Molecular phylogeny, taxonomy and evolution of the land snail genus Iberus (Pulmonata: Helicidae)
Filogenia molecular, taxonomía y evolución del género Iberus (Pulmonata, Helicidae)
Abstract
enPartial DNA sequences of two mitochondrial genes [cytochrome oxidase subunit I (COI) and 16S rRNA] from 59 specimens of Iberus were used to test the validity of the described morphospecies of this genus, and examine genetic divergences within and between main phylogenetic groups. Both gene fragments showed phylogenetic concordance. The COI gene was found to be faster evolving than the 16S gene and was fully protein-coding with no insertions or deletions. 16S rRNA was more informative than COI for resolving basal nodes. Both individual and combined analyses of the two gene fragments revealed five main phylogroups. These five groups are genetically unique lineages that are allopatrically distributed and considered to have full species status. Further subdivisions were also considered. Shell morphology was suitable for delimiting species boundaries, but several incongruences between morphology and mtDNA phylogeny were observed. These incongruences were considered consequence of hybridization between Iberus cobosi and Iberus marmoratus, and the result of shell shape polymorphism in Iberus rositai. According to spatial patterns of sequence divergence, life habits and shell morphology may be concluded that the keeled-flat shelled snails independently originated several times within Iberus and they could represent cases of similar shell adaptation to a karstic arid environment.
Resumen
frSe han analizado dos fragmentos del ADN mitocondrial (COI y 16S rRNA) de 59 ejemplares de Iberus con el fin de determinar la validez de las morfoespecies de tamaño medio descritas para este género, así como para analizar las divergencias genéticas existentes entre los principales grupos filogenéticos identificados. Los resultados obtenidos para ambos genes fueron similares. La tasa evolutiva fue más rápida en el COI, fragmento codificante, no presentando inserciones ni deleciones. Por otra parte, el fragmento del 16S fue más informativo en la resolución de los nodos basales. Tanto los análisis individuales de ambos fragmentos como el análisis combinado permitieron identificar cinco linajes genéticos únicos que presentan una distribución alopátrica y son considerados como cinco especies diferentes. Además, se contemplan otras subdivisiones adicionales. Se ha observado que existe concordancia entre las morfoespecies y los filogrupos obtenidos, indicando que la concha es un criterio taxonómico válido para la delimitación de especies en este grupo. No obstante, se han observado varias incongruencias entre la morfología y la filogenia obtenida mediante secuenciación del ADNmt. Estas incongruencias parecen ser consecuencia de procesos de hibridación entre I. cobosi e I. marmoratus, pero parecen ser resultado del polimorfismo de la concha en I. rositai. Teniendo en consideración los filogramas obtenidos, el modo de vida y la morfología de la concha, se puede concluir que las conchas aquilladas y aplanadas se originaron varias veces independientemente en Iberus y que parecen constituir ejemplos de adaptación de la concha a ambientes kársticos áridos.
Introduction
The two major goals of systematics are delimiting species and reconstructing their phylogenetic relationships (Mayr and Ashlock 1991; Coyne and Orr 2004). Species are routinely used as fundamental units of analysis in biogeography, ecology, macro- and microevolution and conservation biology (Avise 2000; Goldstein et al. 2000; Hey et al. 2003; Weiss and Ferrand 2007). Delimiting species is important in the context of understanding many evolutionary mechanisms and processes (Sites and Marshall 2003). Over- or under-resolved species boundaries, along with taxonomic misidentification, will probably lead to questionable inferences about speciation in studies aimed at understanding these processes (Thacker and Hadfield 2000; Coyne and Orr 2004). The knowledge comprising these fields is based on previous resolution of the taxonomy and phylogenetic relationships of the species in question.
As is true of many invertebrates, molluscs include poorly known groups, for which species delimitation and relationships between species are not clear. Taxonomic inferences in molluscs are often hindered by a lack of morphological diversification between different lineages, as occurs in cryptic species or species showing overlapping variability (Wilding et al. 2000; Liu et al. 2003; Pfenninger et al. 2003, 2006; Pinceel et al. 2004; Geenen et al. 2006; Pfenninger and Schwenk 2007). In other cases, taxonomy is ambiguous because of the high phenotypic plasticity of shell morphology or other traditional taxonomic characters (Giusti and Manganelli 1992; Goodacre 2001; Uit de Weerd 2004). Molecular studies are increasingly being used to test species-level taxonomies based on morphological characters. Molecular tools have enabled objective and rigorous genetic analysis of differences at the population and higher taxonomic levels, and have provided essential data to address many issues concerning speciation (Coyne and Orr 2004). Our knowledge of this field has been greatly improved by phylogenetic studies that have explored interrelationships between species of land snail genera such as Partula (Johnson et al. 2000), Cerion (Gould 2002), Mandarina (Chiba 2002), Mastus (Parmakelis et al. 2003, 2005), Albinaria and other Alopiinae (Uit de Weerd 2004), Ainohelix (Tongkerd et al. 2004), Arianta (Gittenberger et al. 2004), Napaeus (Alonso et al. 2006) and Humboldtiana (Mejía and Zúñiga 2007).
The genus Iberus is one of the main representatives of the helicid land snails of the Iberian Peninsula (Puente 1994), and comprises several forms of medium to large-sized snails, including the largest Iberian land snails. As far as we are aware, Iberus species are anatomically cryptic while variation in shell structure is high. As a consequence, shell morphology is the only character that has been used to diagnose the different taxa (Puente 1994). In effect, characters such as shell size and colour, banding pattern, sculpture, whorl expansion rate and peristome reflection, together with the relative width of the umbilicus are commonly used to describe variation among them. The definitions of many taxa are based on the shape of the periphery, whether the edge of the shell is rounded or angular, or whether it forms a keel in extremely flattened shells. It is, however, known that the morphological classification of a snail’s shell is sometimes arbitrary and misleading for correct species delimitation (Giusti and Manganelli 1992; Pfenninger et al. 2006). Sometimes, morphological variation in snails may be the result of phenotypic plasticity in response to different environmental conditions, whereas in other species, it is the history of the population that gives rise to the morphologies observed (Chiba 1999; Tongkerd et al. 2004; Parmakelis et al. 2005).
The taxonomic validity of the morphospecies of Iberus described remains largely unresolved. Traditional classifications conflict with each other mostly in terms of the number and delineation of species, indicating that species descriptions based on shell morphology are difficult, unreliable and should be supplemented by other analytical methods, such as DNA sequencing (Puente 1994). The molecular taxonomy and phylogenetic relationships of the large forms of the genus (IGC: Iberus gualtieranus complex) has been assessed separately (Elejalde et al. 2008). In the present paper, we examine interrelationships between the remaining Iberus morphospecies including all the medium-sized morphospecies: Iberus angustatus (Rossmässler, 1854), Iberus cobosi (Ibáñez and Alonso, 1978), Iberus guiraoanus (Rossmässler, 1854), Iberus loxanus (Rossmässler, 1854), Iberus marmoratus (Férussac, 1821), Iberus ortizi (García San Nicolás, 1957) and Iberus rositai (Fez, 1950). In addition, we assessed phylogenetic relationships with the larger forms of the genus [Iberus alonensis (Férussaac, 1821), Iberus campesinus (Ezquerra in Pfeiffer, 1846), Iberus carthaginiensis (Rossmässler, 1854), I. gualtieranus (Linnaeus, 1758) and related species].
With the exception of I. gualtieranus, which is listed in the red book of endangered invertebrates of Andalusia (Southern Spain), the other members of the genus Iberus are not formally protected under Spanish state or regional laws, but the group is of conservation concern. In an effort to improve the monitoring of natural populations and manage a captive rearing facility, we sought to better understand the phylogenetic relationships among the extant species of this group of endemic snails, several of whose taxa are restricted to small geographic areas. The genus is subdivided into several geographically localized morphospecies, mainly distributed across eastern Andalusia.
The genus Iberus is potentially an excellent model for understanding speciation, shell adaptation and biogeography in a terrestrial group of snails. Herein, we try to infer the evolutionary history of the genus by almost fully sampling its known species. Partial DNA sequences of two mitochondrial genes [cytochrome oxidase subunit I (COI) and 16S rRNA] from 59 Iberus specimens were used to: (1) establish genetic divergences within and between main phylogenetic groups; (2) test the validity of morphologically defined species; (3) provide a preliminary overview of the geographic distribution of the major genetic phylogroups; (4) genetically identify unique lineages in need of protection and (5) assess the phylogenetic concordance of the two gene fragments. Once we have established this framework, it will be possible to address issues such as the biogeography and evolution changes in morphological characters within Iberus. The identification of genetically unique populations in need of protection is essential for designing strategies for their conservation. Besides providing key data for the conservation of invertebrates, information regarding the extent and current patterns of divergence within and among species and populations along with the geographical distribution of genetic variation, will improve our understanding of speciation processes in a biodiversity ‘hot spot’ such as the one examined here.
Materials and Methods
Sample collection
Fifty-nine specimens of Iberus were collected in Andalusia. Iberellus minoricensis and Otala lactea were used as outgroups. The sampling sites and corresponding specimens appear in Table S1 and are represented in Fig. 1. The collected material was classified according to García San Nicolás (1957). Whenever possible, topotypes were also included in the analysis.
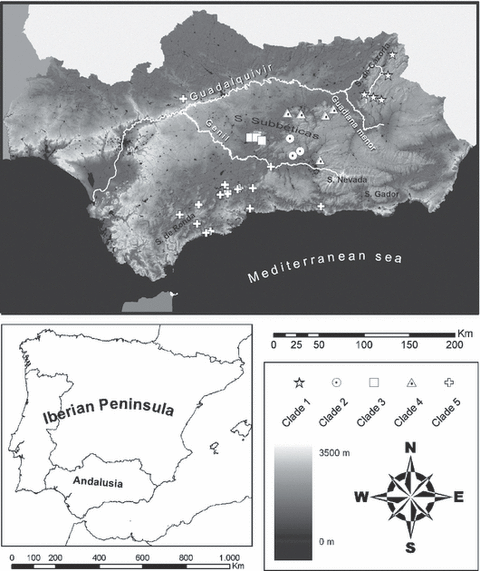
Map of the sampled localities of the Iberus species. Symbols indicate the different clades represented in the Fig. 3
DNA extraction, fragment amplification and sequencing
DNA was extracted from foot muscle of each specimen using the DNAeasy Tissue kit (QUIAGEN, Hilden, Germany). Polymerase chain reaction (PCR) was used to amplify the mitochondrial genes COI and 16S rRNA using universal primers (Palumbi et al. 1991; Folmer et al. 1994, respectively). The PCR conditions were: denaturation at 93°C, 2 min; annealing at 54°C, 1 min (for COI) or 57.7°C, 1 min (for 16S rRNA), elongation at 93°C, 45 s; total 40 cycles. The cycling ended with an extension phase at 72°C for 7 min. Reaction products were run on 1.5% agarose gels and stained with ethidium bromide to verify positive amplifications. Amplicons were sequenced using the dRhodamine Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, CA, USA) run on an ABI PRISM Model 3100 Avant Genetic Analyzer (Foster City, CA, USA). The sequences were deposited in GenBank (Table S1).
Sequence analyses
Sequences were aligned prior to phylogenetic analyses using ClustalX (Thompson et al. 1997) and the alignments were refined manually. Standard DNA polymorphism measures were calculated in DNAsp version 4.00 (Rozas et al. 2003). Results are provided in Table S2. Saturation levels of the COI fragments were determined using the program dambe version 4.2.13 (Xia and Xie 2001). First, second and third codon positions were analysed separately.
The ‘partition homogeneity’ test implemented in paup 4.0b3 (Swofford 2002) was used to establish compatibility between the COI and 16S rRNA data. This test assesses the null hypothesis that the ‘partitions’ indicated by the two data sets reflect the same phylogeny, under parsimony criteria. To obtain the distribution of random partitions, 1000 replications were performed with 10 random additions.
Phylogenetic analyses using paup were performed for both gene fragments and the combined data set. The best model of sequence evolution was selected using the Akaike information criterion implemented in modeltest v3.06 (Posada and Crandall 1998). The final models selected were: K81uf + I+ G (Kimura 1981) for the COI data, TVM + I + G for the 16S rRNA data and TrN + I + G for the combined data set (Tamura and Nei 1993).
For the maximum parsimony (MP) analyses, gaps were treated as missing data. A heuristic search was performed, with 10 random addition replicates, using the tree bisection reconnection option generating multiples trees to determine the most parsimonious. Parsimony bootstrap support values were calculated through 1000 bootstrap replicates. The weight of transversions and transitions was varied depending on the fragment and estimated by maximum likelihood. Weighting was 6 : 1 for the COI gene and 2 : 1 for the 16S rRNA gene.
Neighbour-joining (NJ) (Saitou and Nei 1987) trees were constructed using K81uf and TVM for each gene-sequence data set (COI and 16S rRNA, respectively) and TrN for the combined data set. Uncorrected pairwise p distances were also calculated for the three data sets (see Table S3). Bootstrap confidence estimates for each node were based on 1000 replicates (Felsenstein 1985).
Bayesian analyses (BA) were performed using the MrBayes v3.0 package (Huelsenbeck and Ronquist 2001). The GTR model was used on the three data matrices and rate variation across sites was modelled using gamma distribution, with a proportion of the sites being variants. The Markov chain Monte Carlo search was run with four chains for 2 million generations, with trees sampled every 100 generations (the first 2000 trees were discarded as ‘burnin’). In the combined analyses, variation was partitioned among genes.
Results
Sequence characteristics and genetic divergences
The number of mtDNA haplotypes (H), haplotype diversity (Hd), nucleotide diversity (π) and number of nucleotide differences (k) were calculated for each gene fragment and for the combined data set (Table S2).
The aligned COI fragment comprised of 628 base pairs (bp). The 61 aligned sequences were A : T rich (72.19%), with nucleotide composition of: T (44.22%), C (13.65%), A (27.97%) and G (14.15%). A total of 233 (37.1%) characters were parsimony informative. Using the Drosophila mitochondrial genetic code, five (2.39%) of the 209 amino acids were non-synonymous substitutions, 167 (79.9%) amino acids had synonymous nucleotide substitutions and 37 (17.7%) had no substitutions. The results of the saturation analysis (Fig. 2) indicated that the third codon position in the COI gene showed some saturation when genetic distances were over 6%. No mutational saturation was evident at first and second codon positions. Phylogenetic reconstruction did not differ when third codon positions of COI were considered or excluded. These were thus included in the analyses because of the phylogenetic information provided within the main phylogroups.
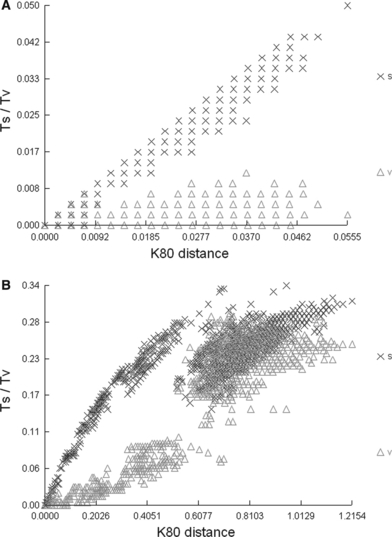
Relationships between the number of substitutions (△, transversions; x, transitions) and genetic distance (K80-distance) in COI fragment, considering: A, first and second positions; B, third position
The aligned 16S rRNA fragment consisted of 395 bp. These sequences were A : T rich (69.47%), with nucleotide compositions of: T (32.87%), C (13.25%), A (36.6%) and G (17.28%). A total of 127 (32.15%) characters were parsimony informative. The combined data set of the COI and 16S rRNA fragments consisted of 1023 bp. Six hundred and twenty-three (60.89%) characters were constant, 40 (3.91%) variable characters were parsimony uninformative and 360 (35.19%) characters were parsimony informative.
Phylogenetic analyses
Independent analysis of the COI and 16S rRNA data rendered slight different topologies, mainly in terms of their basal nodes (Fig. 3).
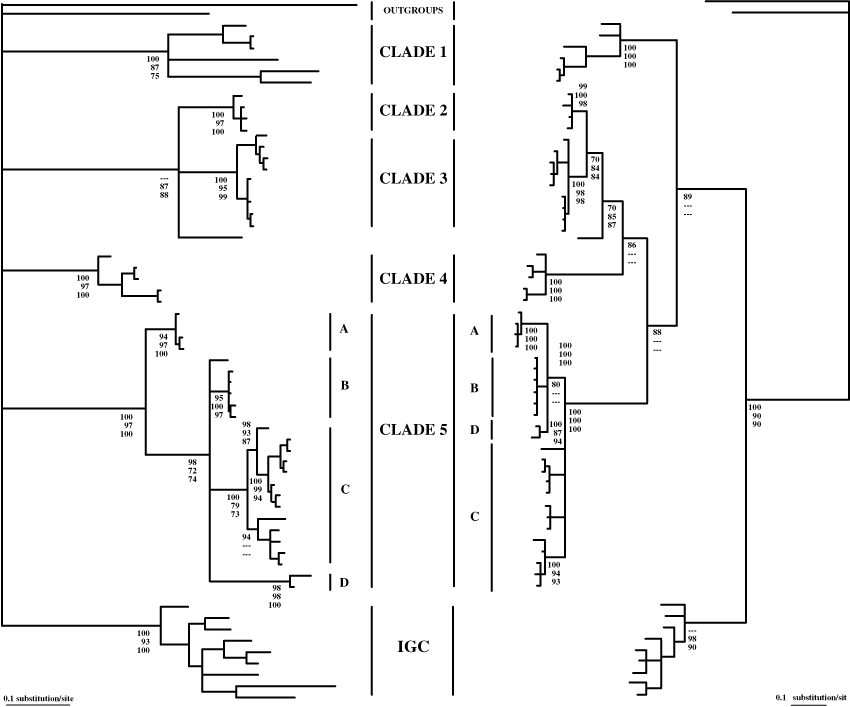
Molecular phylogenies of COI (left) and 16S rRNA (right) by MrBayes analyses. Bootstrap values under MrBayes, neighbour-joining and maximum parsimony are given at each node (MB/NJ/MP) when >70%
The ribosomal 16S gene had better resolving power for the deeper nodes than COI. The level of sequence divergence observed among phylogroups was very different for these two genes owing to the faster evolving sequences of COI. The lower the genetic divergence the greater the differences observed between COI and 16S rRNA p-genetic distances. Thus, for genetic divergences of <10–12%, the COI gene showed p-genetic distances that were nearly double the 16S gene divergence values. Highly reduced differences in p-genetic divergence between COI and 16S for percentages above 14% in the former gene, clearly indicate substitutional saturation in COI, which was consistent with the saturation of third codon positions established by DAMBE for this protein-coding gene.
In separate analyses, combination of the two mitochondrial genes, COI and 16S rRNA, yielded a well-supported consensus topology giving greater confidence to the phylogeny of Iberus shown by the strict consensus tree (Fig. 4).
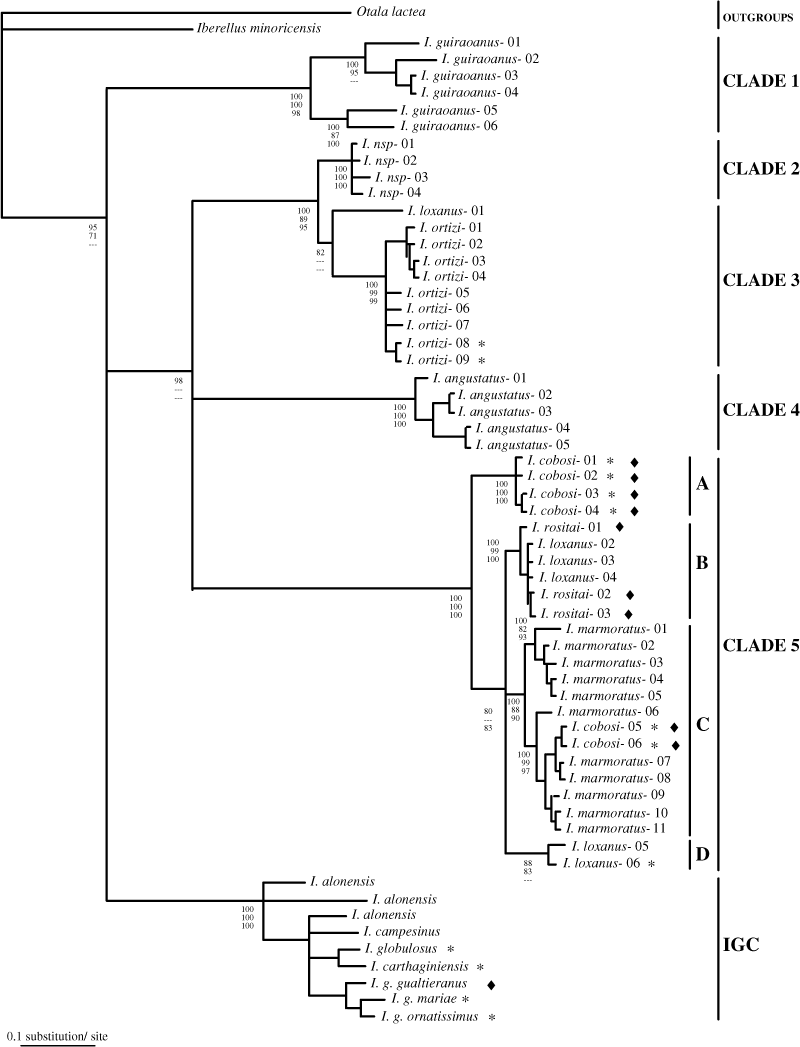
Molecular phylogeny of Iberus as revealed by MrBayes analysis for the combined data set of the COI and 16S rRNA genes. Bootstrap values under MrBayes, neighbour-joining and maximum parsimony are shown at each node (MB/NJ/MP) when >70%. Black rhombuses (♦) indicate specimens with a keeled, flattened shell. The rest of the haplotypes correspond to rounded shell specimens. Asterisks (*) indicate topotypes
As many as five main genetic phylogroups were identified within the medium-sized snails of Iberus. All the five clades had molecular divergences above 3.7% for 16S and 8.0% for COI, and were supported by bootstrap values above 95% in the NJ, MP and BA. They comprised all but one of the haplotypes obtained. The large specimens of the genus were grouped in a separate clade designated IGC. Basal nodes were only resolved in the independent analysis of the 16S locus. COI and concatenated data set analyses revealed a polytomy joining the larger phylogenetic clades.
Clade 1 joined all the specimens classified as I. guiraoanus. The monophyly inferred for the large umbilicated specimens of medium-sized snails (Rossmässler 1854) indicated that they really comprise a natural group. Iberus guiraoanus was structured into two very divergent subclades according to both genes (8.2–10.08% for 16S; 14.33–15.61% for COI). No diagnostic characters based on shell morphology to discriminate between these two groups could be found. Nevertheless, the two lineages were also geographically differentiated, suggesting the possibility of cryptic species.
In all the analyses, clades 2 and 3 clustered with an ungrouped haplotype within a large monophyletic clade. All haplotypes of these two clades corresponded to snails with a closed umbilicus shell. Clade 3 included the specimens collected from Cabra, the type locality of I. ortizi (García San Nicolás 1957). The other specimens included in clade 3 were also assigned to this species and, consequently, we also consider I. ortizi as a natural group. Clade 2 seems to represent an undescribed species. The ungrouped haplotype corresponded to one specimen of the I. loxanus morphotype (denoted I. loxanus 01). This haplotype was very divergent from the five clades identified, showing nucleotide substitution percentages with respect to the closest haplogroups >6% for 16S and 12% for COI. More intensive sampling in the east of Malaga could provide more information about the phylogenetic relationships of this unique population.
All the specimens identified as I. angustatus based on shell morphology, and particularly on the presence of a narrow umbilicus (Rossmässler 1854), constituted clade 4, indicating that I. angustatus is another natural group.
Clade 5 grouped together the remaining haplotypes. This was the most polymorphic clade in terms of shell morphology and brought together as many as four different morphotypes: I. cobosi, I. rositai, I. marmoratus and I. loxanus. Nevertheless, there were several incongruences between shell morphology and molecular systematics that will be discussed later. None of these four morphospecies represented a monophyletic haplogroup.
Discussion
Phylogeny and classification of Iberus
The cluster patterns and arrangements of individual haplotypes indicated by the 16S and COI gene fragments were highly consistent, although the former gene had better resolving power at the deeper nodes. The combined data set was good at resolving phylogenetic relationships among and within phylogroups. Monophyly was confirmed for the IGC-complex. Nevertheless, the monophyly of medium-sized species was only recovered by BA of the 16S gene. Combined analysis of both mitochondrial genes did not serve to confirm the monophyly of this group of snails, which appeared within a polytomy together with the IGC clade. Both individual and combined analyses of the COI and 16S genes indicated that all but one haplotype of the medium-sized species of Iberus could be attributed to five main clades. The phylogenetic analyses and the non-parametric bootstrapping test performed here were conclusive regarding the monophyly of these five basal clades.
All five clades represent geographically confined clades distributed across the Betic region in SE Andalusia (Fig. 1). With the exception of one population assigned to clade 5, all of them occur south of the Guadalquivir River valley. Clade 1 appears in the NE Betic region and is separated from the other clades by the Guadiana Menor River. Clade 5 exhibited the widest distribution range and is confined to an area west of the Genil River with some populations living near the coast. Clades 2–4 are limited by the Guadalquivir River to the north, the Guadiana Menor River to the NE and the Genil River to the SW.
Our analysis of the COI and 16S rRNA genes revealed a high degree of genetic variation in the mtDNA of Iberus, even within main clades. Genetic divergences among basal clades were >4.5% for 16S and 10% for COI. Although intraspecific divergences in molluscs are typically no higher than 5% for both the genes examined here, higher maximum intraspecific levels have been generally reported for freshwater and terrestrial snails (Dillon and Frankis 2004). Thus, it is very difficult to assign a taxonomic status to phylogroups based on sequence divergence levels.
Haplotype phylogenies based on DNA sequence data are increasingly being used to test traditional species-level taxonomies based on morphology (Brower 1999; Templeton 2001; Sites and Marshall 2003). The DNA phylogenies of Iberus were highly consistent with morphological data, especially the specimens assigned to clades 1–4 which corresponded to proper morphospecies. In addition, basal clades were strongly supported by non-parametric bootstrapping and were consistent with geography. Finally, the DNA haplotype phylogenies suggested no apparent gene flow between populations of basal lineages. According to the explicit tree-based species delimitation protocol recommended by Wiens and Penkrot (2002), we could conclude that the five basal clades should be considered different species: I. guiraoanus, I. ortizi, I. angustatus, I. marmoratus and an unnamed species (here designated Iberus nsp.). A formal description of this species, together with the designation of type material is in preparation. The taxonomy and morphology of I. guiraoanus, I. angustatus and I. marmoratus have been well-established (Rossmässler 1854; García San Nicolás 1957). Since early times, I. ortizi has been considered a minor form of I. alcarazanus (Ortiz de Zárate inGarcía San Nicolás 1957) although García San Nicolás (1957) assigned it a subspecific status: I. alcarazanus ortizi. However, the data provided by a recent taxonomic revision (Martínez-Ortí et al. 2004) indicate that I. alcarazanus should be considered synonymous of I. alonensis. Consequently, we consider I. ortizi a valid name for clade 3. Further subdivisions of clade 5 cannot be ruled out.
Incongruences between morphology and mtDNA phylogeny
Numerous studies have critically assessed the significance of morphological characters in the taxonomy of land snails (Giusti and Manganelli 1992; Schilthuizen and Gittenberger 1996; Uit de Weerd et al. 2004; Holland and Hadfield 2007). It has been repeatedly noted that shell morphology in molluscs can be greatly influenced by environmental conditions (Alonso et al. 1985; Chiba 1999; Pfenninger et al. 2006). Thus, species identification based only on shell morphology can lead to mistakes. As molecular phylogenetic studies increasingly gain ground, more examples of incongruence between morphology and phylogeny emerge. These inconsistencies include shell form (Chiba 1999, 2003; Pfenninger et al. 2006), reduced apertural lamellae complexity (Tongkerd et al. 2004), the clausilium apparatus of clausilids (Van Moorsel et al. 2000; Uit de Weerd et al. 2004), body colour (Pinceel et al. 2004) and some characters of the anatomical reproductive system and spermatophore morphology (Hershler 1994; Parmakelis et al. 2003; Steinke et al. 2004; Uit de Weerd 2004; Alonso et al. 2006). Several main incongruences between morphology and mtDNA phylogeny can be attributed to the highly adaptive nature of morphological characters, as has been pointed out in many studies of land snail species (Chiba 1999; Goodacre 2001; Holland and Hadfield 2007).
Our molecular phylogeny showed that I. loxanus was polyphyletic, indicating that the shell characters of medium-sized snails, i.e. their rounded periphery, flattened shell spire and no umbilicus (Rossmässler 1854), are homoplasic and should not be used as diagnostic characters. Similarly, I. alonensis is defined by the presence of a large, non-umbilicated and globose shell of rounded periphery, which is considered a plesiomorphic and conservative condition of the IGC group (Elejalde et al. 2005). The haplotypes of the loxanus form were included in three different clades (5b, 5d and clade 2 + 3) suggesting shell convergence. Probably, the study of more populations of the loxanus shell type from east Malaga would give more information about the geographical distribution and phylogeny of this group. The name I. loxanus should be restricted to the subclade 5d in which topotypes of this taxon were included.
A further incongruence is the keeled, flattened and intensely ornamented shell type. Apart from the IGC group (Elejalde et al. 2005, 2008), keeled shells only appeared in clade 5. Haplotypes of keeled-shelled snails (I. cobosi and I. rositai morphospecies) nested in the same subclades (clades 5b and 5c) and were interspersed with specimens with globular shells. Clade 5b brought together the keeled and rounded-shell snails from the ‘Torcal de Antequera’ and surrounding areas, with haplotypes of both shell morphotypes intermixing with each other. Traditionally, snails with rounded shells have been classified as I. loxanus, while keeled-shelled snails with marked rib ornamentation in this mountain area were designated I. rositai (Alonso and Ibáñez 1978; Puente 1994). However, our phylogenetic analysis indicates that both snail morphotypes from this area constitute a single polymorphic species. This finding is in agreement with the morphological observations of Alonso and Ibáñez (1978), who pointed out the appearance of a complete shell shape gradient between I. rositai and I. loxanus. These authors added that I. loxanus shell type specimens from Torcal de Antequera had slight ribs on the shell surface, resembling I. rositai (Alonso and Ibáñez 1978). Hence, based on both phylogenetic and morphological data, we consider that snails of both shell types, rounded- or keeled-shelled, from ‘Torcal de Antequera’ should be classified as I. rositai. We have no evidence of gene flow between I. rositai s. l. (rounded- and keeled-shelled snails of subclade 5b) and I. loxanus s. str (subclade 5d). Minimum genetic divergence was low for the 16S gene but not for COI (2.4 and 10.6%, respectively). Since they are the closest relatives observed, we cannot exclude the possibility that more extensive sampling of I. loxanus s. str. especially in zones close to ‘Torcal de Antequera’ could render even shorter genetic distances to those observed here. The relationships of both taxa with I. marmoratus should be also further investigated. Based on the available evidence, we cannot infer whether I. rositai s. l. (clade 5a), I. loxanus s. str. (clade 5d) and I. marmoratus should be regarded as valid species or as three subspecies of a single species.
Hybridization evidences between Iberus marmoratus and Iberus cobosi
Two specimens of the I. cobosi shell type were assigned to the I. marmoratus clade 5c. These were collected from the type locality, together with the other I. cobosi specimens grouped in the clade 5a. Such sharing of mtDNA haplotypes can be explained in two ways. Shared haplotypes could be the result of incomplete lineage sorting of ancestral polymorphism or hybridization events. Incomplete lineage sorting has been documented in molluscs (Wilding et al. 2000; Wilke et al. 2005). Theoretically, the ancestral mtDNA polymorphism can persist when speciation is a recent and rapid process and split populations had insufficient time to progress to reciprocal monophyly (Avise 2000). Alternatively, ancestral DNA polymorphism could persist within a species over longer periods of time when the population number is very large. The present results indicate that the I. cobosi (clade 5a) and I. marmoratus (clade 5c) lineages diverged for a fairly long time as reflected by the genetic distances showed by both clades (above 5 and 10% for 16S and COI, respectively). Besides, I. cobosi is restricted to a reduced geographical area (<1 km2) and, consequently, the population size could not be very large. Incomplete lineage sorting is thus not consistent with the pattern observed here.
Introgression of mtDNA from I. marmoratus to I. cobosi is a better explanation for the nested position of several I. cobosi haplotypes in the I. marmoratus phylogroup. Intermediate shell forms have also been identified in the field by the present authors, but the expanse of the hybrid zone is unknown. Iberus cobosi is restricted to a small valley within the geographical range of I. marmoratus where they live as ecological vicariants, with the former restricted to the karstic hills of the ‘Valle de Abdalajís 30SUF48’. Speciation is an ongoing evolutionary process with many intermediate stages between conspecificity and complete reproductive isolation. Species pairs, which are not completely reproductively isolated, could hybridize in contact zones. During speciation and reinforcement, individuals may appear that are intermediate between two well-delineated forms, but these intermediates disappear when reproductive isolation becomes complete (Coyne and Orr 2004). The genetic differentiation observed between I. cobosi and I. marmoratus lineages (clades 5a, 5c) indicates historic isolation between them. Thus, in this case, hybridization can hardly be considered a transient phase of divergent evolution during speciation. Two plausible situations could explain the existence of these tentative hybrids: (1) the populations of snails with globular and keeled shells only recently met because of the expansion of I. marmoratus as a consequence of secondary contact, and hybridizing taxa might be currently in the process of fusing into a single species, or (2) hybrid snails could be less fit (due to diminished outbreeding) such that they are maintained by the equilibrium between selection against them and gene flow (tension zone, Barton and Hewitt 1985). Further research is needed to quantify the introgression level of these two taxa as well as to identify whether interbreeding may affect the whole I. cobosi range or whether it is a rare event limited to a particular hybrid zone. Studies concerning nuclear DNA could provide additional details about the hybridization cases between these lineages. The considerable morphological and ecological differences between I. cobosi and I. marmoratus suggest that current level of introgression is insufficient to threaten their genetic integrity.
Evolution of the Iberus keeled-flat shelled snails
Three independent lineages of Iberus include keeled-flat shelled snails: I. g. gualtieranus, I. cobosi and I. rositai, all of which are also characterized by the presence of conspicuous shell ornamentation. These three keeled shell forms have been well differentiated by taxonomists as separate taxa, based on their very different size, shell thickness and/or sculpturing. Nevertheless, several authors have also considered these three species as the most closely related of the genus (Ibáñez and Alonso 1978). Our molecular analysis clearly indicates that the keeled, flattened and markedly ornamented shell of these three taxa is a homoplasic combination of characters that has evolved repeatedly at different times and in different areas and lineages. In effect, in Iberus this has occurred at least three times independently. Helix gualinoi Michaud is a fossil form from the Miocene (Royo-Gómez 1922) that probably belongs to the genus Iberus, indicating that similar keeled-flat and heavily ornamented shells could have arisen at other times in the past within this genus.
Both options, the depressed- and globular-derived hypotheses, have been described in terrestrial snails as two evolutionary shell form tendencies. The former situation was reported by Teshima et al. (2003) for Ainohelix editha, and the second for Arianta arbustorum (Gittenberger et al. 2004). Elejalde et al. (2005) indicated that the presence of a keeled, flattened shell with prominent ornamentation is synapomorphic in I. g. gualtieranus. Thus, the depressed-derived theory is also the condition in this subspecies. Both hypotheses are equally parsimonious for I. cobosi, which appeared as the sister group of the rounded forms within clade 5. Nevertheless, we think that the depressed-derived hypothesis is the most plausible for the origin of I. cobosi, owing to the absence of keeled-shelled forms in the other basal phylogroups (clades 1–4) of medium-sized snails. The lack of genetic differentiation between rounded and keeled forms in I. rositai s. l. determines that keeled-shell snails should be regarded here as a local and recent adaptation, emerging also from the rounded shells of this species. The shell forms of I. g. gualtieranus and I. cobosi are genetically inherited (Elejalde et al. 2005; present work). The question remains, however, as to whether shell form in I. rositai is also genetically determined or is the result of intense developmental plasticity, with shells becoming rounded or keeled in response to environmental conditions and selection pressures. This question needs to be addressed in field surveys.
One of the main dilemmas regarding the evolution of keeled-flat shelled snails is trying to decipher the mechanisms that have originated these extraordinary forms. Habitat preferences and shell morphology are clearly linked in Iberus, with the keeled-shelled snails always living in arid or semi-arid environments in karstic mountains. These snails find shelter within big and deep vertical crevices protected from the sun and wind during dry weather and periods of starvation (Alonso and Ibáñez 1978; Moreno-Rueda 2006). The keeled-flat snails of I. rositai inhabit the ‘Torcal Alto’ (highest part) of the ‘Torcal de Antequera’ mountain, a bare stone karstic area with large deep crevices, while rounded shelled snails of the same species prefer adjacent areas (Alonso and Ibáñez 1978) with larger amounts of sedimentary material on the ground. The same occurs with I. cobosi, which is restricted to the stone karstic hills of the ‘Valle de Abdalajís’, while I. marmoratus inhabits lower lying areas of this region (Ibáñez and Alonso 1978; A. Ruiz, personal communication). Similarly, karstic mountains are home to the keeled-flat I. g. gualtieranus, while the phylogenetically close rounded forms live in surrounding clay areas where they can bury themselves for protection. Land snails are very susceptible to dehydration especially in an environment as arid as the South Iberian Peninsula. According with López-Alcántara et al. (1983, 1985) and Moreno-Rueda (2002, 2007), flattened shells in Iberus are considered an adaptive morphological trait for easier access to rock crevices where the snails are protected against dehydration and predators when inactive.
Conclusions
Our phylogenetic analysis indicates that the five basal clades identified represent five divergent, allopatric lineages with no known intergradation zone. Our interpretation of the available evidence is that the medium-sized snails of the genus Iberus constitute at least five species, four of which should be designated I. guiraoanus, I. angustatus, I. ortizi and I. marmoratus. The fifth species (clade 2) needs a detailed morphological study for its description as a new taxon. There is some evidence of gene flow among the basal lineages of clade 5, with haplotypes of I. cobosi appearing in the I. marmoratus subclade. As far as we know, I. cobosi, I. loxanus s. str. and I. rositai s. l. represent monophyletic groups, but their taxonomic status (species or subspecies of I. marmoratus) remains to be stated. For the time being, we suggest to maintain their specific nomenclature.
The keeled, flattened and strongly ornamented shells of Iberus have arisen three times independently. This shell form is genetically inherited in I. g. gualtieranus and I. cobosi, but more studies are needed to know if it is determined by the environment or regulated by genes in I. rositai. The depressed-derived hypothesis is considered the most valid to explain the repeated parallel development of the keeled-flat shells of this genus.
Acknowledgements
We thank A. Martínez-Ortí and especially A. Cárcaba and A. Ruiz for their field collection. M.A. Elejalde was sponsored by a predoctoral fellowship of the University of the Basque Country (UPV-EHU). This work received financial support from projects of the University of the Basque Country (1/UPV 00076.125-E-13713/2001) and of the Spanish Ministerio de Ciencia y Tecnología (REN2002-00716). This research is part of the project entitled ‘Programme for Conservation and Sustainable Snail Exploitation in Andalusia’ supported by the Consejería de Medio Ambiente de la Junta de Andalucía.




