extracted from formaldehyde-preserved eggs using QIAamp MiniKit DNA tissue Kit (Qiagen). The eggs were washed with PBS to remove the formalin prior to Qiagen protocol. The total extracted volume of DNA eluted using this kit was 200 μl per egg, performed in two 100-μl elution steps.
The genetic markers employed to identify Trachurus eggs had been previously tested in a large number of adult individuals of all three species to confirm the universality of the markers (Karaiskou et al. 2003a). PCR was employed to amplify a segment of around 370 bp of the mitochondrial cytochrome b gene. PCR amplification of the cytochrome b was performed using the universal primers H15149 and L14841 (Kocher et al. 1989). PCR conditions were as described by Karaiskou et al. (2003a). Specifically, double-stranded DNA amplification was performed in 10 μl reaction volumes containing 1 unit of Taq polymerase (Gibco-BRL), 1 μl of 10x reaction buffer, 2 mm MgCl2, 0.25 mm dNTPs, 25 pmol of each primer, 0.1% BSA and 4 μl of DNA. Thermal cycling amplification conditions were as follows: initial denaturation at 95 °C for 4 min, followed by 31 cycles of strand denaturation at 94 °C for 45 s, annealing at 51 °C for 45 s, and primer extension at 72 °C for 45 s and a final 5 min elongation time at 72 °C. PCR amplification of a shorter DNA fragment (180 bp) within the cytochrome b gene was performed using the conditions previously described. The newly designed primers were: CytbF: 5′-ATCTGCCGGGACGTAAACTA-3′ and CytbR: 5′-CGAAGGCAGTTCCYATAAGT-3′.
PCR products (3 μl) were subjected to restriction endonuclease digestions with 10 units of the appropriate enzymes (NlaIII and NciI) that allow the discrimination between the three Trachurus species (Karaiskou et al. 2003b), in a final reaction volume of 6 μl. Incubation temperature and duration of reaction were according to the manufacturers’ protocol (New England Biolabs, Hertfordshire, UK). Digested samples were electrophorized in 10% acrylamide gel (29:1 ratio of acrylamide/bis-acrylamide) for 6 h at 260 V using 1x TBE buffer. The sizes of the resulting DNA fragments were estimated by comparison with a commercial ϕχ174/HaeIII ladder (New England Biolabs, Hertfordshire, UK). DNA restriction fragments were visualized by silver staining DNA method (Karaiskou et al. 2003b). Genetic analyses were repeated twice per individual to confirm reproducibility of the results.
Results and Discussion
In the present study, DNA was extracted from artificially fertilized eggs of T. trachurus. PCR amplifications were performed successfully in 100 ethanol-preserved eggs, on a single egg basis. Genetic identification of eggs was accomplished using PCR-RFLP methodology of the cytochrome b gene. Specifically, the PCR product was digested with two restriction enzymes (Fig. 1) that are useful for identifying Trachurus species: NlaIII and NciI (Karaiskou et al. 2003b). With NciI digestion, T. trachurus provides a pattern of three fragments of around 145, 135 and 90 bp, while with the NlaIII restriction enzyme a pattern of 175 and 195 bp, is expected. Acrylamide electrophoresis revealed that the restriction fragments obtained by the digestion of ethanol-preserved eggs were identical with the T. trachurus species with 100% reproducibility.
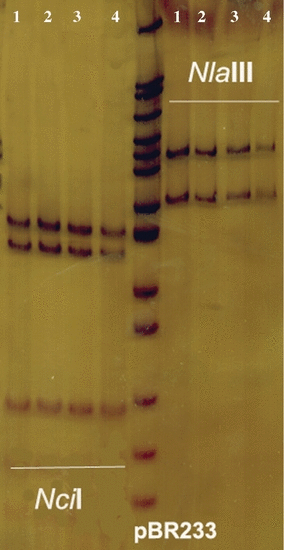
Digestion of the amplified cytochrome b from Trachurus eggs with NlaIII and NciI restriction enzymes. The obtained patterns are in accordance with the patterns described by Karaiskou et al. (2003b). 1: adult individual of T. trachurus; 2, 3, 4: Trachurus eggs with pattern similar to T. trachurus.
Thus, we provide an efficient methodology to amplify successfully a mitochondrial DNA segment from individual eggs in early developmental stages. The protocol described in the present study was successfully applied to many individuals and can be considered for intensive routine surveys employing simple techniques. Additionally, robotic extraction methods and modern molecular techniques like DNA chip technology (Kiesling et al. 2002) may optimize and speed up the process of egg identification in plankton surveys.
The same procedure was followed for formalin-preserved eggs (50 eggs in total) of T. trachurus obtained from artificial fertilization. Those formalin-preserved eggs whose DNA was successfully amplified and digested provided genetic patterns 100% compatible with the real species (T. trachurus), identical to the patterns obtained for ethanol-preserved eggs. However, the success of DNA extraction and PCR amplification of the cytochrome b gene for formalin-preserved eggs was less than 20%, much lower than the success obtained for ethanol-preserved eggs (100%).
The eggs sorted from ichthyoplankton samples (150 eggs in total) visually identified as Trachurus spp. were genetically identified as Trachurus trachurus eggs. The same comment about low success of DNA extraction + PCR amplification in formalin-preserved eggs stated above can be applied to the plankton eggs analysed. Various modifications were carried out in DNA extraction protocols (e.g. changes in Chelex concentration use of Qiagen kit extraction protocols) and PCR amplification conditions (changes in the concentration of template DNA, decrease in annealing temperature, use of different polymerases). However, the percentage of PCR amplification success was always similarly low (20%).
A possible reason for the low amplification success obtained for formalin-preserved samples could be the size of the amplified fragment in the marker of choice, 370 bp when employing the universal primers. A new pair of primers was designed for amplification of a shorter DNA fragment (180 bp). The amplified fragment included all the restriction sites useful for Trachurus species identification. Employing these primers, PCR amplification of ethanol-preserved eggs was 100%. The success of PCR amplification of formalin-preserved eggs was still low (20%).
Additionally, the nuclear 5S rDNA gene, which accurately discriminates between the three Trachurus species (Karaiskou et al. 2003b), was tested as an alternative marker for identifying Trachurus eggs. PCR amplification was not obtained for both ethanol and formaldehyde-fixed eggs, probably because of much fewer copies of the nuclear genes compared with mitochondrial ones.
These results are not surprising. Various studies reported problems in PCR using DNA derived from formaldehyde-preserved tissues. O’Leary et al. (1994) examined the success of amplifying a β-globine gene fragment from tonsil tissue samples preserved in a variety of fixatives. They were successful in amplifying DNA extracted from formaldehyde-fixed samples but with variable degrees of reproducibility. Fixatives containing mercuric chloride caused failure in PCR. They noted that formaldehyde cross-links the histones that coat DNA molecules and suggested that treatment of samples using a proteinase digest step was essential prior to DNA purification and PCR. They also stated that formaldehyde might cause nicks and breaks in the DNA template leading to spurious amplification sequences. Similar problems have been reported by De Giorgi et al. (1994) in the nematode Caenorhabditis elegans. They found that fixation in 2.5% formaldehyde reduced the reliability of PCR for 16S rRNA gene to around 20%. In addition, when sequenced, the PCR products contained several replication mistakes.
On the other hand, some studies reported success in PCR amplification from formalin-preserved tissue, ranging from around 60% (France & Kocher 1996) to 80% (Chase et al. 1998; Kirby & Reid 2001) depending on the species under study. However, in all cases PCR amplification was performed in larvae or in adult individuals. The first successful identification of formaldehyde-fixed fish eggs was reported by Perez et al. (2005), who genetically identified hake and megrim eggs with 85% of success using a partial segment (165 bp) of the 16S rRNA gene. Lower PCR amplification success in Trachurus eggs could be explained by smaller quantity of biological tissue. Trachurus eggs are as small as 0.6 mm diameter, much smaller than hake and megrim eggs (around 0.9–1 mm diameter) and thus they may have smaller quantity of DNA. Another possible reason for lower success of DNA extraction and PCR amplification for formalin-preserved horse mackerel eggs with respect to hake eggs could be the target gene (e.g. formaldehyde may cross-link stronger with the segment of cytochrome b amplified), as well as different concentration of formaldehyde, or the pH of the buffered formalin. According to Steedman (1976), buffers such as sodium acetate are not adequate for formaldehyde, because although the pH can be initially raised to above 7 it declines slowly to below 7 after some months or years.
We concede that our approach lacks control eggs of other Trachurus species. However, it was impossible to find artificially fertilized eggs from T. mediterraneus and T. picturatus so as to test the genetic approach in already accurately identified eggs. During this study, additional eggs were provided from another ichthyoplankton survey. These eggs were assigned to T. mediterraneus and T. picturatus, based on their morphological characteristics though not with high certainty. The genetic analysis revealed that the eggs belonged to T. trachurus species underlying the difficulty to sample and morphologically discriminate the eggs of T. mediterraneus and T. picturatus species. For this reason, the horse mackerel ichthyoplankton samples used in the present study were visually identified only as belonging to genus Trachurus.
The main conclusion of this study is the development of a methodology that can be applied to identify genetically the Trachurus eggs present in ichthyoplankton samples. The results can be visualized employing an easy and relatively cheap method based on acrylamide gel electrophoresis. A simpler and cheaper approach that can be applied in field surveys, with the minimum equipment, is the use of agarose gel electrophoresis. This technology was also applied in the present study but the discriminatory capacity of the agarose gel is lower for smaller fragments (e.g. for the 90 bp fragment from NciI digestion). Therefore, for routine survey in the field, acrylamide electrophoresis and silver staining can be replaced by a simple agarose gel electrophoresis and ethidium bromide staining technique but only for a quick first step identification.
The relevance of this finding for assessing stocks by plankton studies should be seriously considered by managers in charge of conservation and exploitation of marine fish species. Even relatively new applied techniques (Checkley et al. 1997) that are used to estimate spawner biomass by the daily egg production rely on automated optical identification of the eggs that is based on the shape and the dimension of the target egg species. In the case of Trachurus eggs, all three species have eggs with similar morphological characteristics and so the problem of accurate optical identification still remains. Genetic techniques offer a good solution to the problem, and give the opportunity to the scientist in a field survey to identify in cheap and fast way with basic equipment (PCR machine, acrylamide or agarose electrophoresis system) the collected eggs to species before applying the methodology for biomass estimation.
However, both visual and genetic approaches are not possible for identifying the same egg samples, because formaldehyde (that allows the preservation of morphological features for long time) seriously reduces PCR amplification success, at least at the cytochrome b gene. Thus, if this marker is employed for identification of Trachurus eggs in plankton, it would be recommended to preserve a part of the plankton samples in ethanol to achieve accurate species identification for purposes of stock assessment methods.
Acknowledgement
This project is financially supported by the EU QLKS-CT 1999-01157 Research Contract (MARINEGGS).




