The cyprinodont fish Rivulus (Aplocheiloidei: Rivulidae) in Trinidad and Tobago: molecular evidence for marine dispersal, genetic isolation and local differentiation
El pez Cyprinodontiforme Rivulus (Aplocheiloidei: Rivulidae) en Trinidad y Tobago: evidencia molecular de dispersión marina, isolamento genético y diferenciación local
Abstract
enMitochondrial DNA sequences (756 bp) were obtained from the cytochrome b gene of 36 Rivulus individuals collected from 10 sites in Trinidad and one site in Tobago. Eight haplotypes were identified. Low genetic divergence (0.5%) between one western Trinidad (Blue Basin) haplotype and Rivulus hartii from north-western Venezuela (Paria peninsula) and high genetic divergence (c. 11%) between these and the remaining other Trinidad and Tobago haplotypes suggests that the islands were colonized by two lineages. The commoner haplotype is distributed throughout lowland Trinidad, possibly a reflection of flooding of the Orinoco River leading to high dispersal between watersheds. Rivulus from higher altitude (Northern Range) localities that would not have been affected by such flooding show high genetic divergence between sites. The genetic differentiation between northern and southern watersheds suggests isolation between some of these drainages.
Resumen
esSecuencias mitocondriales de ADN (756 bp) fueron obtenidas del gen citocroma b de 36 individuos de Rivulus colectados de diez localidades en Trinidad y de una localidad en Tobago. Ocho haplotipos fueron identificados. Baja variabilidad genética (0.5%) entre un haplotipo del oeste de Trinidad (Blue Basin) y R. hartii del noroeste de Venezuela (Peninsula de Paria) y alta divergencia genética (c. 11%) entre estos y el resto de haplotipos provenientes de Trinidad y Tobago sugiere que las islas fueron colonizadas por dos linages. El haplotipo mas común se distribuye por zonas bajas, posiblemente a consecuencia de las inundaciones del rio Orinoco que facilitaron la dispersión de individuos entre rios. Rivulus provenientes de localidades a elevada altitud (Cordillera del Norte) no afectados por estas inundaciones muestran alta divergencia genética entre localidades. La diferencia genética entre rios del sur y del norte de Trinidad sugiere isolamento entre estos sistemas fluviales.
Introduction
The island of Trinidad lies east of the South American mainland (off northern Venezuela), separated by an 11-km-wide strait, while the island of Tobago lies about 36 km further east. It has long been recognized that the rivulidine freshwater fishes of these islands may have come from South America, perhaps when the sea level was higher than it presently is, or by crossing the straits at times of reduced marine salinity when the Equatorial current causes high, wet season discharges from the Amazon and Orinoco rivers to flow past the islands (Kenny 1978; Alkins and De Souza 1984; Boos 1984; Kenny 1995).
The inter-relationships of many South American, Antillean and Caribbean species of Rivulidae have been explored using cyt b, COI, 12S and 16S rDNA mitochondrial sequences (Murphy and Collier 1996; Collier et al. 1998; Murphy et al. 1999). Two phylogenetic studies have shown that Rivulus hartii from Trinidad (Murphy and Collier 1996) and from Paria peninsula, northern Venezuela (Murphy et al. 1999), are closely related to other Rivulidae from Venezuela and Guianas. Collier et al. (1998) included both of these specimens in a wider but modest R. hartii population study. Their results show high genetic divergence between R. hartii from the mainland and Trinidad, suggesting isolation at these localities. There has been no attempt in this or other prior work on these fish in Trinidad and Tobago to assess the level of within-species variation or its possible relationship to geographical and ecological factors. Here, we present new cyt b sequences from 36 Rivulus individuals from 10 separate localities around Trinidad and one locality in Tobago, and analyse them both phylogenetically and in relation to other factors.
Biological background
Rivulus hartii (Boulenger, 1890), the jumping guabine, is a euryhaline fish known from eastern Colombia, the northern coasts of Venezuela, and the Caribbean islands of Trinidad, Tobago, Margarita and Grenada (Boeseman 1960; Robins et al. 1991; Huber 1996). It is the most widespread freshwater fish in Trinidad (Kenny 1995), where it occurs in estuaries, rivers both below and above rapids and waterfalls, in mountain streams, lowland swamps as well as in temporary, shallow rain pools. It grows up to 10 cm in length (Price 1955; Kenny 1995; Bührnheim and Fernandes 2003). Previous studies in Trinidad have shown that the highest population densities of R. hartii are found in habitats that are inaccessible to other fish (Liley and Seghers 1975; Fraser and Gilliam 1992; Gilliam et al. 1993; Fraser et al. 1995, 1999; Reznick et al. 1997; Gilliam and Fraser 2001). In Trinidad, it also is the only fish that can ascend or bypass waterfalls and rapids, and is known to move from rivers to steep rocky mountain streams (Gilliam et al. 1993; Fraser et al. 1995, 1999; Fraser et al. 2001; Gilliam and Fraser 2001). Notably, individuals are capable of jumping out of the water and may survive on, and travel substantial distances across, damp leaf-litter (Seghers 1978), so that many populations are found some distance away from rivers, in nearby pools (Reznick 1982; Costa 1987).
Materials and Methods
Taxon sampling
Rivulus specimens were captured using handnets during July to August 2002 and 2003. Sites sampled are shown in Fig. 1 and listed in Table 1. After collection, they were killed in 0.01% benzocaine and preserved in 95% ethanol. Freshwater fish surveys and reviews (Price 1955; Boeseman 1960; Kenny 1995) have shown that Rivulus hartii is the only rivulidine species present in Trinidad and thus made identification of individuals simple (specimen museum numbers are listed in Table 1). Despite careful search the only other fish found in the sampled localities was the Guppy, Poecilia reticulata. These were present only at Blue Basin (in the river below the waterfall, not in the steep stream where Rivulus was captured), and at Lopinot.
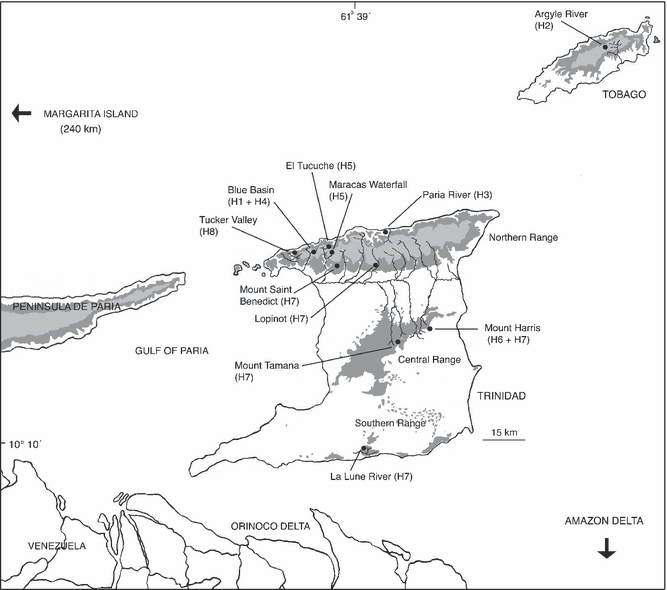
Map representing the locations mentioned in the text and haplotypes. Dark shading: elevations over 25 m; light shading: elevations over 300 m. Black circles: sampled localities
| Location | Coordinates | Altitude | Glasgow numbers (GLAHM) | Field and laboratory numbers | GenBank accessions | Haplotype |
|---|---|---|---|---|---|---|
| Trinidad (Northern Range) | ||||||
| Blue Basin | 61°33′W, 10°44′N | 500 m | 130048 | T538 | AY619607 | H1 |
| 128971 | T461 | AY619606 | H1 | |||
| 130037 | T527 | AY619604 | H1 | |||
| 128972 | T462 | AY619605 | H1 | |||
| 130040 | T530 | AY619621 | H4 | |||
| Tucker Valley | 61°37′W, 10°43′N | Sea level | 130046 | T536 | AY619635 | H8 |
| 130045 | T535 | AY619636 | H8 | |||
| El Tucuche | 61°24′W, 10°44′N | 300 m | 130026 | T516 | AY619628 | H5 |
| 130044 | T534 | AY619627 | H5 | |||
| 130043 | T533 | AY619625 | H5 | |||
| Maracas Waterfall | 61°24′W, 10°43′N | 300 m | 130016 | T506 | AY619631 | H5 |
| 130002 | T492 | AY619630 | H5 | |||
| 128929 | T384 | AY619629 | H5 | |||
| 128965 | T455 | AY619626 | H5 | |||
| Mount Saint Benedict | 61°21′W, 10°39′N | 150 m | 130014 | T504 | AY619608 | H7 |
| 128944 | T399 | AY619615 | H7 | |||
| 128969 | T459 | AY619616 | H7 | |||
| 128947 | T402 | AY619618 | H7 | |||
| Lopinot | 61°19′W, 10°41′N | 150 m | 128968 | T458 | AY619638 | H7 |
| 130039 | T529 | AY619639 | H7 | |||
| 130038 | T528 | AY619623 | H7 | |||
| Paria River | 61°15′W, 10°47′N | Sea level | 130001 | T491 | AY619620 | H3 |
| 50 m | 130047 | T537 | AY619614 | H3 | ||
| 129000 | T490 | AY619617 | H3 | |||
| Trinidad (Central Range) | ||||||
| Mount Tamana | 61°11′W, 10°27′N | 30 m | 128970 | T460 | AY619609 | H7 |
| 128973 | T463 | AY619613 | H7 | |||
| 128999 | T489 | AY619619 | H7 | |||
| Mount Harris | 61°06′W, 10°30′N | 10 m | 130027 | T517 | AY619624 | H6 |
| 130042 | T532 | AY619622 | H7 | |||
| 130041 | T531 | AY619637 | H7 | |||
| Trinidad (Southern Range) | ||||||
| La Lune River | 61°20′W, 10°05′N | 30 m | 130024 | T514 | AY619632 | H7 |
| 130025 | T515 | AY619634 | H7 | |||
| 130036 | T526 | AY619633 | H7 | |||
| Tobago (Main Ridge) | ||||||
| Argyle River | 60°36–35′W, 11°17′–16′N | 400 m | 128964 | T454 | AY619612 | H2 |
| 128967 | T457 | AY619610 | H2 | |||
| 130012 | T502 | AY619611 | H2 | |||
- All R. hartii specimens are deposited at the Glasgow University Hunterian Museum (GLAHM).
- Note: Tucker Valley is located in north-western Trinidad but does not form a part of the hill formations. The approximate altitudes are taken from topographical maps.
Laboratory protocols
DNA extraction protocols were similar to those described by Sambrook et al. (1989). Mitochondrial DNA (mtDNA) was extracted from caudal tail muscle and purified using standard phenol/chloroform protocols. The primers L14724 (Kocher et al. 1989) and H15557 (Hillis et al. 1996) were used to amplify a fragment of approximately 830 bp under standard PCR conditions: 1× (94°C 2 min), 30× (93°C 1 min; 52°C 1 min and 72°C 1 min), 1× (72°C 10 min, 30°C 1 min and 4°C hold) using commercial reagents and protocols similar to those recommended by the manufacturer (Promega, UK). Amplified cyt b fragments were purified by gel electrophoresis and recovered by silica/chaotrope spin column (Qiagen, Crawley, UK). These cyt b sequences are approximately twice as long as those previously reported (Collier et al. 1998). Moreover, 12S rDNA sequences were also obtained from a few animals, but showed no variation.
Sequencing of both strands was performed by the in-house sequencing unit with the amplification primers and dideoxy chain terminators (BigDyes, PE Biosystems, Warrington, UK), and analysed in an ABI 377 apparatus (Applied Biosystems, Perkin Elmer, Warrington, UK). After removing PCR primers and incomplete terminal sequences, 756 base pairs (252 codons) were available for analysis. For DNAs T528 and T531 poor quality internal reads (respectively 62 and 81 nt long) were replaced by Ns. All nucleotide sequences could be aligned without gaps and, when translated into amino acids using the vertebrate mitochondrial code, stop codons were absent.
Phylogenetic analysis
Analyses were performed with paup*4.b.10 (Swofford 2002) and MrBayes 3.0 (Huelsenbeck and Ronquist 2001). The PTP test (Faith and Cranston 1991) was used to assess the presence of non-random structure in the data, based on a heuristic search of 100 randomisations. Trees were constructed by maximum parsimony (MP; heuristic search with TBR branch exchange), maximum likelihood (ML) and Bayesian maximum likelihood (BML) optimality criteria but only ML and BML results are shown because MP did not contribute useful information. The Akaike Information Criterion (Posada and Buckley 2004) best-fit ML model was identified using Modeltest 3.06 (Posada and Crandall 1998) for the ML analysis, and MrModelTest 2.0 (Nylander 2004) for the Bayesian analyses.
Clade support in MP and ML was inferred by bootstrapping and in BML by the frequency with which a clade appeared in the saved trees. BML analyses were performed with default priors and Markov chain settings, and with random starting trees. The gamma shape parameter and proportion of invariant sites were estimated from the data and a codon-based model of site-specific rate heterogeneity was implemented. Trees were sampled every 100 generations for 1 000 000 generations. The log-likelihood scores plateau were reached at about 5000 and 1000 generations (2, 3, respectively) and a consensus tree was constructed from the last 1000 (Fig. 2) and 100 (Fig. 3) trees (100 000 and 10 000 generations, respectively). The likelihood ratio test (Huelsenbeck and Crandall 1997) was used to decide whether ML-clock and ML + clock trees differed significantly. Relative rate tests were performed with RRtree 1.1.13 (Robinson et al. 1998).
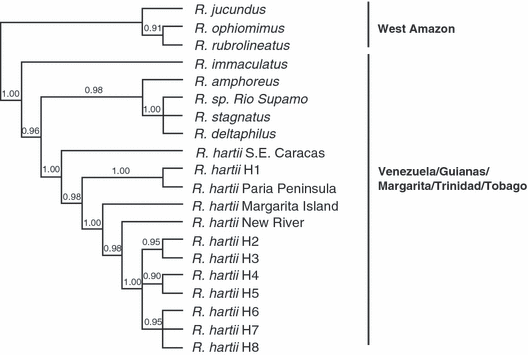
Bayesian 50% majority rule consensus tree of the last 1000 trees saved. Numbers above branches indicate posterior probabilities recovered from the Bayesian analysis. Rivulus species names and GenBank accession numbers used in this figure are R. amphoreus; U41777; R. stagnatus, U41774; R. sp. Rio Supamo, AF002495; R. deltaphilus, AF002494; R. immaculatus, U41779; R. hartii (Paria Peninsula), U41778; R. hartii (New River, Trinidad); AF002496; R. rubrolineatus, AF002493; R. ophiomimus, AF002492; R. jucundus, AF002491. Two specimens (R. hartii. S.E. Caracas and R. hartii Margarita Island are unpublished sequences used in Collier et al. 1998)
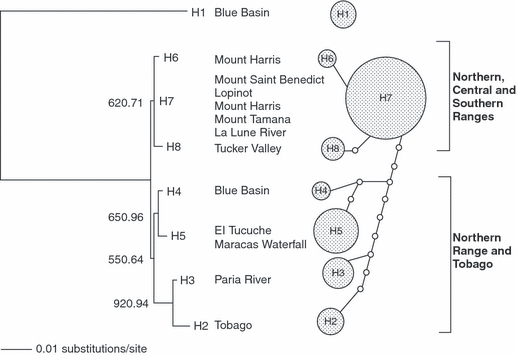
Maximum likelihood phylogram (-lnL = 1406.0414) under a TrN + I model of evolution. The values indicate bootstrap frequencies (%) of 1000 pseudoreplicates from the ML analysis and posterior probabilities recovered from the last 100 trees saved of the Bayesian analysis. The minimum spanning network of all eight Rivulus hartii haplotypes shows the frequency of each haplotype. The size of the circles corresponds to the haplotype frequency. Small circles indicate undetected intermediate haplotype states
Saturation of substitutions was evaluated by plotting (in Excel) transition against transversion ‘p’ distances and fitting the linear or power regression that gave the highest r2 value. TCS 1.18 (Clement et al. 2000) was used to reduce the sequences to haplotypes, and because of the observed low variability and the large number of identical sequences within and between localities, all phylogenetic analyses were performed on the resulting eight haplotypes.
Statistical analysis of population diversity and structure
The geographical structuring of genetic variation was evaluated with Φ-statistics, using the analysis of molecular variance (amova) in ARLEQUIN (Excoffier et al. 1992; Schneider et al. 2000). The significance of variance components and Φ-statistics were assessed with 10 000 data permutations. Haplotype frequencies and a minimum spanning network (MSN) were estimated using TCS 1.18 (Clement et al. 2000), following the method of Templeton et al. (1992).
Results
Sequence characteristics
As in other fish, base composition shows an anti-G bias (A = 27%; C = 27%; T = 32%; G = 14%) (Cantatore et al. 1994; Zhu et al. 1994; Martin and Bermingham 1998; Lee et al. 2001; Doadrio and Dominguez 2003; Peng et al. 2004). Base composition heterogeneity was absent (Chi-squared test, p = 0.96). The scatter-plot of transition versus transversion ‘p’ distances showed a linear relationship (r2 = 0.96), indicating that there was no detectable saturation. There were 77 variable nucleotides (69 parsimony-uninformative and eight informative); 63 variable sites were third codon positions, five were second codon positions and one was a first codon position, but none resulted in an amino acid substitution.
Among the 36 R. hartii individuals sampled, eight haplotypes were found (Table 1, Fig. 3). Six haplotypes occurred in samples from the Northern Range: Blue Basin (H1 and H4); Paria River (H3); El Tucuche and Maracas Waterfall (H5); Mount Saint Benedict and Lopinot (H7); and Tucker Valley (H8). One haplotype (H7) occurred in both of the two Northern Range localities (Mount Saint Benedict, Lopinot) and in the Central (Mount Tamana, Mount Harris) and southern (La Lune River) ranges. A single haplotype was recovered from every sample site except Blue Basin and Mount Harris, at each of them two haplotypes were found (H1 + H4 and H6 + H7, respectively). In Tobago, only one haplotype (H2) was found.
Phylogenetic analyses
The eight haplotype sequences were aligned with the previous cyt b sequences (each of 360 nt, obtained from Genbank) derived from specimens from western Brazil and Venezuelan or Guianese locations. The 360 nt region present in all specimens was analysed by BML to identify a suitable local outgroup for analysis of the 756 nt ingroup alignment. This analysis (Fig. 2) clearly identifies the existence of a derived R. hartii clade containing sister sub-clades, one clade comprising the specimen from south-eastern Caracas, the haplotype H1 and the sequence from the mainland Paria peninsula locality, a specimen from the island of Margarita and yet another comprising the remaining island haplotypes. This result indicates that H1 is an appropriate local outgroup for the main analyses that follow. The levels of divergence associated in this alignment with the various named species of Rivulus are given in Table 2.
| Haplotypes | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (1) | (2) | (3) | (4) | (5) | (6) | (7) | (8) | (9) | (10) | (11) | (12) | (13) | (14) | |
| (1) R. hartii (H1) | – | |||||||||||||
| (2) R. hartii (H7) | 12.4 | – | ||||||||||||
| (3) R. hartii (Paria Peninsula) | 0.5 | 12.4 | – | |||||||||||
| (4) R. hartii (New River) | 12.4 | 1.4 | 12.4 | – | ||||||||||
| (5) R. hartii (Margarita) | 12.0 | 3.0 | 12.0 | 2.7 | – | |||||||||
| (6) R. hartii (S.E. Caracas) | 14.1 | 12.4 | 14.1 | 12.0 | 13.6 | – | ||||||||
| (7) R. sp. (Rio Supamo) | 29.2 | 29.8 | 29.2 | 33.1 | 34.6 | 26.4 | – | |||||||
| (8) R. immaculatus | 32.0 | 26.8 | 31.2 | 29.2 | 28.0 | 23.7 | 20.5 | – | ||||||
| (9) R. stagnatus | 34.0 | 33.1 | 33.9 | 36.7 | 37.3 | 30.4 | 4.9 | 24.9 | – | |||||
| (10) R. amphoreus | 26.5 | 23.6 | 26.5 | 25.7 | 27.9 | 21.1 | 13.9 | 19.3 | 17.5 | – | ||||
| (11) R. deltaphilus | 34.1 | 33.6 | 34.1 | 36.8 | 38.7 | 27.2 | 2.7 | 22.4 | 5.7 | 15.6 | – | |||
| (12) R. ophiomimus | 37.8 | 35.6 | 37.8 | 38.5 | 36.7 | 37.8 | 32.0 | 31.6 | 39.0 | 34.8 | 34.3 | – | ||
| (13) R. rubrolineatus | 36.7 | 32.7 | 37.9 | 35.4 | 32.4 | 36.2 | 30.6 | 26.3 | 35.8 | 34.2 | 31.6 | 15.5 | – | |
| (14) R. jucundus | 38.3 | 28.4 | 38.3 | 30.0 | 32.2 | 32.1 | 35.0 | 29.2 | 38.4 | 39.2 | 36.5 | 21.6 | 21.8 | – |
- Of all the closely related R. hartii haplotypes found, only the commonest (H7) is included in the matrix.
The 360 and 756 nt alignments have strong non-random structure (PTP test, p = 0.01, respectively). From the 756 nt alignment ML and BML analyses both recovered two moderately well-supported clades, shown in Fig. 3. One clade comprises haplotypes H6, H7 and H8, from low-altitude localities in the Northern, Central and Southern Ranges, while the second comprises two sub-clades, one uniting the Tobago haplotype H2 with H3 from the nearest Trinidad locality (Paria River), the other uniting the adjacent Northern Range localities Blue Basin (H4) and El Tucuche/Maracas Waterfall (H5). Thus this analysis provides evidence for both geographical differentiation between Rivulus lineages in different watersheds and for communication between now-isolated sites.
Although there are no direct calibrations of molecular evolution rates in Rivulidae, the customary estimate of approximately 1.5% divergence per million years (Bermingham and Avise 1986; Bernatchez et al. 1991; Bernatchez and Dobson 1991; Zardoya and Doadrio 1999; Machordom and Doadrio 2001; Doadrio et al. 2002; Doadrio and Dominguez 2003) was used to obtain a rough idea of the likely ages of the divergences observed, given that relative rate tests found no significant differences between lineages (p > 0.05) in the rate of change in sequences from all the localities. Divergence estimates used for the molecular clock analysis are given in Table 3.
| Haplotypes | ||||||||
|---|---|---|---|---|---|---|---|---|
| H1 | H2 | H3 | H4 | H5 | H6 | H7 | H8 | |
| H1 | – | 12 | 11.6 | 11.4 | 11.6 | 10.8 | 10.9 | 11.4 |
| H2 | 69 | – | 0.6 | 1.3 | 1.5 | 1.5 | 1.3 | 1.3 |
| H3 | 67 | 5 | – | 0.9 | 1.0 | 1.0 | 0.9 | 1.2 |
| H4 | 66 | 10 | 7 | – | 0.4 | 0.6 | 0.5 | 0.8 |
| H5 | 67 | 11 | 8 | 3 | – | 0.8 | 0.6 | 0.9 |
| H6 | 63 | 11 | 8 | 5 | 6 | – | 0.1 | 0.4 |
| H7 | 64 | 10 | 7 | 4 | 5 | 1 | – | 0.2 |
| H8 | 66 | 10 | 9 | 6 | 7 | 3 | 2 | – |
Analysis of variance
Because of the high sequence divergence observed between H1 and the ingroup, and the large contribution that this haplotype must make to variability, it was excluded from all amova analyses. These located 72% of the variation between and 28% within populations (Table 4). amova also revealed a significant subdivision between the northern (Tobago and Paria River) and southern drainages (all other localities) and between low- and high-altitude populations (<300 versus >300 m above sea level). Other variables, such as mountain ranges, faunal groups (Kenny 1995) or river drainages, showed no significant differentiation (p > 0.05).
| Structure analysed | Among groups | Within groups | Within populations | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Va | % | p | Fct | Vb | % | p | Fsc | Vc | % | p | Fst | |
| All populations | 6.27 | 72.06 | 2.43 | 27.94 | <0.001 | 0.72 | ||||||
| Northern versus southern watersheds | 2.81 | 63.62 | <0.05 | 0.63 | 1.57 | 35.69 | <0.001 | 0.98 | 0.03 | 0.69 | <0.001 | 0.99 |
| High altitude (>300 m) versus all other populations (excluding Tobago) | 2.81 | 51.36 | <0.05 | 0.51 | 2.63 | 47.99 | <0.001 | 0.98 | 0.03 | 0.65 | <0.001 | 0.99 |
| High altitude (>300 m) versus all other populations | 1.10 | 34.36 | <0.05 | 0.34 | 2.07 | 64.69 | <0.001 | 0.98 | 0.03 | 0.95 | <0.001 | 0.99 |
| Mountain ranges | 0.54 | 18.99 | NS | 0.18 | 2.28 | 79.94 | <0.001 | 0.98 | 0.03 | 1.06 | <0.001 | 0.99 |
| Kenny's (1995) divides | 1.19 | 38.70 | NS | 0.38 | 1.86 | 60.31 | <0.001 | 0.98 | 0.03 | 0.99 | <0.001 | 0.99 |
| River drainages | 1.38 | 46.69 | NS | 0.46 | 21.16 | 52.29 | <0.001 | 0.98 | 0.67 | 1.03 | <0.001 | 0.99 |
- By Northern (Paria River, Tobago) versus Southern watersheds (all other localities)
- By localities 300 m above sea level (Blue Basin, El Tucuche, Maracas Waterfall, Tobago) versus below 300 m above sea level (all other localities). By mountain ranges (Tobago Main Ridge, Trinidad Northern, Central and Southern Ranges).
- By Kenny's (1995) faunal distribution (Antillean; Paria River and Tobago, Western Trinidad; Tucker Valley, Blue Basin, El Tucuche, Maracas Waterfall, Mount Saint Benedict, Lopinot, Mount Tamana, Eastern Trinidad; Mount Harris, Southern Trinidad; La Lune River).
- By river drainages; Caroni drainage (Mount Saint Benedict, Lopinot, El Tucuche, Mount Tamana), Blue Basin, Tucker Valley, Oropuche drainage (Mount Harris), La Lune River drainage, Tobago drainage (Argyle River) and Paria River drainage. Non-significant values are shown as NS.
- Values in bold indicate significance (P ≥ 95) among groups.
Discussion
The results provide useful new information on the genetic differentiation of Rivulus in Trinidad and Tobago. The level of divergence between haplotype H1 and the remaining haplotypes (c. 11%) is greater than that between some named pairs of mainland species (e.g. R. stagnatus–R. deltaphilus; 5.7%, R. sp. Rio. Supamo–R. stagnatus; 4.9%, R. sp. Rio. Supamo–R. deltaphilus; 2.7%). Thus, the islands appear to have been colonised by at least two different lineages, now represented on the one hand by the specimens from the Paria peninsula on the mainland and from Blue Basin on Trinidad, and, on the other, by the remaining specimens. The approximately 11% divergence suggests that the lineages split circa 7 My, and it seems likely that H1 evolved in isolation on the mountains of either the Paria peninsula or of northern Trinidad, both of which rise over 600 m and would have provided refugia during marine high-stands. The high divergence between H1 and the remaining Trinidad and Tobago samples, if associated with diagnostic morphological variation, would probably be sufficient to justify the description of a new species. However, the morphological and meristic characters of the specimens involved have not yet been studied.
The low genetic differentiation found between H1 and R. hartii from the Paria peninsula may be the result of a recent dispersal from northern Venezuela to western Trinidad, perhaps when the Orinoco River wet season discharges reduce salinity levels in the Paria Gulf from 30% to 5–15% (Alkins and De Souza 1984; Read 1987; Kenny 1995). Similarly, some Venezuelan species of freshwater fish occur in the south-western peninsula of Trinidad, and are thought to have dispersed there during rainy season periods of reduced salinity (Price 1955; Alkins and De Souza 1984). The close genetic similarity between the Margarita Island and Trinidad and Tobago haplotypes (Fig. 2) and the colonization of Trinidad and Margarita from the mainland probably result from the dispersal facilitated by coastal currents (especially the Orinoco River counter current) that flow in a circular motion around northern Venezuela, from western Trinidad to Margarita (Kenny 1995).
Apart from H1, all the other Trinidad and Tobago specimens are closely related to one another, and may reasonably be regarded as R. hartii. However, they differ substantially (mean ML distance: H2-8 – mainland species; 31.1 ± 0.5%) from all other sampled mainland species, and presumably represent genotypes derived from a common ancestor with H1 or from H1 itself, having been isolated from H1 for circa 7 My. As indicated by the analysis of genetic variance, divergence among these haplotypes shows some geographical and ecological structure associated mainly with isolation by northern and southern watersheds (Kenny 1995), or by altitude (Table 4). Because of complex historical fluctuations in the sea level, much of lowland Trinidad had been repeatedly flooded during the past few hundred thousand years (Murphy 1997), and this provides a reasonable explanation for relative lack of divergence between most low-altitude samples. Five populations from three low-altitude mountain range river drainages (east, west and south drainages, Fig. 1) share identical haplotypes (H7) which suggests that the present distribution of Rivulus in lowland Trinidad may be the consequence of the fragmentation of a once large widespread population during sea level low-stands, leading to the isolation of individuals in separate different river drainages. Although Tucker Valley is at the westernmost side of the Northern Range, Kenny (1995) argues that the faunal distribution of this valley is unlike that of the Northern Range, and is more similar to other southern regions. This claim fits well with the low genetic differentiation between the Tucker Valley haplotype (H8) and other southern haplotypes (Table 3). It is notable, however, that the Tobago haplotype is most closely associated with the one found at the closest northern Trinidad locality sampled, suggesting that Tobago may have been colonized by fish crossing from Trinidad. Fajen and Breden (1992) found a similar close relationship between Tobago and Paria River samples in the guppy Poecilia reticulata. They suggested that for this fish, the ancestral mtDNA haplotype from Venezuela could have also colonized Tobago. However, more samples from eastern Trinidad and from Tobago would be required before the direction and number of colonization events involving Rivulus could be constrained.
This study originated as a part of an investigation of interactions between the endemic Trinidadian frog, Mannophryne trinitatis and its predators (Downie et al. 2001; Jowers and Downie 2005; Jowers et al. 2006) that was extended to include the phylogenetic relations of the Golden Tree Frog, Phyllodytes auratus and M. trinitatis (M. J. Jowers unpublished data). Both of these studies found evidence of the long-term isolation of populations in areas or peaks of the Trinidad Northern Range mountains and, combined with the present study, they emphasize the importance of mountain refuges as a factor in speciation.
Acknowledgements
We wish to thank the Wildlife Section of the Trinidad Government for permission to carry out this work. This study was aided by several members of the University of Glasgow Trinidad Expeditions 2002 and 2003: in particular, Ben Mitchell, Roisin Campbell-Palmer, Graham Stirling, Damian Tobin and Celia Langhorne. We are very grateful to Timothy Page for the advice and guidance given with the population structure and molecular clock analyses. We thank Glen Collier for the R. hartii unpublished sequences. MJJ was supported by a UK Natural Environmental Research Council postgraduate studentship.




