Avian higher-level phylogeny: well-supported clades and what we can learn from a phylogenetic analysis of 2954 morphological characters
Die Großgruppensystematik der Vögel: gut begründete Monophyla und was wir aus einer phylogenetischen Analyse von 2954 morphologischen Merkmalen lernen können
Abstract
enIt has been shown that increased character sampling betters the accuracy of phylogenetic reconstructions in the case of molecular data. A recently published analysis of avian higher-level phylogenetics based on 2954 morphological characters now provides an empirical example to test whether this is also true in the case of morphological characters. Several clades are discussed which are supported by multiple analyses of mutually independent molecular data (sequences of nuclear genes on different chromosomes and mitochondrial genes) as well as morphological apomorphies, but did not result from parsimony analysis of the large morphological data set. Incorrect character scorings in that analysis notwithstanding, it is concluded that in the case of morphological data, increased character sampling does not necessarily better the accuracy of a phylogenetic reconstruction. Because morphological characters usually have a strongly varying complexity, many simple and homoplastic characters may overrule fewer ones of greater phylogenetic significance in large data sets, thus producing a low ratio of phylogenetic signal to ‘noise’ in the data.
Zusammenfassung
deIm Fall molekularer Daten verbessert eine Berücksichtigung von mehr Merkmalen die Genauigkeit phylogenetischer Rekonstruktionen. Eine kürzlich publizierte, auf 2954 morphologischen Merkmalen basierende phylogenetische Analyse der Großgruppensystematik der Vögel stellt nun ein empirisches Beispiel dar, um zu testen, ob dies auch im Fall von morphologischen Merkmalen zutreffend ist. Einige Monophyla werden besprochen, die durch zahlreiche Analysen voneinander unabhängiger molekularer Daten (Kerngensequenzen auf unterschiedlichen Chromosomen und mitochondrielle Gene) sowie morphologischer Apomorphien gestützt werden, aber nicht aus der Parsimonie-Analyse des umfangreichen morphologischen Datensatzes resultierten. Ungeachtet zahlreicher falscher Merkmalskodierungen in jener Studie, wird daraus gefolgert, dass im Fall morphologischer Daten die Berücksichtigung von mehr Merkmalen nicht notwendigerweise die Genauigkeit einer phylogenetischen Rekonstruktion verbessert. Weil morphologische Merkmale üblicherweise eine stark unterschiedliche Komplexität haben, können in großen Datensätzen viele einfache und homoplastische Merkmale wenige von größerer phylogenetischer Bedeutung überlagern und dadurch ein niedriges Verhältnis von phylogenetischem Signal zu “Rauschen” in den Daten produzieren.
Introduction
Recent molecular studies of avian higher-level phylogeny agree in several clades which depart from traditional classifications (Cracraft et al. 2004; Fain and Houde 2004; Ericson et al. 2006). In particular, the Bayesian analysis of Ericson et al. (2006), based on sequences of five nuclear gene loci on different chromosomes, resulted in a well-resolved phylogeny with high posterior probability values for many clades in the analysis of the combined data, and a high level of congruence concerning some clades in the separate gene trees (Ericson et al. 2006: electronic supplementary material). This study not only supports novel groupings which were proposed in the last years from analyses of morphological and/or molecular data, but also provides evidence for several previously undetected clades.
The data set of Ericson et al.’s (2006) analysis includes 4408 base pairs of aligned nuclear sequences for 91 avian taxa. Until recently, no morphological character matrix of comparable size has been analysed. However, this situation has now changed with publication of a phylogeny based on 2954 morphological characters for 150 neornithine taxa (Livezey and Zusi 2006, 2007). Livezey and Zusi (2006: 440) noted that the ‘richness of characters (…) is significantly higher than any previous qualitative characterization of a group within Tetrapoda, let alone any limited to Aves’. This certainly is true, but to which extent do the results of this analysis coincide with those of Ericson et al. (2006), which appeared too late to be considered by Livezey and Zusi (2007), and which of the conflicting results better reflect the true phylogeny?
Several clades were recovered in the analyses of both Ericson et al. (2006) and Livezey and Zusi (2007) (Fig. 1), but most of these are uncontroversial and meanwhile generally accepted, such as Galloanseres [Galliformes (landfowl) + Anseriformes (waterfowl)], Neoaves (all neognathous birds except Galloanseres) and Suloidea [Sulidae (gannets and boobies) + Phalacrocoracidae (cormorants) + Anhingidae (anhingas)].
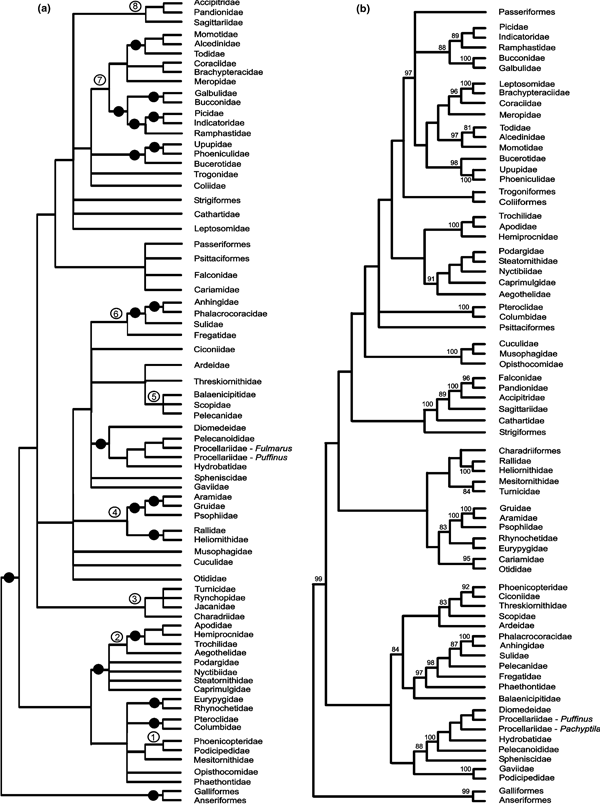
Phylogeny of neognathous birds resulting from (a) a Bayesian analysis of five nuclear genes (β-fibrinogen, c-myc, RAG-1, myoglobin and ornithine decarboxylase; after Ericson et al. 2006: fig. 1), and (b) a parsimony analysis of 2954 morphological characters (simplified after Livezey and Zusi 2007: figs 13–18). Nodes which received posterior probability below 95% are collapsed in (a), and only bootstrap support values above 80% are given in (b). The black dots in (a) indicate nodes that were also recovered in the analysis of Livezey and Zusi (2007); the numbered nodes in (a) are discussed in the text and were also retained with parsimony jackknifing (Ericson et al. 2006: fig. ESM-8)
Some of the controversial nodes received high support values in both analyses. For example, sister group relationship between Podicipedidae (grebes) and Phoenicopteridae (flamingos) was supported with a Bayesian posterior probability above 95% in the analysis by Ericson et al. (2006), whereas Livezey and Zusi (2007) recovered the clades [Podicipedidae + Gaviidae (loons)] and [Phoenicopteridae + Ciconiidae (storks)] with bootstrap values of 100% and 92%, respectively (Fig. 1).
In the case of molecular data, it has been shown that increased character sampling, i.e. adding of sequence data, betters the accuracy of phylogenetic reconstructions (e.g. Poe and Swofford 1999; Braun and Kimball 2002; Simmons and Miya 2004). Concerning morphological data, however, Scotland et al. (2003: 539) argued that ‘rigorous and critical anatomical studies of fewer morphological characters, in the context of molecular phylogenies, is a more fruitful approach’ and ‘preferable to compiling larger data matrices of increasingly ambiguous and problematic morphological characters’. Livezey and Zusi's (2007) study now provides an empirical example to evaluate whether an increased character sampling also leads to more accurate phylogenetic reconstructions in the case of morphological characters.
Except for laboratory strains, it is of course not possible to know the phylogenetic history of any group of organisms with absolute certainty. We can only assess the likelihood of phylogenetic hypotheses and, as noted by Miyamoto and Fitch (1995: 64), ‘[t]rees of natural taxa, well supported by many independent lines of evidence, should be used […] as standards for evaluating the accuracy of different phylogenetic methods’.
Several higher-level clades of birds are now recovered by independent analyses of mitochondrial sequences and/or nuclear sequences from gene loci on different chromosomes. As detailed below, many of these clades, which for the first time are summarized in the present study, can also be supported with morphological apomorphies. Concerning the molecular evidence, identification of these well-supported clades is straightforward, as there are no alternative phylogenies with a congruent support from independent molecular data.
A detailed review of Livezey and Zusi (2007) is beyond the scope of this study. However, it should be noted that, in addition to many misspellings of scientific avian names both in the figures and the text1, numerous misquotations2 and erroneous statements concerning fossil taxa3, the study is flawed by incorrect4 or very doubtful5 character scorings, and unacceptable generalizations on character distribution based on unverified assumptions6. The results of the phylogenetic analysis are further presented in an astonishingly uncritical way and form the basis of a curious classification, in which even paraphyletic taxa not recovered in the analysis are named [e.g. Archaeopteryx and Confuciusornis are united in the ‘Superorder Archaeornithes’, all Mesozoic birds grouped into the ‘Parvclass Palaeoaves’, and Megapodiidae and Cracidae classified into the ‘Suborder Craci’; note that Livezey and Zusi (2007: 88) also incorrectly state that the International Code of Zoological Nomenclature does not cover suprageneric taxa, as article 35 regulates naming of ‘family-group nominal taxa’].
Well-supported clades recovered in sequence analyses of different gene loci and their morphological apomorphies
[Phoenicopteridae + Podicipedidae] (Fig. 1a: node 1)
Molecular support
(1) Combined sequences of c-mos proto-oncogene exon, G3PDH intron 11, 12S rRNA, tRNAVal, 16S rRNA (van Tuinen et al. 2001); (2) ZENK (Chubb 2004); (3) c-myc (Cracraft et al. 2004: fig. 27.4); (4) combined sequences of c-myc and RAG-2 exon (Cracraft et al. 2004: fig. 27.8); (5) RAG-2 exon and mitochondrial sequence data (Cracraft et al. 2004: fig. 27.6); (6) RAG-1 exon (Ericson et al. 2006: fig. ESM-2); (7) myoglobin intron 2 (Ericson et al. 2006: fig. ESM-3).
Morphological apomorphies
(1) Eleven (instead of ten) primaries (Mayr 2004); (2) eggshell covered with chalky layer of amorphous calcium phosphate (Mayr 2004); (3) at least 23 praesacral vertebrae (Mayr 2004); (4) prominent caudolateral projections on ventral side of cervical vertebrae (processus ventrolaterales) (Manegold 2006); (5) at least four thoracic vertebrae fused to a notarium; (6) phalanx proximalis digiti majoris elongate and craniocaudally narrow (Mayr 2004); (7) ungual phalanges nail-like (Manegold 2006).
Comments
Molecular evidence for sister group relationship between Phoenicopteriformes and Podicipediformes comes from analyses of both nuclear and mitochondrial gene sequences. However, and as noted above, Livezey and Zusi's (2007) analysis resulted in sister group relationship between Podicipediformes and Gaviiformes (loons) and Phoenicopteriformes and Ciconiidae (storks), with high bootstrap support for each of these groupings (Fig. 1b). These findings correspond with the traditional classification of birds (e.g. Wetmore 1960), but are not supported by any molecular study. Eleven of the 17 characters ‘diagnostic or highly supportive’ of a sister group relationship between loons and grebes are from the pelvic girdle (Livezey and Zusi 2007: 47). Livezey and Zusi (2007: 26) correctly anticipated that sister group relationship between Podicipediformes and Gaviiformes will ‘engender concerns of artefactual pairing by convergence’ of these foot-propelled divers, whereas the very different way of living of Phoenicopteriformes and Podicipediformes makes it difficult to explain by convergence the shared derived similarities of these birds (Mayr 2004, in press).
Aegothelidae + [Trochilidae + (Apodidae + Hemiprocnidae)] (Fig. 1a: node 2)
Molecular support
(1) Combined sequences of c-myc, RAG-1 exon and myoglobin intron 2 (Mayr et al. 2003); (2) combined sequences of c-myc and RAG-2 exon (Cracraft et al. 2004: fig. 27.8); (3) β-fibrinogen intron 7 (Ericson et al. 2006: fig. ESM-4); (4) c-myc (Cracraft et al. 2004: fig. 27.4, Ericson et al. 2006: fig. ESM-1); (5) RAG-1 exon (Ericson et al. 2006: fig. ESM-2, Barrowclough et al. 2006); (6) myoglobin intron 2 (Ericson et al. 2006: fig. ESM-3). Owlet nightjars and apodiform birds share a duplication of 12 base pairs in the c-myc gene (Mayr et al. 2003: 234) and a ‘15 base synapomorphy’ in the RAG-1 gene (Barrowclough et al. 2006: 240).
Morphological apomorphies
(1) Os palatinum with strongly protruding angulus caudolateralis (Mayr 2002); (2) processus basipterygoidei reduced (Mayr 2002); (3) quadratum, presence of pneumatic foramina on caudal surface of processus oticus (Mayr 2002); (4) coracoid, extremitas omalis hooked and processus lateralis greatly reduced (Mayr 2002); (5) sternum, incisions in caudal margin closed or completely reduced (Mayr 2005a: fig. 5); (6) cruciform origin of musculus splenius capitis (Mayr 2002); (7) absence of caeca (Mayr 2002).
Comments
Sister group relationship between Aegothelidae (owlet-nightjars) and apodiform birds [Trochilidae (hummingbirds), Apodidae (swifts) and Hemiprocnidae (tree swifts)] was first proposed by study of morphological characters (Mayr 2002), and resulted from all subsequent molecular analyses including these taxa. By contrast, Livezey and Zusi (2007) listed five diagnostic apomorphies in order to support monophyly of the traditional ‘Caprimulgiformes’ [i.e. a clade including owlet-nightjars, Podargidae (frogmouths), Steatornithidae (oilbird), Nyctibiidae (potoos) and Caprimulgidae (nightjars)]. I only checked two of these: character 280 (Livezey and Zusi 2006: 67) concerns beak morphology which in all ‘Caprimulgiformes’ was considered of ‘(1) distinctly triangular dorsoventral and lateromedial form, (2) dorsoventral compression, (3) variably prominent medial carina, (4) short but strong terminal hamulus, (5) mediocaudal portion composed of triangular-shaped os maxillare and (6) arcus jugalis with variable, largely lateral orientation’. It is incomprehensible, why the beak of Podargidae and Steatornithidae was assigned the same character state as that of Caprimulgidae and Aegothelidae, whereas the beaks of Apodidae and Hemiprocnidae, which are extremely similar to that of owlet-nightjars in all of the above features, were not (see Mayr 2002: fig. 3). Character 2921 (Livezey and Zusi 2006: 437) refers to the alleged presence of a tapetum lucidum, which was coded as present in all ‘caprimulgiform’ birds and absent in all other taxa included in the study. Livezey and Zusi (2006): 437) list a reference in which this character was described for Caprimulgidae and stated that it was ‘extended to Caprimulgiformes by Holyoak (2001)’. However, the only statement about this character in Holyoak (2001: 11) is that ‘[t]apeta lucida may be unique to Caprimulgiformes’, and the character scoring in Livezey and Zusi (2006) per contra, there exists no evidence that a tapetum lucidum is present in ‘caprimulgiform’ birds other than nightjars and, perhaps, the oilbird (Thomas 1999: 247). Clearly, Livezey and Zusi's (2006) scoring of this character as present in all ‘caprimulgiform’ birds is thus an unacceptable generalization [see also Evans and Martin 1993: 598 who noted that a tapetum lucidum has been ‘recorded in birds only in the retina of goatsuckers (Caprimulgidae)’].
Livezey and Zusi's (2007) analysis further resulted in sister group relationship between Apodidae and Trochilidae, a result which conflicts with virtually all traditional hypotheses on the interrelationship of apodiform birds (see Sibley and Ahlquist 1990) and all molecular studies which consistently support sister group relationship between Hemiprocnidae and Apodidae (e.g. Cracraft et al. 2004; Ericson et al. 2006).
Position of Turnicidae within Charadriiformes (Fig. 1a: node 3)
Molecular support
(1) RAG-1 (Paton et al. 2003); (2) combined sequences of c-myc and RAG-2 exon (Cracraft et al. 2004: fig. 27.8); (3) combined sequences of 14 mitochondrial genes (Paton and Baker 2006); (4) β-fibrinogen intron 7 (Fain and Houde 2004; Ericson et al. 2006: fig. ESM-4); (5) myoglobin intron 2 (Ericson et al. 2006: fig. ESM-3).
Morphological apomorphies
(1) Coracoid, extremitas sternalis forming three pointed projections (e.g. Mayr 2000a: fig. 1); (2) humerus without pneumatic foramina in fossa pneumotricipitalis; (3) os carpi ulnare with tubercle at insertion area of ligamentum humerocarpale (Ericson 1997: character 63); (4) fourth phalanx of fourth toe shorter than third phalanx (Hesse 1988: pl. 1).
Comments
A position of buttonquails within charadriiform birds is supported by analyses of both nuclear and mitochondrial genes, and is in concordance with the charadriiform overall morphology of fossil stem group representatives of the Turnicidae (Mayr 2000a; Mayr and Knopf in press).
Buttonquails resulted as sister taxon of the Mesitornithidae (mesites) in the analysis of Livezey and Zusi (2007), a grouping already proposed by Fürbringer (1888: pls. 27 and 28). Livezey and Zusi (2007) did not list apomorphies of this clade and failed to mention any evidence for charadriiform affinities of the Turnicidae, despite excessive citation of less relevant literature in many other instances.
[(Rallidae + Heliornithidae) + (Gruidae + Aramidae) +Psophiidae] (Fig. 1a: node 4)
Molecular support
(1) RAG-2 exon (Cracraft et al. 2004: fig. 27.7); (2) β-fibrinogen intron 7 (Fain and Houde 2004; Ericson et al. 2006: fig. ESM-4); (3) c-myc exon 3 (Ericson et al. 2006: fig. ESM-1); (4) RAG-1 exon (Ericson et al. 2006: fig. ESM-2); (5) myoglobin intron 2 (Ericson et al. 2006: fig. ESM-3); (6) ornithine decarboxylase (Ericson et al. 2006: fig. ESM-5).
Morphological apomorphies
(1) Caudal end of mandible with narrow, dorsally projecting hook-like projection (except Psophiidae, very weakly developed in Heliornithidae); (2) praefrontale with narrow, caudally projecting processus supraorbitalis (except Psophiidae in which the corresponding area shows an apomorphic morphology owing to the formation of small supraorbital ossicles); (3) pterygoid, rostral end markedly widened (e.g. Beddard 1898: fig. 167); (4) sternum elongate and very narrow (except Heliornithidae); (5) pelvis, cristae iliacae dorsales fused over entire length with crista spinosa of synsacrum, thus forming a completely closed canalis iliosynsacralis [except Heliornithidae and Fulicinae (Rallidae)]; (6) pelvis, recessus caudalis fossae deeply excavated.
Comments
Based on study of morphological characters, a clade including Gruidae (cranes), Aramidae (limpkin), Psophiidae (trumpeters) and Rallidae (rails) has been proposed by various earlier authors (e.g. Wetmore 1960; Hesse 1990) and the clade {[Rallidae + Heliornithidae (sungrebes)] + [(Gruidae + Aramidae) + Psophiidae)]} was recovered in an analysis of morphological data by Livezey (1998). By contrast, Livezey and Zusi's (2007) analysis supported a clade including all taxa traditionally assigned to ‘Gruiformes’, with the exception of Mesitornithidae (mesites) and (Rallidae + Heliornithidae) (the latter clade was recovered as sister taxon of Charadriiformes). Livezey and Zusi listed a single diagnostic apomorphy of this clade, i.e. ‘[e]xtremitas proximalis tibiotarsi, caput tibiotarsi, facies (gastrocnemialis) medialis, crista (interna) medialis (…) markedly textured by impressiones, jugae, et concavitas subcristalis’ (Livezey and Zusi 2006: 329; 2007: tab. 2, character 2111), which, however, occurs in a variety of taxa with enlarged cnemial crests [e.g. the charadriiform Laridae (gulls)].
(Pelecanidae + Balaenicipitidae + Scopidae) (Fig. 1a: node 5)
Molecular support
(1) Combined sequences of 12S rRNA and16S rRNA (Scopidae not included; Hedges and Sibley 1994); (2) combined sequences of 12S rRNA,16S rRNA and cytochrome B (Scopidae not included; Siegel-Causey 1997); (3) combined sequences of c-mos proto-oncogene exon, G3PDH intron 11, 12S rRNA, tRNAVal, 16S rRNA (van Tuinen et al. 2001); (4) c-myc (Scopidae not included; Cracraft et al. 2004: fig. 27.4); (5) combined sequences of RAG-2 exon and mitochondrial sequence data (Cracraft et al. 2004: fig. 27.6); (6) RAG-2 exon (Cracraft et al. 2004: fig. 27.7); (7) β-fibrinogen intron 7 (Fain and Houde 2004); (8) RAG-1 exon (Ericson et al. 2006: fig. ESM-2); (9) myoglobin intron 2 (Ericson et al. 2006: fig. ESM-3); (10) ornithine decarboxylase (Ericson et al. 2006: fig. ESM-5).
Morphological apomorphies
Pelecanidae and Balaenicipitidae share several derived and perhaps synapomorphic features (e.g. coracoid with foramen nervi supracoracoidei, furcula fused with apex carinae of sternum; Cottam 1957; Mayr 2003), and there is also a number of derived features which may unite Pelecanidae, Scopidae, Balaenicipitidae and the traditional ‘Pelecaniformes’ except Phaethontidae (e.g. eggshell covered with layer of amorphous calcium carbonate, furcula, extremitas omalis with strongly developed, laterally protruding facies articularis acrocoracoidea; Mayr 2003, 2005b). However, only two characters could be identified which are exclusively shared by Pelecanidae, Scopidae and Balaenicipitidae, and none is unique to these taxa: (1) os palatinum, pars choanalis very deep in dorsoventral direction, ossa pterygoidea very short; (2) musculus ambiens extremely vestigial or absent (McKitrick 1991; Mayr 2003).
Comments
See clade below.
Fregatidae + [Sulidae + (Phalacrocoracidae + Anhingidae)] (Fig. 1a: node 6)
Molecular support
(1) Combined sequences of 12S rRNA, 16S rRNA and cytochrome B (Siegel-Causey 1997); (2) combined sequences of RAG-2 exon and mitochondrial sequence data (Cracraft et al. 2004: fig. 27.6); (3) c-myc (Cracraft et al. 2004: fig. 27.4); (4) β-fibrinogen intron 7 (Fain and Houde 2004; Ericson et al. 2006: fig. ESM-4).
Morphological apomorphies
(1) Palatinum with well-developed angulus caudolateralis (e.g. Mickoleit 2004: fig. 427); (2) recessus tympanicus dorsalis greatly enlarged and situated rostrally to the articular facets of the quadrate (except Phalacrocoracidae and Anhingidae); (3) tarsometatarsus greatly abbreviated, measuring about half the length of the carpometacarpus or less (except Phalacrocoracidae); (4) trochlea metatarsi II protruding farther distally than trochlea metatarsi III; (5) claw of third toe distinctly pectinate on its medial side (e.g. Mayr 2003).
Comments
Molecular analyses of both nuclear and mitochondrial sequences consistently support polyphyly of the traditional ‘Pelecaniformes’. Although Livezey and Zusi (2007: tab. 2) listed four characters as diagnostic apomorphies of the traditional ‘Pelecaniformes’ [including Phaethontidae (tropicbirds) but excluding Balaenicipitidae (shoebill)]. Of these, character 335 (presence of a ‘microapertura nasi ossea’; Livezey and Zusi 2006: 75) is absent in Phaethontidae but present in Balaenicipitidae (contra Livezey and Zusi 2006; see Mayr 2003: appendix II, character 2 – note also that this reference has been misquoted by Livezey and Zusi 2006: 75 who referred to both, a wrong reference and character number). Character 1832 (presence of ‘bilaterally symmetrical spinae emerging perpendicularly and dorsal to processes [sic] transverses [sic] synsacri’; Livezey and Zusi 2006: 289) was considered non-comparable for Pelecanidae by Livezey and Zusi (2006: 289).
Livezey and Zusi's (2007) study and most other analyses of morphological data result in sister group relationship between Pelecanidae (pelicans) and Suloidea (e.g. Cracraft 1985; Mayr 2003; see, however, Mayr 2005b). By contrast, molecular analyses identify Fregatidae (frigatebirds) as the sister group of Suloidea and recover a clade including Pelecanidae and the ‘ciconiiform’ Scopidae (hamerkop) and Balaenicipitidae. Although there are only few morphological apomorphies which support the molecular results, a close relationship between pelicans and the shoebill was first proposed from study of morphological data (Cottam 1957). The fact that this hypothesis is congruent with all molecular analyses including Pelecanidae and Balaenicipitidae strongly suggests that the traditional ‘Pelecaniformes’ are not monophyletic (contra Livezey and Zusi 2007), and that the derived characters shared by Pelecanidae, Fregatidae and Suloidea are homoplastic (e.g. the totipalmate feet, the gular pouch and the strongly developed acromion of the scapula).
[Piciformes + (Coraciiformes sensu stricto + Alcediniformes)] (Fig. 1a: node 7)
Molecular support
(1) Combined sequences of c-myc, RAG-1 and myoglobin intron 2 (Mayr et al. 2003); (2) β-fibrinogen intron 7 (Fain and Houde 2004; Ericson et al. 2006: fig. ESM-4); (3) myoglobin intron 2 (Ericson et al. 2006: fig. ESM-3); (4) c-myc (Cracraft et al. 2004: fig. 27.4); (5) ornithine decarboxylase (only Ramphastidae included; Ericson et al. 2006: fig. ESM-5).
Morphological apomorphies
Piciformes, Coraciiformes sensu stricto and Alcediniformes share four deep incisions in the caudal margin of sternum, but the polarity of this feature is uncertain, and the above clade cannot be convincingly supported with derived morphological characters. There are, however, derived characters which support a more inclusive clade also comprising Bucerotes: (1) the mandible of the hatchling distinctly projects beyond the upper beak (Manegold 2005); (2) the fossa parahypotarsalis medialis (tarsometatarsus) is very marked and the proximal part of the margo medialis forms a sharp ridge (Mayr et al. 2003; Manegold 2005); (3) the greater ventral coverts of the secondaries (tectrices secundariae ventrales majores) are reduced (Manegold 2005; this character is also present in passeriform birds).
Comments
In contrast to virtually all recent analyses of molecular and morphological data, Livezey and Zusi's (2007) study supported monophyly of the traditional ‘Coraciiformes’, i.e. a clade including Bucerotes [Upupiformes (hoopoes and wood-hoopoes) and Bucerotiformes (hornbills)], Coraciiformes sensu stricto [Coraciidae (rollers) and Brachypteraciidae (ground rollers)], Leptosomidae (cuckoo-roller) and Alcediniformes [Meropidae (bee-eaters), Todidae (todies), Momotidae (motmots) and Alcedinidae (kingfishers)].
Livezey and Zusi (2007) listed a single apomorphy to support this clade, i.e. ‘[e]xtremitas distalis tarsometatarsi, trochlea metatarsale [sic] II, prominent distomedial extent (…), defining by linear prominence a distinct, asymmetrical angulus with margo medialis tarsometatarsi’ (Livezey and Zusi 2006: 366, character 2360). Clearly, this character has been incorrectly coded: on the one hand, it does not distinguish the very similar trochleae metatarsi II of Coraciidae and, for example, Podargidae, whereas on the other hand the shape of the trochlea metatarsi II is extremely variable within ‘coraciiform’ birds, being very narrow in, for example Todidae where it forms hardly any angle with the medial margin of the tarsometatarsus (e.g. Mayr 1998: fig. 20).
Certainly one of the most disturbing results of Livezey and Zusi's (2007) analysis is a 100% bootstrap support for sister group relationship between the Madagascan Leptosomidae and Brachypteraciidae, given the fact that Leptosomidae are utterly different from rollers in their morphology. Leptosomus discolor has been assigned to cuckoos (Cuculidae) in the 18th and early 19th century, until Sclater (1865) carried out the first detailed anatomical studies and considered it to be most closely related to rollers. Almost all subsequent authors followed this classification, but no derived characters have been presented which convincingly support monophyly of a clade including Leptosomidae, Coraciidae and Brachypteraciidae. Sibley and Ahlquist (1990: fig. 359) showed Leptosomus as sister taxon of rollers in the summary tree of their DNA–DNA hybridization study, but such a position is not supported by their figure 70 where the melting curve of Leptosomus is widely separated from that of Eurystomus (Coraciidae) (Sibley and Ahlquist 1990: 350 note that the ‘position of Leptosomus compared with that of Eurystomus is interesting, but its significance is unknown’). Leptosomus was not included in the phylogenetic analyses of Cracraft et al. (2004); nevertheless these authors also followed tradition and considered it to be the sister taxon of Coraciidae and Brachypteraciidae in their summary tree (Cracraft et al. 2004: fig. 27.10). Coraciiform affinities of Leptosomus were doubted by Herremans and Louette (1992) and Mayr (1998), and were not supported by subsequent cladistic analyses of morphological data by Mayr et al. (2003); Mayr (2005c, 2006 and Manegold (2005). I have checked the scoring of two of the most conspicuous characters which separate Leptosomidae and Brachypteraciidae, and both were incorrectly coded by Livezey and Zusi (2006) for Leptosomidae: character 2334, a trochlea accessoria on the trochlea metatarsi IV, is coded as absent despite being one of the most distinct features of the cuckoo-roller (e.g. Cracraft 1971: fig. 13), and likewise a foramen nervi supracoracoidei (character 1286) is coded as absent, although Leptosomidae are among the few ‘higher landbirds’ which actually possess such a foramen (e.g. Cracraft 1971: fig. 9). There are numerous other incorrect character scorings for Leptosomus in the character matrix (i.e. many characters are incorrectly assigned the same state as in Brachypteraciidae), and either the voucher specimen of Leptosomus discolor used by Livezey and Zusi (2006) does not belong to that species but is a ground roller, or there has been a profound mix-up of data.
[Sagittariidae + (Pandionidae + Accipitridae)] (Fig. 1a: node 8)
Molecular support
(1) Combined sequences of c-myc and RAG-2 exon (Cracraft et al. 2004: fig. 27.8); (2) β-fibrinogen intron 7 (Fain and Houde 2004; Ericson et al. 2006: fig. ESM-4); (3) RAG-1 exon (Ericson et al. 2006: fig. ESM-2); (4) myoglobin intron 2 (Ericson et al. 2006: fig. ESM-3); (5) ornithine decarboxylase (Ericson et al. 2006: fig. ESM-5).
Morphological apomorphies
(1) Syrinx morphology (see Griffiths 1994); (2) absence of musculus plantaris (‘F’ muscle in the formula of George and Berger 1966: Tab. IX.1; see George and Berger 1966: 442).
Comments
The above clade also resulted from the DNA–DNA hybridization studies of Sibley and Ahlquist (1990), whereas Livezey and Zusi's (2007) analysis supported a clade {Accipitridae [hawks and allies] + [Falconidae (falcons) + Pandionidae (osprey)]}. Although a clade including these three taxa corresponds with traditional classifications (e.g. Wetmore 1960), sister group relationship between falcons and the osprey has never been proposed, despite the high bootstrap value for this grouping in Livezey and Zusi's (2007) phylogeny (Fig. 1b). The authors did not discuss this curious clade, nor did they list any apomorphies of it. Instead, and with any further justification, they stated that Fain and Houde (2004) who recovered the clade advocated in the present study ‘failed to resolve relationships among the diurnal raptors’ (Livezey and Zusi 2007: 35).
Discussion
None of the well-supported clades discussed above resulted from the analysis of Livezey and Zusi (2007). However, to cite this study as evidence for the failure of morphological data to resolve avian higher-level phylogenetics would be a distortion of facts.
Although there is a widespread belief among molecular systematists that morphological data are inferior to molecular ones regarding the reconstruction of phylogenetic relationships (e.g. van Tuinen 2002; Scotland et al. 2003), this assumption certainly is wrong since several of the clades discussed above were originally proposed from morphological studies (see also Jenner 2004; Wiens 2004). Very likely, however, systematists using morphological data are more often misled by homoplastic characters than students of molecular sequences.
Incorrect character scorings notwithstanding, Livezey and Zusi's study further exemplifies severe shortcomings of analyses of large sets of morphological characters in recovering accurate phylogenetic relationships. Such numerical cladistic approaches may work well for molecular data, where only four more or less equivalent characters, the nucleotides, are analysed. Morphological characters, however, usually have a strongly varying complexity, and in large data sets many simple and homoplastic characters may overrule fewer ones of greater phylogenetic significance, thus producing a low ratio of phylogenetic signal to ‘noise’ in the data. This is especially problematic in the case of short internodes on which there has been little time for the formation of phylogenetically informative apomorphies. Accordingly, even Livezey and Zusi's (2007) analysis of almost 3000 morphological characters results in a tree in which most of the critical nodes, i.e. those grouping ‘supraordinal’ neoavian taxa, receive very low support values. By contrast, nodes which in all likelihood are incorrect (see above) are strongly supported, not least because taxa which underwent extensive convergent evolution are likely to group together in analyses in which as many characters as possible are included (see also the discussion concerning grebes and loons in Mayr and Clarke 2003: 535).
Phylogenetic hypotheses of extant taxa which are derived from morphological data can be tested with molecular studies. The likelihood that congruent results are independent and reflect the true phylogeny is especially high, if the morphological hypothesis has been proposed before the molecular evidence was available, as in the case of some of the clades discussed in the present study. For fossil taxa, however, morphology usually constitutes the only source of phylogenetic information, which makes an independent assessment of phylogenetic accuracy more difficult.
As exemplified by Livezey and Zusi (2007) analysis, high support values are not necessarily accompanied with phylogenetic accuracy, stressing importance of a critical assessment of the apomorphies supporting conflicting hypotheses. Unfortunately, this has not be done by Livezey and Zusi who did not discuss any character evidence for the proposed phylogeny at all (space limitation certainly could not have been a reason therefore, given a reference list of 31 pages and nine pages with a dispensable classification). Surprisingly, the authors even failed to discuss, or only mention other than as character numbers in a table, the most significant result of their study, i.e. identification of the absence of a phallus and associated features as the first morphological apomorphy of Neoaves (Livezey and Zusi 2007: tab. 2, characters 2502, 2893, 2895, 2896, 2900; see also King 1981).
The well-supported clades discussed in the present study may serve as a framework to test the accuracy of future phylogenetic reconstructions. Further insight into the avian higher-level phylogeny can be gained by comparison of two well-resolved trees resulting from analyses of subsets of the data of Ericson et al. (2006), i.e. the β-fibrinogen sequence and combined sequences of c-myc, RAG-1, myoglobin, and ornithine decarboxylase (Fig. 2). Analysis of the β-fibrinogen gene suggests a basal dichotomy of Neoaves into two clades, termed ‘Metaves’ and ‘Coronaves’ by Fain and Houde (2004), which as yet were, however, only retained in analyses including sequences of this gene (Fig. 2b; Fain and Houde 2004; Ericson et al. 2006).
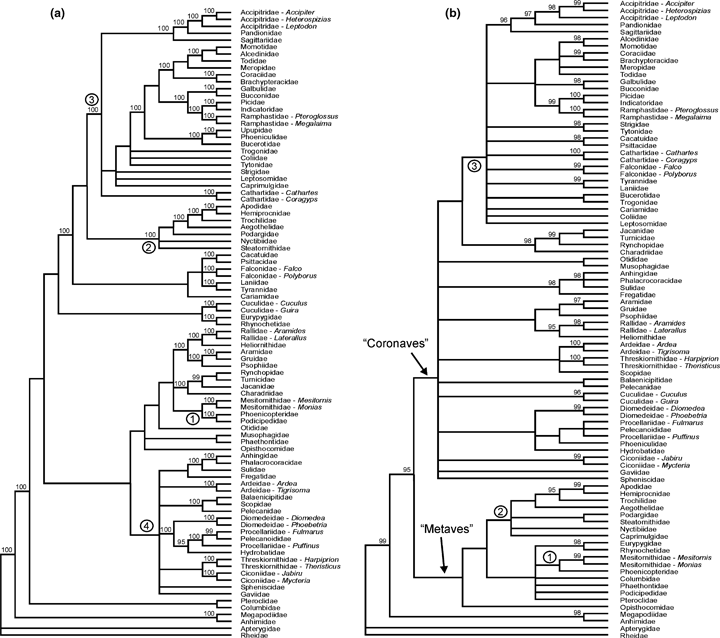
Comparison of two gene trees from the Bayesian analysis of Ericson et al. (2006). (a) Combined sequences of c-myc exon 3, RAG-1 exon, myoglobin intron 2 and ornithine decarboxylase introns 6 and 7 with intercepting exon 7 (after Ericson et al. 2006: fig. ESM-6); (b) β-fibrinogen intron 7 (after Ericson et al. 2006: fig. ESM-4). Nodes which received are posterior probability below 50% are collapsed, and only those with a posterior probability of 95% or more are labelled. The numbered nodes are discussed in the text
Both analyses congruently recover a novel clade comprising Mesitornithidae and Phoenicopteridae (Phoenicopteridae and Podicipedidae in the analysis of the four genes; Fig. 2a,b: node 1), as well as a clade including the paraphyletic ‘Caprimulgiformes’ and apodiform birds (Fig. 2a,b: node 2) which was also retained in the analysis of Livezey and Zusi (2007), albeit, and as detailed above, with different internal relationships (Fig. 1b). Both analyses further support a clade which includes ‘higher’ landbirds (see Mayr et al. 2003), Strigiformes (owls), Falconiformes (diurnal birds of prey), Psittaciformes (parrots) and Cariamidae (seriemas) (Fig. 2a,b: node 3; see also Ericson et al. 2006). However, the position of the ‘Caprimulgiformes’/Apodiformes clade was not unambiguously resolved, as it grouped with higher landbirds in the analysis of the four genes (Fig. 2a) but within ‘Metaves’ in that of the β-fibrinogen sequence (Fig. 2b). Analysis of the combined sequences without β-fibrinogen further resulted in a clade including various aquatic and semi-aquatic taxa (Fig. 2a: node 4), which is also supported by an analysis of whole mitochondrial genome sequences (Gibb et al. 2007). A similar ‘waterbird clade’ was recovered by Livezey and Zusi (2007), but also included Phoenicopteridae and Podicipedidae in the analysis of these authors (Fig. 1b).
Figure 3 depicts a summary cladogram with the well-supported clades discussed above, together with those which are recovered in both gene trees in Fig. 2a,b. Although this tree still contains many polytomies, recent progress in avian phylogenetics motivates some optimism that many of these will be resolved by ongoing molecular analyses (e.g. Harshman et al. 2006), and future studies of morphological characters. In particular, it is to be hoped that molecular systematists intensify their search for congruence among results of independent data sets (e.g. mitochondrial and nuclear sequences), and that morphologists concentrate on the identification and discussion of apomorphies rather than analyses of ever-increasing data sets.
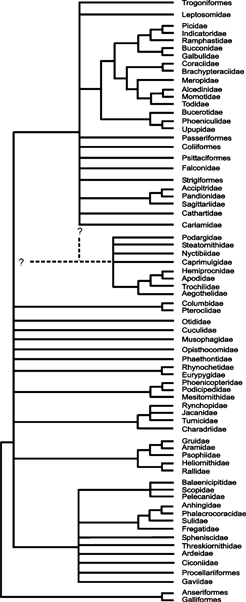
Summary tree for neognathous birds with the well-supported clades discussed in the present study and those which are recovered in both gene trees in Fig. 2a and b
Footnotes
Acknowledgements
I thank two anonymous referees for their comments on the manuscript.




