Identification of the sibling species of the Drosophila willistoni subgroup through the electrophoretical mobility of acid phosphatase-1
Taxonomische Identifizierung der verwandten Arten der Drosophila willistoni-Untergruppe durch die elektrophoretische Mobilität der sauren Phosphatase-1
Abstract
enThe Drosophila willistoni group consists of 23 species of which six are sibling species and belong to the D. willistoni subgroup: D. willistoni, Drosophila equinoxialis, Drosophila tropicalis, Drosophila insularis, Drosophila pavlovskiana and Drosophila paulistorum. These sibling species are abundant in the Neotropical region and can hardly be differentiated by the usual taxonomic traits. Four of them (D. willistoni, D. equinoxialis, D. tropicalis and D. paulistorum) cover extensive geographic distribution areas overlapping in places while two of them are endemic (D. insularis and D. pavlovskiana). In this study, we presented a method for the identification of five sibling species of the D. willistoni subgroup based on the allozyme variation of acid phosphatase-1 (Acph-1) in acrylamide gel electrophoresis. Our work showed that Acph-1 allozyme differences can be used for species-diagnostic characterization. This method was shown to be a more efficient tool for species identification than others because it is both quicker and produces reliable results.
Zusammenfassung
deDie Drosophila willistoni-Gruppe umfasst dreiundzwanzig Arten, von denen wieder die folgenden sechs Arten zur Drosophila willistoni-Untergruppe gehören: D. willistoni, D. equinoxialis, D. tropicalis, D. insularis, D. pavlovskiana und die Superspezies D. paulistorum, welche selbst wieder in sechs Semispezies aufgegliedert werden kann. Die Arten der Drosophila willistoni-Untergruppe sind in der neotropischen Region sehr häufig zu finden, können aber durch gewöhnliche taxonomische Merkmale kaum voneinander unterschieden werden. Vier von ihnen (D. willistoni, D. equinoxialis, D. tropicalisund D. paulistorum) zeigen ein weites überlappendes geografisches Verbreitungsgebiet, zwei Arten (D. insularis and D. pavlovskiana) sind endemisch. In dieser Arbeit wird eine Methode beschrieben, durch die fünf der verwandten Arten der Drosophila willistoni-Untergruppe an Hand der Wanderungsgeschwindigkeit der sauren Phosphatase-1 (Acph-1) in der Stärkegel-Elektrophorese voneinander unterschieden werden können. Die Untersuchung zeigte, dass die Unterschiede im Muster der Acph-1 Allozyme als verlässliches diagnostisches Kriterium verwendet werden können. Das Verfahren erweist sich als ein wirksames Werkzeug zur Arterkennung und liefert im Vergleich zu bisher vorhandenen Methoden brauchbare Ergebnisse in weniger Zeit.
Introduction
The Drosophila willistoni group consists of 23 species of which six are siblings and belong to the D. willistoni subgroup: D. willistoniSturtevant, 1921, Drosophila equinoxialisDobzhansky, 1946, Drosophila tropicalisBurla et al., 1949, Drosophila insularisDobzhansky et al., 1957, Drosophila pavlovskianaKastritsis and Dobzhansky, 1967 and Drosophila paulistorum Dobzhansky and Pavan (in Burla et al., 1949), which include six semispecies. This subgroup is one of the most outstanding and widespread groups of drosophilids in the Neotropical region and the cytological, morphological and behavioural aspects have been extensively studied. The species are so similar in their morphology that it is difficult to tell them apart by studying only the external morphology (Burla et al. 1949).
The geographical distribution of the species of D. willistoni subgroup overlaps in extensive areas but D. pavlovskiana and D. insularis are endemic in small defined places as shown by Kastritsis and Dobzhansky (1967) for D. pavlovskiana and Dobzhansky et al. (1957) for D. insularis– the former in Guyana and the latter in some islands of Lesser Antilles. The other four sibling species are sympatric from Guatemala, through Central America and northern South America, down to central Peru and Brazil (Spassky et al. 1971; Dobzhansky and Powell 1975). Drosophila willistoni presents the widest geographical distribution in Neotropical regions. It is the dominant species in most hot and humid forests, despite considerable seasonal fluctuations (Dobzhansky 1957; Da Cunha et al. 1959; Spassky et al. 1971). Its ecological versatility is clearly shown by the variable range of breeding sites successfully exploited, the most important of which are fermented fruits (Carson 1965; Valente and Araújo 1986). The D. paulistorum is, in fact, a superspecies, comprising six morphologically indistinguishable semispecies: Andean-Brazilian, Centroamerican, Orinocan, Amazonian, Transitional (Dobzhansky and Spassky 1959) and Interior (Pérez-Salas et al. 1970). In many parts of tropical South America it is the second most abundant species of the D. willistoni subgroup. Individuals of different semispecies do not cross in nature when sympatric, thus behaving as completely isolated species. In the laboratory, however, they may, in some cases, produce hybrids. Distribution of Drosophila equinoxialis extends from central Mexico through Central America, the Greater Antilles, and the northern half of continental South America. Drosophila tropicalis is distributed from Central America to the centre of South American continent. Although in recent years many authors have published a reasonable number of ecological studies with the D. willistoni subgroup (Tidon et al. 1994; De Toni and Hofmann 1995; Saavedra et al. 1995; Valiati and Valente 1996; Vilela and Mori 1999; Martins 2001; Medeiros and Klaczko 2004; Silva et al. 2005) only a few of these studies present the identification at the species level (Valiati and Valente 1996; Vilela and Mori 1999; Medeiros and Klaczko 2004). This fact is explained by the abundance of the sibling species in the collections, their extreme morphological similarity and the lack of a practical method for the identification of the species. The methods currently used to separate the species of the D. willistoni subgroup are the inspection of external genitalia of males (Burla et al. 1949; Malogolowkin 1952; Spassky 1957), the detailed study of the band patterns of polytene salivary gland chromosomes (Rohde et al. 2006), intercrossing tests (Cordeiro and Winge 1995) and the analyses of the song produced by males during the courtship (Ritchie and Gleason 1995). However, these methods are very laborious to identify large numbers of flies.
Here a method is presented for the identification of five species of D. willistoni subgroup which is much more efficient than those traditionally used. This involves determining the allozyme variation of Acid phosphatase-1 (Acph-1, EC3.1.3.2) locus in acrylamide gel electrophoresis in a representative number of individuals. The results obtained prove that such measurements of Acph-1 can distinguish between these sibling species in spite of their great morphological similarity.
Materials and Methods
Fifty-three population samples of the D. willistoni subgroup (Table 1) were analysed. Of these, 26 were of D. willistoni (including 5148 individuals), 21 of D. paulistorum (904 individuals), four of D. equinoxialis (699 individuals), one of D. tropicalis (127 individuals) and one of D. insularis (132 individuals). Unfortunately, D. pavlovskiana could not be included in the study because no sample strain was available from any of the suppliers of Drosophila stock. However, this has no great effect on the study results because this species is a rare endemic in Guyana. As indicated in Table 1, eight populations of D. willistoni and eight of D. paulistorum correspond to recently collected samples that were analysed as isofemale lines making a total of 5059 and 842 individuals of each species respectively. The other populations correspond to laboratory stocks maintained as mass cultures for varying periods of time. There is no doubt about the identification of the species investigated. They were confirmed by the analysis of the banding patterns of the polytene chromosomes, examination of the genitalia and outcrosses with known species strains (data not shown).
| Species | Semispecies | Population sample | Place of collection | Source |
|---|---|---|---|---|
| D. willistoni | wilAPA | Apazapan, Veracruz, Mexico | 1 | |
| wilPAR | Belém, PA, Brazil | 2 | ||
| wilWIP | Salvador (Ipitanga), BA, Brazil | 3 | ||
| wilCIP | Serra do Cipó, Santana do Riacho, MG, Brazil | 4 | ||
| wilIQG* | Queimada Grande Island, SP, Brazil | 5 | ||
| wilRIB | Ribeirão Preto, SP, Brazil | 6 | ||
| wilMEL | Ilha do Mel Island, PR, Brazil | 6 | ||
| wilISC | Santa Catarina Island, SC, Brazil | 7 | ||
| wilARV | Arvoredo Island, SC, Brazil | 7 | ||
| wilMLC* | Morro da Lagoa da Conceição, SC, Brazil | 8 | ||
| wilCMP | Campeche Island, SC, Brazil | 7 | ||
| wilRAT | Ratones Grande Island, SC, Brazil | 7 | ||
| wilBEN | Bento Gonçalves, RS, Brazil | 9 | ||
| wilDLA | Dois Lajeados, RS, Brazil | 6 | ||
| wil17A2 | Eldorado do Sul, RS, Brazil | 9 | ||
| wilMSA | Porto Alegre (Morro Santana), RS, Brazil | 10 | ||
| wilRMT* | Porto Alegre (Mário Totta Street), RS, Brazil | 10 | ||
| wilPGK* | Porto Alegre (Gabriel Knijnik Park), RS, Brazil | 10 | ||
| wilJBO* | Porto Alegre (Botanic Garden), RS, Brazil | 10 | ||
| wilPFA* | Porto Alegre (Farroupilha Park), RS, Brazil | 10 | ||
| wilITA* | Itapuã Park, Viamão, RS, Brazil | 11 | ||
| wilSPE | São Pedro, Osório, RS, Brazil | 3 | ||
| wilLAG | Laguna Negra, Rocha, Uruguay | 12 | ||
| wilCOR | Coronilla, Uruguay | 12 | ||
| wilTER* | National Park Santa Teresa, Rocha, Uruguay | 6 | ||
| wilGUA | Guadeloupe Island, Lesser Antilles | 13 | ||
| D. paulistorum | Andean Brazilian | pauECU | Jaton Sacha, Ecuador | 1 |
| Andean-Brazilian | pauMES | Mesitas, Colombia | 14 | |
| Andean-Brazilian | pauMAN | Manaus, AM, Brazil | 2 | |
| Andean-Brazilian | pauPAR | Belém, PA, Brazil | 2 | |
| Andean-Brazilian | pauIQG* | Queimada Grande Island, SP, Brazil | 5 | |
| Andean-Brazilian | pauRIB | Ribeirão Preto, SP, Brazil | 9 | |
| Andean-Brazilian | pauJAI | São Paulo, SP, Brazil | 5 | |
| Andean-Brazilian | pauALC* | Alcatrazes Island, SP, Brazil | 5 | |
| Andean-Brazilian | pauMLC* | Morro da Lagoa da Conceição, SC, Brazil | 8 | |
| Andean-Brazilian | pauRAT | Ratones Grande Island, SC, Brazil | 7 | |
| Andean-Brazilian | pauITC* | Itacorubi, SC, Brazil | 15 | |
| Andean-Brazilian | pauMSA | Porto Alegre (Morro Santana), RS, Brazil | 16 | |
| Andean-Brazilian | pauRMT* | Porto Alegre (Mário Totta Street), RS, Brazil | 10 | |
| Andean-Brazilian | pauPGK* | Porto Alegre (Gabriel Knijnik Park), RS, Brazil | 10 | |
| Andean-Brazilian | pauJBO* | Porto Alegre (Botanic Garden), RS, Brazil | 10 | |
| Andean-Brazilian | pauITA* | Itapuã Park, Viamão, RS, Brazil | 11 | |
| Centroamerican | pauC2 | Lancetilla, Honduras | 14 | |
| Transitional | pauT1 | Santa Marta, Colombia | 14 | |
| Amazonian | pauA28 | Belém, PA, Brazil | 14 | |
| Orinocan | pauO11 | Georgetown, Guyana | 14 | |
| Interior | pauI1 | Llanos, Colombia | 14 | |
| D. equinoxialis | equAPA | Apazapan, Veracruz, Mexico | 14 | |
| equPAN | Panama | 6 | ||
| equTEF | Tefé, AM, Brazil | 3 | ||
| equHON | Honduras | 13 | ||
| D. tropicalis | tro0801.0 | San Salvador, El Salvador | 13 | |
| D. insularis | insSTK | Saint Kitts, Lesser Antilles | 3 |
- Numbers refer to the name of fly collectors: (1) Margaret Kidwell, (2) Marlúcia Martins, (3) Antonio Cordeiro and Helga Winge, (4) Carlos Vilela, (5) Hermes Medeiros, (6) Claudia Rohde, (7) Daniela De Toni, (8) Marco Gottschalk, (9)Vera Valente, (10) Ana Lauer Garcia, (11) André Schnorr, (12) Beatriz Goñi, (13) Tucson Stock Center, (14) Lee Ehrman and Yong Kyu Kim, (15) Hermes Schmmitz, (16) Victor Hugo Valiati.
- *Recently collected population samples.
Adult individuals of both sexes were submitted to horizontal electrophoresis for Acph-1 enzyme. Each fly was individually macerated with a drop of distillated water. Electrophoresis of Acph-1 in acrylamide slab gels was carried out by adjusting the method described by Hüettel and Bush (1972). The 6% polyacrilamide gels, containing 95% of acrylamide and 5% bisacrylamide, were prepared with a 80-ml buffer (composed of 0.038 M Tris and 0.0025 M citric acid pH 7.5), 0.8 ml Ammonium persulphate (APS) and 0.08 ml TEMED (Sigma, St. Louis, MO, USA). The buffer in the electrode chamber contained 0.34 M Tris and 0.078 M citric acid pH 7.5. The gel was subject to an electrophoretical run for approximately 4 h at 20 V cm−1 and was stained with 0.05 g of Fast Blue RR stain and 0.05 g of Na α-naphtyl acid phosphate, diluted in 100 ml of 0.2 M acetate buffer pH 5.0 for 1.5 h at 37°C. The gel was soaked in 0.25 M boric acid for 1 h prior to staining. After the appearance of the enzyme bands, the reaction was stopped by washing the gel with water and adding the fixing solution of 5 : 5 : 1, methanol, water and acetic acid respectively.
In each gel, a minimum of three control flies of two species were used: D. willistoni, strain wilSPE, and D. paulistorum, strain pauRMT. They were used as a positive control of the gel conditions and as controls for the two more frequent homozygous allelic forms of Acph-1. Numbers were used to identify the different alleles and the Acph-1 form for D. willistoni was considered the species standard and its allelic was denominated Acph-11.00. The alleles of other species were designated by their relative mobility in relation to the standard Acph-11.00.
Results
Electrophoretic variations in the protein encoded by Acph-1 locus were detected in 7010 individuals, representing 53 populations of five sibling species of the D. willistoni subgroup. The optimal conditions for the separation of the different alleles of Acph-1 were obtained by adjusting the pH to 7.5 and the gel concentration to 6%. The genotypic variants detected for Acph-1 and the characteristic for the species studied were Acph-10.54/0.54, Acph-11.08/0.54 and Acph-11.08/1.08 for D. equinoxialis, Acph-10.64/0.64 for D. tropicalis, Acph-11.00/1.00 for D. willistoni, Acph-11.31/1.31 for D. insularis and Acph-11.42/1.42 for D. paulistorum. All phenotypes are shown in Fig. 1. Uniquely, D. equinoxialis presented two segregating alleles, Acph-11.08 (fast) and Acph-10.54 (slow). None of the allelic forms described here were shared among the species indicating that Acph-1 can be used as species-diagnostic enzyme. Pairs of species were found with near migration alleles for Acph-1 enzyme such as (i) D. paulistorum (Acph-11.42) and D. insularis (Acph-11.31) (ii) D. tropicalis (Acph-10.64) and D. equinoxialis (Acph-10.54). The D. willistoni (Acph-11.00) and D. equinoxialis (Acph-11.08) were the species with the closest migration for their allelic forms. However, even there the variation is still more than sufficient to clearly identify each species (see Fig. 2).
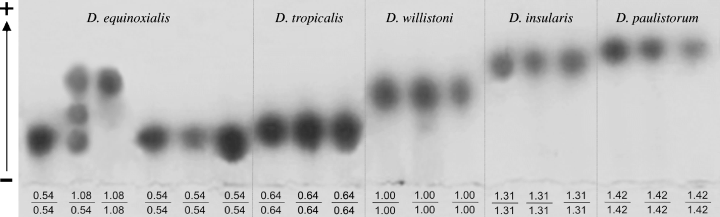
Zymogram of the phenotypic patterns of Acph-1 found in each one of the five species of D. willistoni subgroup. Different homozygous and heterozygous genotypes are indicated. The direction of migration of the protein is up toward the anode according to the arrow
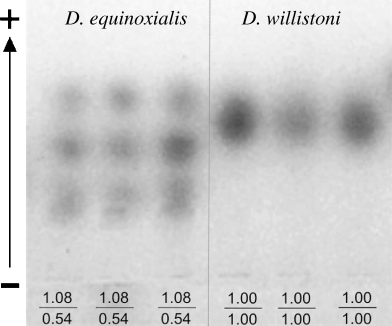
Differences in the phenotypic patterns of Acph-1 found in two species of the D. willistoni subgroup: Acph-11.08/0.54 for D. equinoxialis and Acph-11.00/1.00 for D. willistoni. The direction of migration of the protein is up toward the anode according to the arrow
The efficiency of our diagnostic method to identify species of the D. willistoni subgroup was also confirmed by the absence of intraspecific variation of the Acph-1 allelic forms in each species. The two most exhaustively investigated species were D. willistoni and the Andean–Brazilian semispecies of D. paulistorum. For D. willistoni, for example, the same allele Acph-11.00 in homozygosis is found in all the 26 geographical populations studied, including eight that were recently collected: wilIQG, wilMLC, wilRMT, wilPGK, wilJBO, wilPFA, wilITA, wilTER (see Table 1). These populations corresponded to 5059 individuals (98.3% of all D. Willistoni samples) analysed as parental or F1 generation. In the same way, among 21 populations for Andean–Brazilian semispecies of D. paulistorum, all were homozygous for Acph-11.42. Eight of these populations corresponded to recently collected strains (pauIQG, pauALC, pauMLC, pauITC, pauRMT, pauPGK, pauJBO, pauITA, see Table 1) or to 842 parental or F1 individuals.
Discussion
Despite the considerable number of individuals belonging to the D. willistoni subgroup in Neotropical drosophilid communities, not much data is found in the literature about the frequency and the ecology of each species. Recent studies on the population structure and the ecology of Drosophila species have demonstrated that the D. willistoni subgroup can represent up to 80% of collected specimens from the Amazon area (Martins 2001). In this way, methodologies that make it possible to distinguish rapidly between a large number of individuals of D. willistoni subgroup at the species level can be extremely important for a fuller comprehension of the ecological affinities of each species in particular.
The present study suggests a new methodology to identify species of D. willistoni subgroup based on the pattern of Acph-1 enzyme because each sibling species has it own specific allelic form. This method has the advantage of being faster than other known methods permitting analysis of a larger number of individuals. The localization of the Acph-1 gene in an autosomal chromosome is also favourable for species-specific identification in both sexes.
For many years, researchers have sought investigative methods to separate the sibling species of the D. willistoni subgroup. In this sense, Burla et al. (1949) found several minute morphological differences between sibling species and concluded that ‘the variability is great enough to make identification of species in single individuals hazardous’. A more detailed study of the genitalia of males of the D. willistoni subgroup was made by Malogolowkin (1952) that revealed several additional characteristics, which help to differentiate the sibling species. Spassky (1957) described slight but consistent difference between the external male genitalia that allows a direct identification of single male individuals. Although the females are not themselves distinguishable, they can be identified by the inspection of their male progenies. Nowadays this is a helpful auxiliary method in spite of being limited to one sex.
Since 1969, efforts have been made to detect one enzyme that could be used as species diagnostic for species of the D. willistoni subgroup. Ayala (1975) studied for several years the pattern and the geographical variation of a large number of enzymes by electrophoresis starch gels in this subgroup of flies. However, between 36 loci investigated none conclusively identified the species. The Acph-1 enzyme was also investigated for this purpose, but in contrast to our results the authors found in this loci alleles shared between more than one species of the D. willistoni subgroup. It is believed that the reason for our success in conclusively distinguishing species using Acph-1 enzyme can be explained by differences in our methodology when compared with that used by the former authors. For example, acrylamide gel and electrode buffer conditions were used modifying the method described by Hüettel and Bush (1972) while the earlier investigators used starch gel according methods ascribed to Poulick (1957) (see Ayala et al. 1972).
In relation to D. paulistorum, Richmond (1972a,b) found differences of Acph-1 alleles between the semispecies and between different populations of each one. At this interspecific level, the same allelic form (Acph-11.42) was always found in all the populations and semispecies tested. The best semispecies studied was the Andean–Brazilian with 904 individuals, 93% recently collected and analysed as isofemale lines. Although just one sample of each of Amazonian, Centroamerican, Interior, Orinocan and Transitional semispecies was studied, it is sure that Acph-11.42 is the pattern for D. paulistorum. Considering that semispecies of D. paulistorum are populations in the process of speciation, our results indicate that Acph-1 enzyme diverged between the species of the D. willistoni subgroup but not at the semispecies level.
Acknowledgements
This research was supported by grants and fellowships from Conselho Nacional de Desenvolvimento Científico e Tecnológico/CNPq, Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul/FAPERGS and PROPESQ/UFRGS.




