Character- and tree-based delimitation of species in the ‘Cichlasoma’ facetum group (Teleostei, Cichlidae) with the description of a new genus
Identifikation von Arten der‘Cichlasoma’ facetum Gruppe mit der Berschreibung einer neuen Gattung
Abstract
enThe ‘Cichlasoma’ facetum group is part of the taxonomically complex group of Neotropical cichlid fishes of the tribe Heroini. Many species groups and unplaced species of heroines are still left without a generic name following the revision of the genus Cichlasoma. We describe here the ‘Cichlasoma’ facetum group as a new genus, Australoheros, and provide evidence for its monophyly based on phylogenetic analyses of morphological and mtDNA characters. Australoheros is morphologically characterized by the lowest values in meristic characters among heroines and by three apomorphic characters in coloration pattern. In addition to the three described species of Australoheros, our results of species delimitation based on a combination of tree- and character-based approaches identify seven putatively new species of Australoheros. Several coding schemes of morphological characters are used to recover the intrageneric relationships within the genus, resulting in very similar topologies. Discovery of additional species within the genus is expected once material from the whole distribution area is studied.
Zusammenfassung
deDie ‘Cichlasoma’ facetum Gruppe ist eine der zahlreichen Artengruppen von Amerikanischen Buntbarschen der taxonomisch komplizierten Gruppe der Heroini. Viele Artengruppen der Heroinen haben keinen stabilen Gattungsnamen nach der Revision der Gattung Cichlasoma. Wir beschreiben diese Artengruppe als neue Gattung, Australoheros, und demonstrieren ihre Monophylie anhand einer phylogenetischen Analyse von morphologischen und mtDNA Merkmalen. Morphologisch ist Australoheros durch die niedrigsten meristischen Werte innerhalb der Heroini sowie durch drei apomorphe Merkmale gekennzeichnet. Die Kombination verschiedener Verfahren der Merkmalsanalyse ermöglicht die Identifikation von sieben weiteren Arten neben den drei bereits beschriebenen dieser Artengruppe. Durch verschiedene Kodierungsschemata der morphologischen Merkmale entwickeln wir eine phylogenetische Hypothese der Verwandtschaftsverhältnisse der Arten innerhalb von Australoheros und zeigen, dass verschiedene Kodierungen zu sehr ähnlichen Topologien führen.
Introduction
Cichlasoma facetum is the oldest Neotropical aquarium fish, first brought alive to Europe in 1889 and bred for the first time in captivity before the end of the 19th century (Bade 1897). The first specimens described as Chromis facetusJenyns, 1842, were collected in coastal Uruguay during the famous voyage of Charles Darwin. Chromys oblongade Castelnau, 1855, Heros autochtonGünther, 1862, H. jenynsiiSteindachner, 1869, and H. acaroidesHensel, 1870 are treated as synonyms to C. facetus, known as C. facetum for most of the 20th century. More than two decades ago, Kullander (1983) restricted Cichlasoma to only 12 South American species, most of them described in that revision, and the species is since then referred to as ‘Cichlasoma’ facetum (Jenyns, 1842). The genus level taxonomy and also phylogenetic relationships of Mesoamerican heroines are poorly understood, and after the restriction of Cichlasoma, more than eight species groups including >20 species are without any applicable generic name. In view of the difficulty of a prompt comprehensive analysis of Cichlasoma sensu lato, Kullander (1996) suggested the provisional use of available generic names, an advice that other workers had already widely adopted, in spite of the earlier opinion by Kullander himself (1983), Stiassny (1991), and Miller (1993), that ‘Cichlasoma’– with the quotation marks indicating its informality – should be used for species for which no generic name was applicable. To date, no study has been undertaken to demonstrate monophyletic status of any of those groups. To make the situation even more complicated, new genera are being described for some of the most complex groups (e.g. Allgayer 2001), ignoring results of studies not in agreement (Martin and Bermingham 1998) and without any phylogenetic analysis demonstrating monophyly.
Due to collecting efforts and also recent imports of new cichlids via the pet trade, it has been noted that populations of ‘Cichlasoma’ facetum coming from different areas are frequently distinct and that ‘Cichlasoma’ facetum likely represents a species complex (Lucena and Kullander 1992; Staeck 1998a,b, 2003; Körber and Stawikowski 1999; Casciotta et al. 2003). Two additional species to ‘Cichlasoma’ facetum have been formally described to date, ‘Cichlasoma’ tembe (Casciotta et al. 1995) and ‘Cichlasoma’ scitulum (Říčan and Kullander 2003).
The ‘Cichlasoma’ facetum group is distributed in the southern half of South America in the Río Paraná–Río Paraguay system and their tributaries and in the Río Uruguay system. To the west, the group reaches the foothills of the Andes and to the east occurs in the Atlantic coastal drainages of Argentina, Uruguay and Brazil. The southern distributional border is shared with Crenicichla scottii making it one of southernmost distributed cichlids on the South American continent.
The question of how to recognize species is a heated topic (see e.g. Wheeler and Meier 2000). There is an immense literature concerning what species are and how they are to be discovered. We share the notion of de Queiroz (1998), who suggested that, despite the long history of dispute over species concepts, most species concepts agree fundamentally that species are lineages (Simpson 1961; Wiley 1978; Cracraft 1983; de Queiroz and Donoghue 1988; Frost and Kluge 1994; Baum and Shaw 1995). What previous authors have generally disagreed about are the best criteria for recognizing these lineages (de Queiroz 1998). This trend is particularly apparent when the meager literature on the methodology of species delimitation is contrasted with the extensive body of work on the theory and methods of phylogenetic analysis. Several species criteria or methods for species delimitation have been proposed (e.g. Avise and Ball 1990; Davis and Nixon 1992; Baum and Donoghue 1995; Mallet 1995; Brower 1999; Wiens 1999; Wiens and Servedio 2000; Puorto et al. 2001; Templeton 2001; Wiens and Penkrot 2002), but empirical taxonomists rarely state their criteria for species delimitation explicitly. In this work, we use character-based and tree-based approaches to analyse morphological characters as two tests of species delimitation.
Character-based delimitation
Character-based species delimitation involves finding diagnostic character states that represent seemingly fixed differences between the putative species, or differences that are at least non-overlapping. This approach has been formalized as population aggregation analysis (PAA; Davis and Nixon 1992). However, given finite sample sizes, determining with certainty as to whether traits are truly fixed is virtually impossible (Wiens and Servedio 2000).
Most systematic studies using the character-based delimitation are based on the assumption that diagnosability itself is sufficient evidence for species recognition. This notion heavily rests on the assumption that there is a strong differentiation between species and weak differentiation within species. Very few studies have tested this assumption, and there are studies which suggest the ‘worst-case scenario’ with striking differentiation within species and limited differentiation between species (Wiens and Penkrot 2002).
Many systematists utilize statistical analyses of quantitative morphological characters to test species boundaries, often evaluating the extent to which individuals of a putative species cluster together using principal components or canonical variates analysis. This approach is very useful but lacks the clear relationship to estimated patterns of gene flow that the phylogenetic component of the tree-based approach offers.
Tree-based delimitation
Tree-based delimitation with morphology, although advocated by some authors (e.g. Baum and Donoghue 1995), has rarely been used by empirical systematists (e.g. Hollingsworth 1998; Wiens and Penkrot 2002). A precise methodology for its use has recently been proposed by Wiens and Penkrot (2002), facilitated by methods that allow continuous quantitative characters and polymorphic characters to be included in phylogenetic analyses with little loss of information (e.g. Thiele 1993; Wiens 1999, 2001). Populations rather than individuals are used as terminal units (following Hollingsworth 1998; Wiens and Penkrot 2002) because using individuals will inappropriately treat all polymorphisms shared between populations as homoplasies rather than potential synapomorphies (Wiens 2000).
The tree-based approach provides the parsimonious solution of character distribution, a homology hypothesis, and presents monophyletic groups, which are compared with results of the character-based approach. This two step system, combining character- and tree-based approaches, has multiple advantages over a single step system. Empirical studies show that monophyletic groups without diagnostic characters can receive strong support in the tree-based approach, demonstrating that species as lineages do not need to be defined by obvious character states (Wiens and Penkrot 2002). On the contrary, cases in which intraspecific branch lengths are similar to interspecific branch lengths can be reconciled using the character-based approach providing a priory diagnostic characters if these do not conflict with the tree topology.
Materials and Methods
Morphological methods
Character-based delimitation
Most characters are based on alcohol preserved museum specimens with notes on live coloration if available. Measurements and counts were taken as described by Kullander (1986). Measurements were taken with digital calipers to 0.1 mm and are made point to point except for head length and snout length, which are projections from the anterior tip of the premaxilla to the orbital margin and the posterior margin of the gill cover, respectively. Scale rows are numbered as described by Kullander (1990), i.e. the horizontal row including the lower lateral line is designated as row E0, and the rows are counted as E1, E2, etc. dorsally, and H1, H2, etc. ventrally. Dorsal and anal fin rays and vertebrae were counted on X-radiographs. Vertebral counts include the last halfcentrum. Colour marking terminology follows Kullander (1983, 1986. Bars are counted and numbered in postero-anterior succession (e.g. Kullander 1983; Kullander and Silfvergrip 1991), in this case because of their more stable number in the posterior part of the body. The number of specimens is indicated in parentheses. Institutional abbreviations are as listed in Leviton et al. (1985) and Leviton and Gibbs (1988).
We have used statistical analyses of quantitative morphological characters to test species boundaries, evaluating the extent to which individuals of a putative species cluster together using principal components analysis. The putative species have been formulated based on (i) possession of unique characters/character states; and (ii) based on a unique combination of character states.
Statistical analyses were done in the Systat package (SPSS 2000) and the constrained principal components analysis (RDA) has been performed using the Canoco for Windows program supplemented with CanoDraw to visualize results of the analyses (TerBraak and Šmilauer 2002). The RDA analysis has been performed as described in Lepš and Šmilauer (2003; p. 245–252). Data were log transformed prior to analysis and only specimens of a similar size range were included (>50 mm SL). The multivariate analysis has been performed in an iterative approach. Species separated in the first round of the analysis are removed, and the analysis is repeated without these already separated species. The resolution should improve as the most divergent species condensing the axes of the first analysis have been removed. Characters used in the character-based delimitation include all the following (colour pattern characters not included; HL: head length; SL: standard length): HL/SL, snout L/HL, body depth/SL, orbital diameter/HL, head width/HL, interorbital dist./HL, preorbital dist./HL, caudal peduncle L/caudal peduncle depth, pectoral fin L/SL, ventral fin L/SL, last dorsal fin spine L/SL, and the following counts: scale counts (E0, L1, L2, scales between anterior insertion of the dorsal fin and the upper lateral line, scales between the posterior end of the upper lateral line and the dorsal fin, cheek scale rows), lower ceratobranchial rakers, caudal vertebrae, caudal peduncle vertebrae, anal pterygiophores anteriorly from the first haemal spine, anal fin spines, anal fin rays, anal fin total, dorsal fin spines, dorsal fin rays, dorsal fin total, pectoral fin rays. A discriminant analysis has been performed using Statistica (StatSoft, Inc. 2000).
Tree-based delimitation
Data sets. We have constructed two kinds of data sets for the tree-based approach of species delimitation. In one, we use populations as terminal units (PTU) in tree building to test as to whether the character-based species are monophyletic units on the resulting cladograms.
The second data set includes species as terminal units (STU). Cladograms resulting from this matrix are used to evaluate possible differences in topology compared with the PTU analyses and thus to test the robustness of character coding on the phylogenetic hypotheses (see below).
All characters that have been found during the study were included into the parsimony analyses. For the STU analysis, 21 characters have been scored and all multistate characters are ordered. See Appendix 1 for details.
In the PTU analysis, 35 characters were scored, with most multistate characters ordered. See Appendix 3 for details. The total number of characters is lower in the STU analysis than in the PTU analysis, as some characters are autapomorphies of one species only or because some character complexes could not be split into character states under the coding methods used in the STU analysis.
The terminology for the coding methods used follows Campbell and Frost (1993) and Wiens (1995, 1999). Qualitative characters were coded using the majority approach. Two meristic characters in the STU analysis have been coded using the majority coding. Some characters, such as the number of abdominal bars have been coded using the scaled coding (Campbell and Frost 1993). The states are ordered under the assumption that traits pass through a polymorphic stage between absence and fixed presence. The scaled method is advantageous in that it allows polymorphisms to act as synapomorphies.
Quantitative characters in the PTU analyses have been coded using two coding methods. The first was a modified gap weighting (GW) method of Thiele (1993). Thiele's implementation of GW involves finding, for a given character, the mean value of the trait in each species in the analysis, the range of mean species values among taxa (i.e. the species with the greatest mean value and the species with the lowest), and then dividing this range into smaller ranges or segments equal to the maximum number of character states allowed by the phylogenetic software program (e.g. 32 for paup*). We have used a less fine grained spacing, thus having in most cases <32 states. Species are then assigned states based on these ranges, and the character is ordered. Evolving from low to high mean trait values (or vice versa) therefore requires passing through many intermediate states and requires many steps, whereas smaller changes in trait values involve fewer state changes and fewer steps. An important advantage of the gap-weighting method is that it incorporates information on the distance between states, weighting the changes according to the difference between mean species values.
The second method of coding of quantitative characters in the PTU analyses was the step matrix gap weighting (SMGW) method of Wiens (2001) and Wiens and Etheridge 2003). The SMGW method assigns to each taxon for a given character a unique mean trait value, and the costs of changes between these states are specified with a step matrix, based on the difference in mean trait values between each pair of species. The analyses are constrained by the number of distinct states allowed by the computer software package (>32 for paup or paup*), which does not allow analyses of large numbers of taxa with unique trait means. If the number of taxa with unique means is too large, the gap-weighting method of Thiele (1993), or it's modification as used here is the best approximation. These methods use less-fine-grained information, but have no limits on the number of taxa that can be coded.
We have used the between-state scaling (Wiens 2001) to weight quantitative characters against qualitative characters. This weighting scheme assigns transformations between species with fixed and adjacent values of meristic variables (e.g. 13–14 vertebrae) the same weight as changes in binary variables (0 to 1), and species with intermediate mean values (e.g. 13.5) receive proportionally intermediate weights. Data matrices and descriptions of characters are in Appendices 1–5.
Molecular methods
Sequences of the mitochondrial cytochrome b (cyt b) gene of 49 species (including outgroup taxa) were obtained from Genbank. Additionally, 29 taxa (representing 24 species, including the first published cyt b sequences of the ‘Cichlasoma’ facetum group) were sequenced during this study in order to have a complete coverage of all lineages within heroines, and most lineages of cichlasomatines, the sister group of heroines. The sequenced specimens are mainly from aquarium stocks, except some of the Australoheros sequences (Table 1). DNA was extracted from small pieces of muscle or gill (10–25 mg) using the DneasyTM Tissue Kit (Qiagen). The entire cyt b gene (1.3 kb) was PCR amplified with primers GLuDG.L-TGA CTT GAA RAA CCA YCG TTG (Palumbi et al. 1991) and H15915-AAC TGC CAG TCA TCT CCG GGT TAC AAG AC (Irwin et al. 1991). PCR reactions were carried out with initial denaturation at 94°C for 5 min, followed by 30 cycles with denaturation at 94°C for 1 min, primer annealing at 45–50°C for 40 s and primer extension at 72°C for 1 min. PCR was finished by final extension at 72°C for 5 min. PCR products were purified by ethanol precipitation or using Microcon PCR Filter Units (Millipore) and directly sequenced on an automated DNA sequencer using BigDyeTM Terminator Cycle Sequencing Kit v.3.1 (PE Applied Biosystems). Sequencing reaction products were cleaned by ethanol precipitation or with DyeEx 2.0 Spin Kit (Qiagen) were resolved on ABI Prism 310 Genetic Analyser (Perkin Elmer). Except the amplification primers, the following additional primers were used for sequencing: modified L14952 of Lydeard et al. (1995; TCA TCC GTC GCC CAC AT), modified L15162 of Taberlet et al. (1992; CCA TGA GGA CAA ATA TC), and L15299 (Lydeard and Roe 1997). Chromatograms were assembled and checked by eye for potential mistakes using SeqMan II of the DNAStar software package (http://www.dnastar.com). Edited sequences were aligned using the default settings in ClustalX software (Thompson et al. 1997). The alignment was manually revised in BioEdit (Biological sequence alignment editor v5.0.9, http://www.mbio.ncsu.edu/bioedit/bioedit.html). The alignment includes no gaps.
| Species | Accession number | Catalogue number | Tissue number | Country, province | Drainage | Locality | Coordinates |
|---|---|---|---|---|---|---|---|
| Australoheros sp. jacutinga | AY998658 | NRM 49554 | 1179 | Brazil, Santa Catarina | Río Parana | Aquarium stock, Río Iguaçu | |
| Australoheros sp. uruguai | AY998659 | NRM 49555 | 1180 | Uruguay, Salto | Río Uruguay | Aquarium stock, around town of Salto | |
| Australoheros tembe | AY998660 | Collection O. Říčan | Specimens alive | Argentina, Misiones | Río Parana | Aquarium stock, Arroyo Tirica | |
| Australoheros scitulus | AY998661 | NRM 36435 | 121 | Uruguay, Colonia | Río Rosario, La Plata | Arroyo Colla, the type locality | 34° 19′ 7′′ S 59° 20′ 13′′ W |
| Australoheros scitulus | AY998662 | NRM 41626 | 116 | Uruguay, Colonia | Río Rosario, La Plata | Arroyo Colla, the type locality | 34° 19′ 7′′ S 59° 20′ 13′′ W |
| Australoheros scitulus | AY998663 | NRM 39514, NRM 33048 | Arg. | Argentina, Entre Rios | Río Uruguay | Arroyo El Palmar, 200 m from road RN14 | 31° 53′ 55′′ S 58° 19′ 55′′ W |
| Australoheros facetus | AY998664 | Collection O. Říčan | Specimens alive | Argentina, Entre Rios | La Plata | Aquarium stock | |
| Australoheros facetus | AY998665 | Collection O. Říčan | Specimens alive | Argentina, Entre Rios | La Plata | Aquarium stock | |
| Australoheros facetus | AY998666 | NRM 49538 | 1163 | Uruguay, Maldonado | La Plata | Aquarium stock, around town of Maldonado | |
| Australoheros facetus | AY998667 | NRM 33033 | Arg. | Argentina, Entre Rios | Río Uruguay | Río de La Reconguista, La Reja, below dam |
Phylogenetic analyses
The phylogenetic analyses were performed using paup* 4b.10 (Swofford 2001) and the STU analysis also with NoNa (Goloboff 1993) run from the Winclada interface (Nixon 1999), which was used to map character states onto the tree. Maximum parsimony (MP) analyses were performed with same search strategy both with paup* and NoNa/Winclada (100 random sequence additions, 10 trees kept per addition, search on the saved trees to find all the shortest trees). Bootstrap analyses were done using the same approach, with five random sequence additions per one bootstrap. Bootstrap analyses were run with 1000 replications.
As the sister group of Australoheros is not known, we have used Heroina isonycterina as the outgroup based on several lines of evidence. First, it is the geographically closest heroine species in a clade of heroines related to Australoheros. Secondly, it is morphologically plesiomorphic in most characters within that clade. Thirdly, when a hypothetical outgroup is reconstructed for the clade containing Australoheros based on the commonality principle, Heroina approaches this reconstructed outgroup most closely. For a few characters, the plesiomorphic condition among the clade of heroines containing Australoheros cannot be decided (characters 1, 9, 12, 15, 17, 19 and 20 in Appendices 1 and 2), and these characters are represented by question marks in the character matrices.
To reconstruct the phylogenetic hypothesis testing Australoheros monophyly using cyt b sequences, we have used paup* with the same search strategy and commands as described above. We have weighted all positions and Ti/Tv equally and robustness of the hypothesis has been assessed with bootstrap (1000 pseudorelications). The analysis included all major lineages of heroine cichlids, as well as cichlasomatines and outgroup taxa.
Results
Monophyly of Australoheros
The alignment of the 1143 nucleotide positions of the cyt b gene for 78 species contained 492 parsimony informative characters. The recovered phylogenetic hypothesis strongly supports the monophyly of the genus Australoheros (Fig. 1; length, 4708; n, 36; CI, 0.19; RI, 0.49; bootstrap support 100%). The monophyly of Australoheros is supported by 39 unambiguous nucleotide substitutions. Results of the analysis also strongly support Australoheros as a heroine cichlid genus, in agreement with morphological characters (Fig. 1; see below; BS, 79%). Based on the cyt b phylogeny, Australoheros is nested within the predominantly Mesoamerican heroine clade (Fig. 1; BS, 76%), to which it is more closely related than to the South American genera Pterophyllum, Hypselecara, Hoplarchus, Mesonauta, Uaru, Symphysodon and Heros. The cladogram supports the distinctiveness of the five included species, i.e. Australoheros scitulus, Australoheros facetus, Australoheros tembe, A. sp. jacutinga and A. sp. uruguai. Both A. scitulus and A. facetus, for which multiple individuals from different localities were sequenced form strongly supported clades. The average uncorrected pairwise sequence divergence between the five species is 5.5%. The highest pairwise divergence is between A. scitulus and A. sp. jacutinga (7.0%), the smallest between A. sp. jacutinga and A. sp. uruguai (4.2%). The five included species are diagnosed by 18 (A. facetus, A. sp. uruguay) to 33 (A. scitulus) unambiguous nucleotide substitutions.
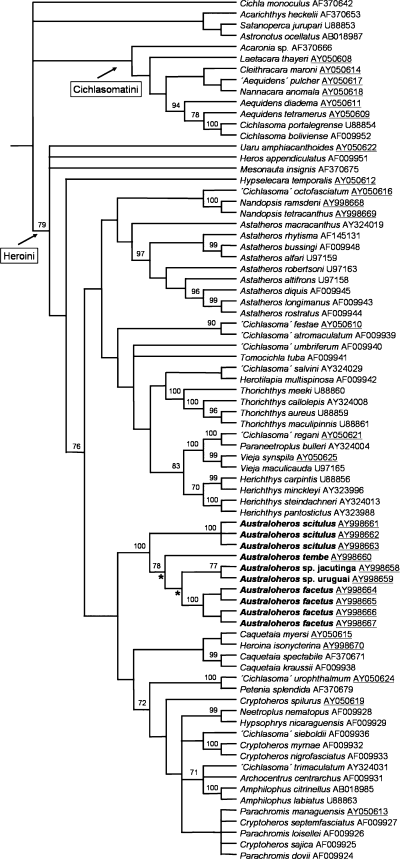
Cladogram depicting relationships among heroine cichlids and showing monophyly of the genus Australoheros. The cladogram shown is the strict consensus of 36 maximum parsimony topologies obtained from a parsimony analysis of the complete cyt b gene. Bootstrap support values shown above nodes (only values >70% are shown). Asterisks below branches show two nodes at which is the cyt b partition strongly in conflict with the morphological partiotion in combined analysis (see Discussion). Combined analysis topology of Australoheros is identical to cyt b topology.
Australoheros, new genus
Diagnosis
A monophyletic group of heroine cichlids having the following synapomorphies: lowest scale counts (modally <25 in E1 scale row); scales on chest of comparable size to flank scales; lowest counts of vertebrae (13 + 13 – 14); unique breeding coloration characteristic in the interruption of the abdominal bars in their middorsal part (Fig. 2); juveniles with distinct xanthophore dots at the base of the caudal fin (Fig. 3); Most species of Australoheros develop four abdominal bars (vs. three), an apomorphic condition among heroines (Říčan et al. in press).
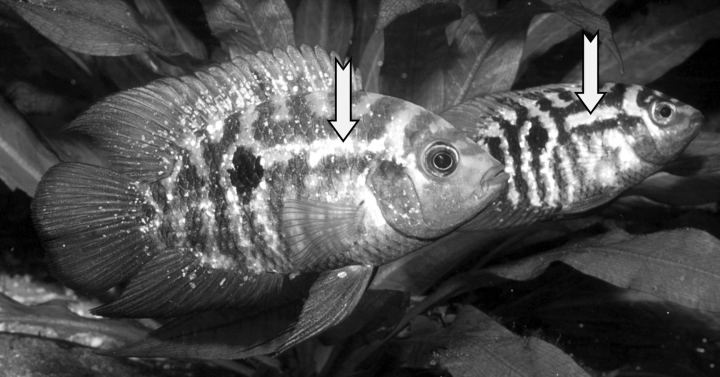
Australoheros facetus pair in breeding coloration showing the genus specific apomorphy of interruption of the dorsal portion of the abdominal bars.

Australoheros facetus juvenile showing the genus specific xanthophore dots at the base of the caudal fin.
The genus is part of the heroine lineage (Fig. 1). Heroines are morphologically diagnosed with the following characters: (i) a single palato-ethmoid articulation (Kullander 1998); (ii) five or more anal fin spines (vs. 4 or less in cichlasomatines and most other Neotropical cichlids; Kullander 1996, 1998); (iii) the palatine bone of heroines is shifted away from the head of the vomer (Kullander 1996, 1998); (iv) midlateral blotch of heroines develops in the fourth ontogenetic bar (vs. in the fifth in all other Neotropical cichlid groups), which is an apomorphic condition (Říčan et al. in press).
Etymology
From the latin word australis, meaning southern, and the name Heros, after the nominotypic genus of the heroini tribe.
Type species
Australoheros facetus (Jenyns, 1842).
Contained species
Australoheros tembe (Casciotta et al., 1995), Australoheros scitulus (Říčan and Kullander, 2003), and at least seven undescribed species (Australoheros sp. forquilha, Australoheros cf. facetus, Australoheros sp. jacui, Australoheros sp. jacutinga, Australoheros sp. paraguay, Australoheros sp. pirapo, and Australoheros sp. uruguai).
Description
The description is based on specimens over 50 mm SL. Body generalized in proportions (see Říčan and Kullander 2003), with variation between the species, variation also in head and mouth shape (short to long snouted, with inferior to superior mouths). American type lips (Kullander 1986: Fig. 12).
Scales on head and chest not distinctly smaller than on flanks. Scales in E1 row 23–26 (95% range 24–25). Upper lateral line scales 15–19 (95% range 16–18), lower lateral line scales 7–10 (95% range 8–9). Scales between upper lateral line and dorsal fin 3–5 anteriorly, one large one small to two large one small posteriorly, scales between lateral lines two. Circumpeduncular scales 16. Cheek scale rows 3–4 (5). Lower lateral line continued on caudal fin by 1 or 2 scales. Scale cover of the dorsal and anal fins varies between the species, as does the extent of scalation of the soft parts of the unpaired fins. Pectoral and pelvic fins without scales. Caudal fin densely scaled, scales ctenoid; interradial scales in single rows.
Fins: Anal fin meristics typically heroine: V–IX, 9–6. Anal fin pterygiophores 11–14, with one to three pterygiophores anteriorly of the first haemal spine. Dorsal fin: XV–XVII, 11–8. Pelvic fin base below pectoral fin base. Pectoral fin rays 12–14.
All teeth caniniform, slightly recurved. Outer row teeth increasing in size symphysiad, upper jaw anterior teeth longest, lower jaw anterior teeth subequal. Jaw teeth with a distinct second cusp in all species examined (contra Říčan and Kullander 2003).
Gill rakers externally on first gill arch 2 epibranchial, 1 in angle, 5–9 ceratobranchial. Microbranchiospines on external side on second to fourth ceratobranchials. Lower pharyngeal tooth plate of varying shape, ranging from very robust (see Říčan and Kullander 2003), over intermediates (e.g. A. sp. paraguay, A. sp. jacui) to very shallow (e.g. A. sp. forquilha), including marked differences in dentition.
Thirteen abdominal and 13–14 (15) caudal vertebrae, with the combination 13 + 13 typical for most species. Caudal peduncle moderate to short, containing none to three vertebrae.
Colour pattern: Coloration includes one of the best distinguishing characters of Australoheros and is also one of the characters showing relationships of the genus. A midlateral blotch of variable size, vertical flank bars, an interrupted midlateral stripe and the caudal fin blotch make up the principal coloration markings. Vertical bars can range from wide to relatively narrow, partly depending on their number in the abdominal area. The majority of Australoheros species have four abdominal bars (i.e. between the bar bearing the midlateral blotch and the opercular cleft), which is an apomorphic condition among heroines. Four species (A. scitulus, A. sp. pirapo and A. sp. jacutinga, A. sp. jacui) do always have only three abdominal bars, while three are the norm in a fifth (A. sp. paraguay). The degree of completeness of development of the four bars varies among the species.
Two bars posteriorly from the midlateral blotch above the anal fin, the posterior one at the border of the caudal peduncle. The caudal peduncle bar can be fused with the caudal spot bar in species with very short-caudal peduncles.
Horizontal markings include an interrupted midlateral band from the opercular cleft posteriad to the midlateral blotch. In some species, the bar continues in the same scale rows posteriad, while in A. sp. pirapo and to a lesser extent in A. scitulus, A. sp. jacutinga and also A. sp. uruguai, the stripe bends upwards posteriorly from the midlateral blotch. Various species specific spotting patterns are observed inside the genus, including an opalescent stripe below the eye in A. sp. forquilha, spotted opercular bones and anterior part of the body, in A. scitulus, or checkerboard spotted membranes of unpaired fins (A. sp. forquilha).
Australoheros juveniles show two very distinct xanthophore dots on the base of the caudal fin, dorsaly and ventraly bordering the caudal fin blotch (Fig. 3). The dots develop at the size of 9 mm TL, are best visible at 11–14 mm TL and become covered by melanophores before 25 mm TL. These spots are known from all three species for which we have complete ontogenetic series (A. facetus, A. scitulus and A. sp. jacutinga). Such spots are not known from any other heroine cichlid (Říčan et al. in press).
Delimitation of Australoheros species
Examination of the variation encountered while examining the material of Australoheros supported the notion that more species than the three described can be diagnosed. Most of the hypothesized new species can be diagnosed based on unique combinations of characters, but at least one putative species also possesses several unique characters. Two tests of this a priori delimitation have been used, i.e. the character- and the tree-based delimitation.
Character-based delimitation: The results of the multivariate RDA analysis support all putative species. The first separated groups are A. sp. forquilha, A. tembe and A. sp. jacui (Fig. 4). The best separating characters of A. tembe and A. sp. forquilha are the number of caudal peduncle vertebrae, those of A. sp. jacui the number of anal and dorsal spines and C1 gill rakers. The best separating characters between A. tembe and A. sp. forquilha are the interorbital and preorbital distances (and coloration characters not included in RDA). Species separated in the second round are A. facetus and A. cf. facetus (Fig. 5). The best separating character between A. facetus and A. cf. facetus and the remaining species is the number of C1 rakers. The best separating characters between A. facetus and A. cf. facetus are the number of caudal peduncle vertebrae, and anal fin spines. The overlap between the four remaining species and A. scitulus is only marginal, and when A. facetus and A. cf. facetus are removed, A. scitulus is clearly separated from the four species and the only overlap remains between A. sp. pirapo and A. sp. jacutinga (Fig. 6). The best separating characters of A. scitulus from the remaining species are the number of dorsal spines, anal spines, E0 scales and caudal vertebrae. When A. scitulus is further removed, all remaining species are separated by the data (Fig. 7). The best separating characters of A. sp. paraguay is the number of C1 gill rakers, interorbital distance, and preorbital distance. A. sp. pirapo is best separated by the number of E0 scales and caudal vertebrae. A. sp. jacutinga and A. sp. uruguai are very similar in most characters, but optically quite different (see above for cyt b divergence of the two species). The largest difference is in body depth, with number of anal fin rays and cheek scales also being significant separating characters. Most important meristic and morphometric characters are summarized as results of a discriminant analysis and shown in Appendix 6.
First round of the constrained principal components analysis (RDA) resulting in separation of A. sp. forquilha, A. tembe and A. sp. jacui.
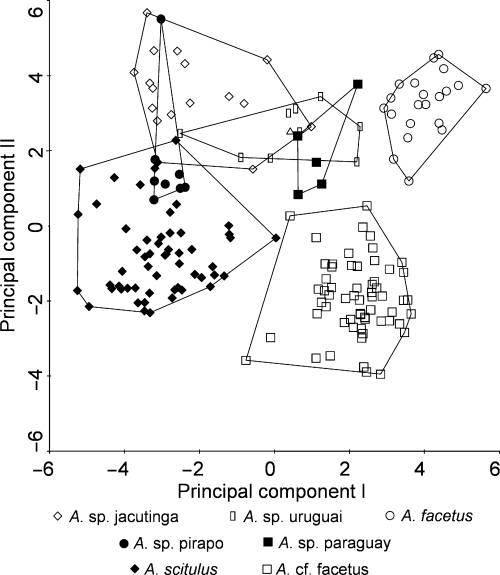
Second round of the constrained principal components analysis (RDA) resulting in separation of A. facetus and A. cf. facetus.
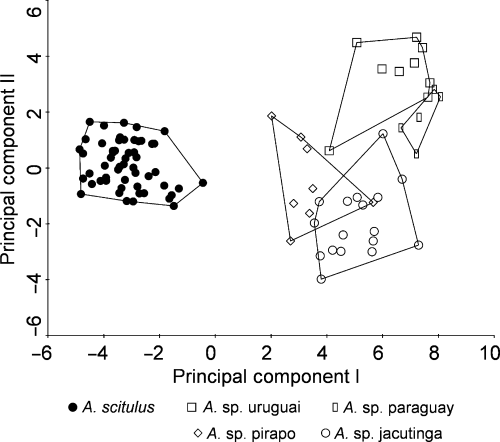
Third round of the constrained principal components analysis (RDA) resulting in separation of A. scitulus.
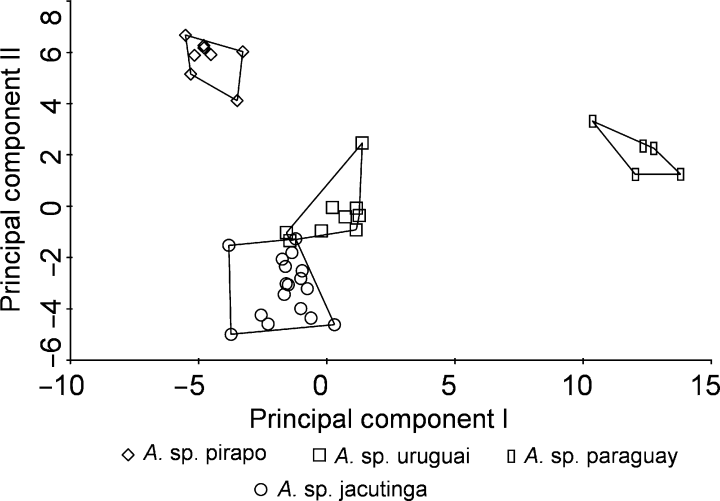
Fourth round of the constrained principal components analysis (RDA) resulting in separation of A. sp. paraguay, A. sp. pirapo, and showing only marginal overlap between A. sp. jacutinga and A. sp. uruguai.
Tree-based delimitation: The character-based species delimitation supports the recognition of 10 species among the studied material of Australoheros. The nominal species A. facetus probably contains two distinct groups, A. facetus and A. cf. facetus. In order to test as to whether these 10 species also form monophyletic lineages, we have constructed a parsimony analysis of 18 populations representing these 10 putative species (see Materials and methods). For four putative species very few individuals are known and their monophyly thus was not tested in the analysis.
(1) Populations as terminal units (PTU) analyses. Phylogenetic analysis of the PTU character matrix (Appendix 4) using the modified coding method of Thiele (1993) and with between-state scaling recovers two shortest trees (L, 619; CI, 0.52; RI, 0.70; Fig. 8a). The MP analysis supports the monophyly of all but one species. All the tested species except A. sp. jacutinga are also well supported using bootstrap support (>90%). A. sp. forquilha is found as non-monophyletic. The Argentinean populations (A. cf. sp. forquilha) form distinct lineage from the Brazilian populations (A. sp. forquilha). One of the two MP cladograms supports A. cf. sp. forquilha as a monophyletic group (65%), while the other MP tree places the two populations as successive sister groups of the remaining species (except A. tembe).
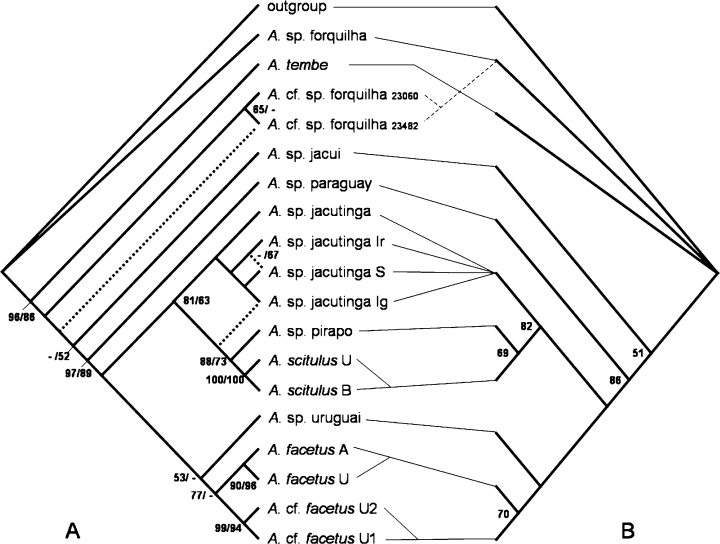
Intrageneric relationships of Australoheros. (a) Results of the populations as terminal units (PTU) analyses. Dotted lines show disagreements in topology of the step matrix gap weighting (SMGW) (Wiens 2001) and the modified gap weighting method (GW) of Thiele (1993). Numbers indicate bootstrap support, first value from left refers to GW, second value to SMGW. (b) Results of the species as terminal units analysis. Numbers indicate bootstrap support. The interrupted line between the PTU and STU cladograms shows the non-monophyletic of A. sp. forquilha in the PTU analyses. Population abbreviations: (a) Argentina; (b) Brazil, Ig, Iguaçu; Ir, Irai; S, Soberbio; U, Uruguai; U1, Uruguai 1; U2, Uruguai 2; 23060: ZSM 23060; 23482: ZSM 23482.
We have also performed an analysis in which we have not used any scaling between quantitative and qualitative characters. This analysis results in eight MP trees (results not shown), with consistency and retention indices 10% lower than in the scaled analysis and a length of 462 steps. The major difference was the non-monophyly of A. sp. jacutinga, which formed a polytomy at the base of the A. scitulus–A. sp. pirapo group. Overall bootstrap proportions were also significantly lower than in the scaled analysis. Moreover, the agreement in topology with the STU analysis is higher in the scaled analysis.
The phylogenetic analysis of the PTU matrix using SMGW (sensu Wiens 2001; Wiens and Etheridge 2003) resulted in one MP tree (L, 169149; CI, 0.50; RI, 0.66; Fig. 8a). The tree topology is identical to tree 2 of the analysis using the GW of Thiele (1993) (Fig. 8a) with one exception, and that is the position of the Iguaçu population of A. sp. jacutinga, which is recovered as the sister group of the A. sp. pirapo–A. scitulus group.
(2) Species as terminal units analysis. Phylogenetic analysis of the STU character matrix (Appendix 2) yielded three shortest trees (L, 55; CI, 0.67; RI, 0.72; 8, 9). The analysis in NoNa results in two shortest trees with the same consensus topology, which is due to a different and more strict collapsing rule algorithm (P.A. Goloboff, personal communication). Otherwise there is only one difference in topology compared with the PTU analyses, and that is the non-monophyly of A. sp. forquilha in the PTU analyses (Fig. 8a).
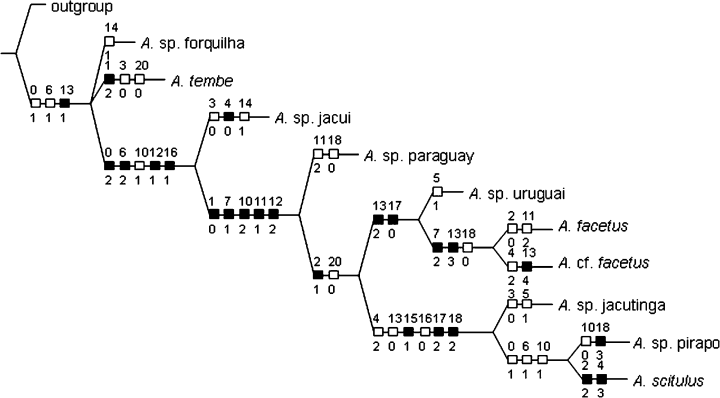
Species as terminal units (STU) analysis showing optimization of character states (see Appendix 1 for character states). Note that four abdominal bars (character 13) are mapped as synapomorphic for the genus (state 1), while the plesiomorphic condition in A. sp. jacutinga, A. sp. pirapo and A. scitulus is interpreted as a reversal. Australoheros is also supported as monophyletic by the possession of 14 or less caudal vertebrae (character 0; state 1), while the majority of species have 13 caudal vertebrae (state 2). A. sp. pirapo and A. scitulus are again interpreted as reversed to the more plesiomorphic condition (state 1). Synapomorhies not showing variation inside the genus are not mapped on the tree (i.e. breeding coloration, juvenile coloration; see diagnosis). A. cf. sp. forquilha is omitted from the figure.
Discussion
We have demonstrated here the monophyly of the ‘Cichlasoma’ facetum group and have provided a suite of apomorphic character states diagnosing the species group as a new genus, Australoheros. Australoheros is one of the few circumamazonian South American heroines, but its affinities are clearly with Mesoamerican heroines, both in morphological as well as mtDNA characters. Despite our increased efforts at elucidating the phylogenetic position of Australoheros, no sister group can be singled out at present.
Our second goal in this paper was to explore the species diversity in the genus Australoheros, based on a combination of character- and tree-based approaches to species delimitation using predominantly morphological characters. A priory, based on possession of unique character states and unique combinations of character states, we have hypothesized that 10 species can be recognized within Australoheros. Two species (A. scitulus and A. sp. forquilha) posses unique characters, the remaining species are diagnosed by unique combinations of characters. The recognition of these 10 species is supported by the results of the characters-based species delimitation using multivariate analysis of meristic and morphometric data. The results of the tree-based species delimitation, utilizing also colour pattern characters, support the recognition of all these species but one (A. sp. forquilha), which is represented by two lineages in the tree-based delimitation (Fig. 8a). The populations from the upper Río Uruguay tributaries in the Brazilian states of Río Grande do Sul and Santa Catarina cluster separately from those around the town of Soberbio in the Argentinean province Misiones (A. sp. forquilha vs. A. cf. sp. forquilha; 8, 10). The situation is complex because of the bad condition of the material from Misiones, for which coloration characters diagnostic for A. sp. forquilha cannot be scored. The material from Misiones is slightly different in a number of meristic characters, but the specimens do posses the diagnostic shortest interorbital and longest preorbital distances of A. sp. forquilha. Australoheros tembe (Casciotta et al. 1995) is to date the only described species of Australoheros from Misiones (Argentina), and all material comes from tributaries of the Arroyo Urugua-í and its tributaries, which form an affluent of Río Paraná. A single specimen referred to as ‘Cichlasoma’ sp. tembe is known from Arroyo Fortaleza (Casciotta et al. 2003), which is a tributary of Río Uruguay. Arroyo Fortaleza is very close to Soberbio, and this specimen is likely to be A. cf. sp. forquilha rather than A. tembe.
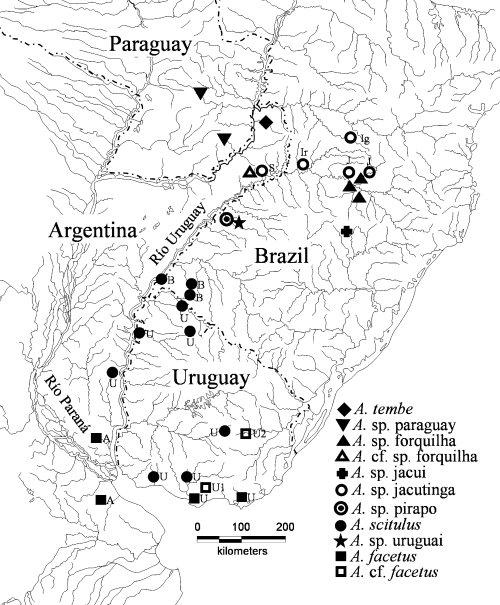
Drainage map of south-eastern South America showing the distribution of Australoheros species along the Río Uruguay. One symbol may indicate more than one adjacent collection locality. Population abbreviations: A, Argentina; B, Brazil, Ig, Iguaçu; Ir, Irai; J, Jacutinga; S, Soberbio; U, Uruguay; U1, Uruguay 1; U2, Uruguay 2.
Australoheros jacutinga has a similar distribution as A. sp. forquilha (i.e. including A. cf. sp. forquilha), occurring both in the upper Río Uruguay tributaries of Santa Catarina as well as in the Argentinean tributaries around Soberbio. The lots from Soberbio in Misiones were actually all a mix of A. sp. forquilha and A. sp. jacutinga. While in the case of A. sp. forquilha the Soberbio samples form a separate lineage in the tree-based analyses, this is not the case with the A. sp. jacutinga samples. The four populations of A. sp. jacutinga do form one monophyletic lineage in the phylogenetic analysis of populations using the GW method (Fig. 8a). In the phylogenetic analysis using the SMGW method the Iguaçu population is found outside the cluster of the three other A. sp. jacutinga populations, but still in very close proximity, as the sister group of the A. sp. pirapo–A. scitulus clade (Fig. 8a). The position of the Iguaçu population is probably changing because of considerable amount of missing data entries (few specimens available). The result of the GW analysis is in agreement with the character-based delimitation, and we prefer to treat all the four populations as representing one species only, i.e. A. sp. jacutinga. The Iguaçu population has an interesting distribution, as the Río Iguaçu tributaries are connected with Río Paraná, but not with Río Uruguay, where all other samples of A. sp. jacutinga come from.
Cyt b data are available only for five species, but they show significant sequence divergences (>4%) between the species, supporting the recognition of these species based on morphological characters. Several heroine species are separated by much smaller divergences in the cyt b gene (e.g. Astatheros alfari and A. bussingi at 2.5%; Loiselle 1997; Martin and Bermingham 1998). Given the relatively slow rate of nucleotide substitution in fish mitochondrial DNA relative to ‘conventional’ rate estimates for vertebrates (Bermingham et al. 1997), these levels of sequence differentiation within species are remarkable and suggest an isolation of several millions of years. The morphological distinctiveness of the species for which DNA was available and those for which it was not is similar, suggesting that also molecular divergences of these species will be significant. While both morphological as well as cyt b data agree that Australoheros includes more species than generally assumed, the relationships between the species are in disagreement depending on the type of characters used (morphology vs. mtDNA). The extent of the conflict is difficult to assess under the unbalanced taxon sampling, but the disagreements cannot be attributed to different rooting position only. In order to test this conflict, we performed a combined analysis of the two data sets with a reduced taxon sampling. This analysis resulted in a topology identical to that based on cyt b only (results not shown; see Fig. 1). The differences between the cyt b and the morphological topologies are statistically significant (tested with the compare-2 test in paup*; MP topology of the combined data forced to morphological topology p = 0.045; morphological topology forced to the cyt b data set p = 0.013). Two nodes (shown by asterisks in Fig. 1) show strong conflict between the two data sets as judged from the partitioned bremer support (results not shown). The most notable difference is that the best recognizable species (A. scitulus) is placed in the cyt b tree as the sister group of all other species and not as a close relative of A. sp. jacutinga, and A. tembe is not placed as a basal species. Contrary to the morphological hypothesis, the highest observed sequence divergence is between A. scitulus and A. sp. jacutinga (7.0%). Morphological cladograms show A. sp. uruguai as the sister group of A. facetus plus A. cf. facetus, but the cyt b tree supports A. sp. uruguai as being most closely related to A. sp. jacutinga. The sister group relationship between A. sp. uruguai and A. sp. jacutinga can be supported from the morphological data by the low number of pectoral fin rays uniquely shared by the two species and the sister group relationship is also supported by the partitioned bremer analysis, where both partitions agree in the combined topology. A. sp. uruguai and A. sp. jacutinga also replace each other along the middle Río Uruguay.
Four of the hypothesized species come from the Brazilian portion of the middle-upper Río Uruguay. The Río Uruguay is known to contain surprisingly many species of fishes for the size of the area. We are not aware of any phylogenetic study published which would include fish faunas of the Río Uruguay, but there are several faunal listings and taxonomic papers dealing with the area. The faunistic studies of Bertoletti et al. (1989a,b, 1990) included 131 fish species and suggested that the faunas of middle and upper Río Uruguay are quite distinct. The middle Río Uruguay region had 100 species of which 50 were not found in the upper Río Uruguay. Whether the difference in species composition between the upper and middle Río Uruguay is through a gradual or abrupt shift is not known. Lucena and Kullander (1992) recognized eleven Crenicichla species in the Brazilian portion of the Río Uruguay drainage. No monophyletic groups have been presented, but the authors hypothesize two species groups and several additional species with an uncertain position. Five species were included in the missioneira group, and the group should be endemic to the middle and upper Río Uruguay. Four Australoheros species delimited here, A. sp. forquilha, A. sp. jacutinga, A. sp. pirapo, and A. sp. uruguai seem to be endemic to the same area as the missioneira group of Crenicichla. A partial exception is A. sp. uruguai, as the southernmost record is from the middle/lower Río Uruguay in Uruguay (collection of T. Litz, cat. no. 495). Whether known records of A. facetus from lower/middle Uruguay are actually all A. sp. uruguai or if the two species occur in sympatry remains to be examined. Contrary to the missioneira group of Crenicichla, the four Australoheros species do not form a monophyletic group in neither of our analyses, at least A. sp. forquilha does not seem to be related to the remaining three species, and A. sp. pirapo is clearly the sister species of A. scitulus. The middle-lower Río Uruguay hosts three Australoheros species, A. facetus, A. scitulus and probably A. sp. uruguai. In the case of Crenicichla, the middle-lower Río Uruguay also hosts three species, C. scottii, Crenicichla lepidota, and Crenicichla vittata. Crenicichla lepidota and C. vittata are widespread Paraguayan/La Plata species, occurring also in the lower-middle Río Uruguay, most likely paralleling the distribution of A. facetus. Crenicichla scottii was placed with Crenicichla gaucho and Crenicichla prenda in the scottii group. If this group is indeed monophyletic, and if A. sp. jacutinga is the sister group of A. sp. pirapo and A. scitulus, these two groups would have the same distribution pattern, crossing the hypothesized boundary between upper and middle-lower Río Uruguay. The cladograms suggest that the coastal areas of Argentina, Uruguay and Brazil were colonized by two different lineages (the A. scitulus and A. facetus groups), probably from the area of the middle Río Uruguay. There are several other cited areas of endemism and biogeographical patterns in the La Plata region (Lucena and Kullander 1992), one of them being the distinction between the species in Río Uruguay drainages and those in the Atlantic drainages of Uruguay and Southern Brazil. This distinction holds true for Crenicichla (Lucena and Kullander 1992) and also for Australoheros.
Acknowledgements
We wish to thank the museum staff of MCP for enabling us to study specimens in their care, to Uwe Werner (Ense-Bremen, Germany) for kindly providing photographs from his extensive collection and permission to use one of his pictures here, to Simona Poláková for help with the RDA analysis, to Miroslav Oborník, Ivan Fiala (České Budějovice), and Mari Källersjö, Pia Eldenäs and Erik Åhlander (Stockholm) for assistance in the laboratories. Important samples and material of Australoheros were kindly provided by Stefan Körber and Thomas Litz (Germany). Financial support was provided by a grant from the HIGH LAT programme for Access to the Swedish Museum of Natural History to OŘ within the European Community programme ‘Improving the Human Research Potential and the Socio-Economic Knowledge Base’ and by the grants of The Ministry of Education of the Czech Republic, grant no. 155/99 and grant no. CEZ:AJ06/98:F1–123100003.
Appendix
Appendices
Material examined
Australoheros sp. forquilha: MCP 13936 (1); MCP 12123 (1); MCP 13389 (1); NRM 13389 (1); MCP 12525 (1); MCP 12777 (1); MCP 18743 (4).
Australoheros cf. sp. forquilha: ZSM 23482 [15/16; A, B, C, E, F, G, H, I, J, K, L, M, N, O; D (C&S)]; ZSM 23060 (6/12; I, J, L, C, G; D C&S); MCP 6262 (9/35; C, D, N, P, Q, R, T, U, V).
Australoheros sp. jacui: ZSM 30441 (18; spec. L,P C&S).
Australoheros sp. pirapo: MCP 13938 (1); MCP 12667 (5); NRM 12667 (2).
Australoheros sp. jacutinga: MCP 13937 (1). MCP 13383 (6; spec. H C&S). MCP 12509 (1). MCP 13011 (6). Fractions denote numbers of specimens in mixed lots including more than one species. Soberbio (Argentina): ZSM 23048 (1/16; spec. P). ZSM 23060 (6/12 E, H, K, F; spec. A,B C&S). Irai (Brazil): MCP 6262 (13/35; A, B, E, F, G, H, I, J, K, L, M, O, S). Arroyo Canoin (Brazil): MCP 12710 (2/11; J, K). Iguaçu (Brazil): Collection Mus. Maringá cat. no. 3683 (1), 3967 (1), 7743 (1), 8861 (1).
Australoheros sp. paraguay: MHNG 2237.58 (1); NRM 33498 (2); NRM 42215 (2); MHNG 2764 (2); MHNG 2237.56 (7).
Australoheros sp. uruguai: MCP 12710 (9); Collection of T. Litz, cat. no. 495 (12; 3 spec. C&S).
Australoheros facetus: NRM 33033 (3); NRM 33035 (2); NRM 33050 (1); NRM 39509 (14); NRM 39552 (1); NRM 43453 (6); NRM 43485 (1).
Australoheros cf. facetus: NRM 47999 (20); NRM 48074 (15); NRM 48078 (39); NRM 36495 (2); NRM 36774 (2); NRM 36848 (1); NRM 39527 (13); NRM 43943 (2); NRM 37035 (1 C&S); NRM 37037 (1 C&S); NRM 37039 (1 C&S).
Australoheros tembe: STRI 2517 (1); STRI 2518 (1); STRI 2524 (1); STRI uncat (2).
Australoheros scitulus: NRM 36647 (3); MCP 13589 (5); MCP 13944 (1); NRM 13588 (2); NRM 33048 (1); NRM 36435 (1); NRM 36465 (1); NRM 36866 (11); NRM 40063 (10); NRM 40122 (1); NRM 36638 (1); NRM 36671 (17); NRM 41626 (1); NRM 39533 (7); NRM 43136 (4); NRM 39580 (1).
Comparative material. MCP 18317 (6), MCP 14387 (106), MCP 13587 (7), MCP 11041 (44), MCP 22440 (13), MHNG 2514.86 (2), MHNG 2514.85 (3), NRM 30953 (2), MNRJ16176 (4), MNRJ 16178 (9), MCP 13732 (4), NRM 42269 (1).
Appendix 1. List of all parsimony informative characters for the species as terminal units (STU) analysis
0. Caudal vertebrae: more than 15 [0]; 14 [1]; 13 [2]; outgroup [0] – ordered
1. Caudal peduncle vertebrae: modally <1 [0]; mode 1.5–2.5 [1]; mode >2.5 [2]; outgroup [?] – ordered
2. Anal fin pterygiophores: three character states using gap coding; 11 (12) pterygiophores with 1 (2) pterygiophore anteriorly from the first haemal spines [0]; (12) 13 with (1) 2 [1]; 14 (13–15) with 2 (3); outgroup [0] – ordered
3. Dorsal fin pterygiophores: 10 [0]; 11 [1]; outgroup [1] – ordered
4. Anal fin count: four character states using gap coding; five spines with eight to nine rays (V, 8–9) [0]; VI, 7–8 (9) [1]; VII, 7–8; [2]; VIII–IX, 7 (6–8) [3]; outgroup [1] – ordered
5. Pectoral fin rays: 13 or more [0]; 12 [1]; outgroup [0]
6. Scales in the E0 scale row: 26 or more [0]; 25 [1]; 24 [2]; outgroup [0] – ordered
7. Length of dorsal fin scale cover: long, reaching anterior insertion of dorsal fin [0]; intermediate, covering the bases of the middle portion of the hard part of the dorsal fin [1]; short, only covering the bases of the two last spines [2]; outgroup [0] – ordered
8. C1 gill rakers: six or less [0]; seven or more [1]; outgroup [1]
9. Scale pattern along the anterior dorsal fin border: scale row terminating with one small scale [0]; scale row terminating with two small scales arranged horizontally [1]; outgroup [?]
10. Scale rows between posterior end of upper lateral line and the dorsal fin: two large one small or more [0]; one large and one of almost the same size, one additional small from 13 to 14th dorsal spine [1]; one large one small, one additional small from 13 to 14th dorsal spine [2]; one large one small, one additional small from 9th spine [3]; outgroup [0] – ordered
11. Scale rows between anterior insertion of the dorsal fin and the upper lateral line: 5 [0]; 4 [1]; 3 [2]; outgroup [0] – ordered
12. Interorbital distance: wide [0]; intermediate [1]; narrow [2]; outgroup [?] – ordered
13. Abdominal bars: three in all developmental steps and also in adults [0]; four in about 50% of juveniles, three in all adults [1]; four in about 50% of juveniles, four about 50% of adults [2]; four in all juveniles, four in >80% of adults, but only in <20% completely separated [3]; four in all juveniles, four in >80% of adults, completely separated in >80% of adults [4]; outgroup [0] – ordered
14. Forquilha head type, including mouth shape: no [0]; yes [1]; outgroup [0]
15. Distinct and dominant midlateral stripe between operculum and midlateral spot, continuous, not fragmented into spots: no [0]; yes [1]; outgroup [?]
16. Large and dominant and well circumscribed midlateral blotch in juveniles and adults: no [1]; yes [0]; outgroup [0]
17. Caudal base spot: distinct, rounded spot [0]; weakly developed [1]; very narrow or completely missing [2]; outgroup [?] – ordered
18. Midlateral stripe posterior from the midlateral blotch: running in scale rows 0 and E1 as anterior of the blotch [0]; the midlateral stripe runs in scale rows E0, E1 and E2 posterior to the midlateral blotch, i.e. the midlateral stripe gets wider posterior of the midlateral blotch [1]; midlateral stripe bend upwards posterior from the midlateral blotch – the blotch posterior to the midlateral stripe is centred in the same scale row as the midlateral bar (i.e. E1 scale row), and the last blotch is high on the body [2]; midlateral stripe bend upwards posterior from the midlateral blotch – the midlateral blotch is centred in the E1 scale row, while the next posterior blotch is centred in the E2 scale row and the blotch in the last body bar is centred in the E3 scale row. The midlateral stripe does not run in the 0 scale row posterior from the midlateral blotch [3]; outgroup [1] – ordered
19. Midlateral stripe: without distinct borders [0]; clearly bordered [1]; outgroup [?]
20. Spots in scales arranged into stripes also ventral from the 0 scale row (at least into one): no [1]; yes, at least in the posterior part of the body [0]; outgroup [?]
Appendix 2
Character matrix for the STU analysis
| 0000000000 | 1111111111 | 2 | |
| 0123456789 | 0123456789 | 0 | |
| outgroup | 0?0110001? | 00?00?0?1? | ? |
| A. sp. forquilha | 1101101011 | 0001100110 | 1 |
| A. sp. jacui | 21000020?1 | 101?101110 | 1 |
| A. tembe | 12001010?0 | 0001000111 | 0 |
| A. sp. pirapo | 1011201100 | 0120010231 | 0 |
| A. sp. jacutinga | 2010212100 | 2120010221 | 0 |
| A. scitulus | 1021301100 | 1120010221 | 0 |
| A. sp. uruguai | 2011112100 | 2122001011 | 0 |
| A. sp. paraguay | 2001102110 | 2221001101 | 1 |
| A. facetus | 2001102210 | 2223001001 | 0 |
| A. cf. facetus | 2011202210 | 2124001001 | 0 |
| ; | |||
| proc/; | |||
| BEGIN ASSUMPTIONS; | |||
| OPTIONS DEFTYPE = UNORD POLYTCOUNT = MINSTEPS; | |||
| TYPESET * default = ORD: 1 - 5 7 -8 11 - 14 20 - 21; | |||
| ENDBLOCK; | |||
Appendix 3. List of all parsimony informative characters for the populations as terminal units (PTU) analysis using the modified gap weighting method (GW) of Thiele (1993)
1. Length of dorsal fin scale cover. states: long, reaching anterior insertion of dorsal fin [0]; intermediate, covering the bases of the middle portion of the hard part of the dorsal fin [1]; short, only covering the bases of the two last spines [2]; outgroup [0] – ordered.
2. Scale pattern along anterior dorsal fin border. states: scale row terminating with one small scale [0]; scale row terminating with two small scales arranged horizontally [1]; outgroup [?].
3. Scale rows between posterior end of upper lateral line and dorsal fin. states. Two large one small or more [0]; one large and one of almost the same size, one additional small from 13 to 14th dorsal spine [1]; one large one small, one additional small from 13 to 14th dorsal spine [2]; one large one small, one additional small from 9th spine [3]; outgroup [0] – ordered.
4. Scale rows between anterior end of dorsal fin and upper lateral line. states. 5 [0]; 4 [1]; 3 [2]; outgroup [0] – ordered.
5. Abdominal bars. states: three in all developmental steps and also in adults [0]; four in about 50% of juveniles, three in all adults [1]; four in about 50% of juveniles, four about 50% of adults [2]; four in all juveniles, four in >80% of adults, but only in <20% completely separated [3]; four in all juveniles, four in >80% of adults, completely separated in >80% of adults [4]; outgroup [0] – ordered
6. Distinct and dominant midlateral stripe between operculum and midlateral spot continuous, not fragmented into spots. states no [0]; yes [1]; outgroup [?].
7. Large, dominant and well circumscribed midlateral blotch in juveniles and adults: no [1]; yes [0]; outgroup [0].
8. Caudal base spot. states: distinct, rounded spot [0]; weakly developed [1]; very narrow or completely missing [2]; outgroup [?] – ordered.
9. Midlateral stripe posterior from the midlateral blotch. states: running in scale rows 0 and E1 as anterior of the blotch [0]; the midlateral stripe runs in scale rows E0, E1 and E2 posterior to the midlateral blotch – i.e. the midlateral stripe gets wider posterior of the midlateral blotch [1]; midlateral stripe bend upwards posterior from the midlateral blotch – the blotch posterior to the midlateral stripe is centred in the same scale row as the midlateral bar (i.e. E1 scale row), and the last blotch is high on the body [2]; midlateral stripe bend upwards posterior from the midlateral blotch – the midlateral blotch is centred in the E1 scale row, while the next posterior blotch is centred in the E2 scale row and the blotch in the last body bar is centred in the E3 scale row. The midlateral stripe does not run in the 0 scale row posterior from the midlateral blotch [3]; outgroup [1] – ordered.
10. Midlateral stripe. states: without distinct borders [0]; clearly bordered [1]; outgroup [?]
11. Spots in scales arranged into stripes (at least one) also ventral from the 0 scale row. states: no [1]; yes, at least in the posterior part of the body [0]; outgroup [?]
12. Opalescent line below the circumorbital series. states: absent [0]; present [1]; outgroup [0].
13. Checkerboard spotted unpaired fins (i.e. soft part of dorsal, caudal and soft part of anal fins); states: absent [0]; present [1]; outgroup [0].
14. Mouth position and size; states: mouth proportionally large, terminal [0]; mouth proportionally large, pointing down, lower jaw proportionally shorter [1]; mouth proportionally large, pointing up, lower jaw projecting in front of upper [2]; mouth very small, terminal or slightly pointing down [3] – unordered.
15. Species develops thick lips. no [0]; yes [1].
16. Anal pterygiophores. Range 11–15. Frequency bins spaced at 0.2; states: 11.0–11.2 [0]; 11.2–11.4 [1]; … [2,3,4,5,6,7,8,9, A,B,C,D,E,F,G,H,J,K] – ordered.
17. Anal spines. Range 5–9. Frequency bins spaced at 0.2; states: 5.0–5.2 [0]; 5.2–5.4 [1]; … [2,3,4,5,6,7,8,9,A,B,C,D,E, F,G,H,J,K] – ordered.
18. Anal rays. Range 6–9. Frequency bins spaced at 0.2; states: 6.0–6.2 [0]; 6.2–6.4 [1]; … [2,3,4,5,6,7,8,9,A,B,C,D,E,F,G, H,J,K] – ordered.
19. Dorsal spines. Range 14–18. Frequency bins spaced at 0.2; states: 14.0–14.2 [0]; 14.2–14.4 [1]; … [2,3,4,5,6,7,8,9,A,B,C,D, E,F,G,H,J,K] – ordered.
20. Dorsal rays. Range 7–12. Frequency bins spaced at 0.2; states: 7.0–7.2 [0]; 7.2–7.4 [1]; … [2,3,4,5,6,7,8,9,A,B,C,D,E, F,G,H,J,K,L,M,N,P,Q] – ordered.
21. Dorsal total. Range 24–27. Frequency bins spaced at 0.2; states: 24.0–24.2 [0]; 24.2–24.4 [1]; … [2,3,4,5,6,7,8,9,A,B, C,D,E] – ordered.
22. Caudal vertebrae. Range 12–15. Frequency bins spaced at 0.2; states: 12.0–12.2 [0]; 12.2-12.4 [1]; … [2,3,4,5,6,7,8,9, A,B,C,D,E] – ordered.
23. Caudal peduncle vertebrae. Range −2–(+3.5). Frequency bins spaced at 0.2; states: –2–(–1.8) [0]; –1.8–(–1.6) [1]; … [2,3,4,5,6,7,8,9,A,B,C,D,E,F,G,H,J,K,L,M,N,P,Q] – ordered.
24. Body depth/SL. Range 0.40–0.53. Frequency bins spaced at 0.1; states: 0.40–0.41 [0]; 0.41–0.42 [1]; … [2,3,4,5,6,7,8,9, A,B,C] – ordered.
25. Head width/HL. Range 0.44–0.64. Frequency bins spaced at 0.2; states: 0.44–0.46 [0]; 0.46–0.48 [1]; … [2,3,4,5,6,7,8,9] – ordered.
26. Interorbital distance/HL. Range 0.22–0.46. Frequency bins spaced at 0.2; states: 0.22–0.24 [0]; 0.24–0.26 [1]; … [2,3,4,5,6, 7,8,9,A,B] – ordered.
27. Preorbital distance/HL. Range 0.10–0.36. Frequency bins spaced at 0.2; states: 0.10–0.12 [0]; 0.12–0.14 [1]; … [2,3,4,5, 6,7,8,9,A,B,C] – ordered.
28. Pectoral fin length/SL. Range 0.24–0.36. Frequency bins spaced at 0.2; states: 0.24–0.26 [0]; 0.26–0.28 [1]; … [2,3,4,5] – ordered.
29. Ventral fin length/SL. Range 0.22–0.48. Frequency bins spaced at 0.2; states: 0.22–0.24 [0]; 0.24–0.26 [1]; … [2,3,4,5,6,7, 8,9,A,B,C] – ordered.
30. Pectoral fin rays. Range 12–14. Frequency bins spaced at 0.2; states: 12.0–12.2 [0]; 12.2–12.4 [1]; … [2,3,4,5,6,7,8,9] – ordered.
31. E0 scales. Range 23–26. Frequency bins spaced at 0.2; states: 23.0–23.2 [0]; 23.2–23.4 [1]; … [2,3,4,5,6,7,8,9,A,B,C, D,E] – ordered.
32. L1 scales. Range 13–19. Frequency bins spaced at 0.4; states: 13.0–13.4 [0]; 13.4–13.8 [1]; … [2,3,4,5,6,7,8,9,A,B,C, D,E] – ordered.
33. L2 scales. Range 6–11. Frequency bins spaced at 0.2; states: 6.0–6.2 [0]; 6.2–6.4 [1]; … [2,3,4,5,6,7,8,9,A,B,C,D,E,F,G,H,J, K,L,M,N,P,Q] – ordered.
34. C1 gill rakers. Range 5–9. Frequency bins spaced at 0.2; states: 5.0–5.2 [0]; 5.2–5.4 [1]; … [2,3,4,5,6,7,8,9,A,B,C,D,E,F, G,H,J,K] – ordered.
35. Opercular spots. states: absent [0]; present [1].
Appendix 4
| 0000000000 | 1111111111 | 2222222222 | 33333 | |
| 1234567890 | 1234567890 | 1234567890 | 12345 | |
| Outgroup | 0?000?0?1? | ?00?0K??CQ | EER9063??? | EEQA0 |
| A. sp. forquilha | 0100100110 | 111106699H | CCP3248126 | ECDC0 |
| A. cf. sp. forquilha 23060 | 0101100110 | 1??101469F | 97H6326335 | 9AJE0 |
| A. cf. sp. forquilha 23482 | 0101100110 | 1??101478E | 89L5227246 | ABEF0 |
| A. sp. jacui | 0110?01110 | 1001011A4F | 54K5355136 | 4A980 |
| A. tembe | 0000100111 | 0000115599 | 49R434612? | 9???0 |
| A. sp. pirapo | 1001010231 | 000307A8BD | A9A6566334 | BAC50 |
| A. sp. jacutinga | 1021010221 | 000306979C | 75D9365242 | 5BA40 |
| A. sp. jacutinga Irai | 1021010221 | 00030596AB | 64C9565360 | 2A820 |
| A. sp. jacutinga Soberbio | 1021010221 | 00030596A9 | 547??????? | ????0 |
| A. sp. jacutinga Iguaçu | ?0220????? | ?00308989B | 633??????? | ???40 |
| A. scitulus | 1011010221 | 00030CH5EA | 9AB6443335 | 9AA51 |
| A. scitulus quarai | 1011010221 | 00030BG5E9 | 9997466345 | 9AF41 |
| A. sp. uruguai | 1021201011 | 0000086CAD | 9496453230 | 58D40 |
| A. sp. paraguay | 1022101101 | 100005599C | 74D9587244 | 49790 |
| A. facetus Argentina | 2022301001 | 0002066A9E | 94C9565248 | 59FD0 |
| A. facetus Uruguay | 2022301001 | 0002055C9F | A6A8454248 | 7ACE0 |
| A. cf. facetus Uruguay2 | 2021401001 | 000208A8BB | 76F6553235 | 677C0 |
| A. cf. facetus Uruguay1 | 2021401001 | 000209A8AB | 65F4542234 | 76BC0 |
| TYPESET * UNTITLED = ord: 1 3-5 8-9 16-34; | ||||
| WTSET * BETWEENSTATE = 5: 1-15 35; | ||||
Appendix 5
| 0000000000 | 1111111111 | 2222222222 | 33333 | |
| 1234567890 | 1234567890 | 1234567890 | 12345 | |
| Outgroup | 0?000?0?1? | ?00?0270B0 | 000??????? | ???00 |
| A. sp. forquilha | 0100100110 | 11110A5531 | 111013E10B | 10460 |
| A. cf. sp. forquilha 23060 | 0101100110 | 1??10G2D22 | 464530AC47 | 63030 |
| A. cf. sp. forquilha 23482 | 0101100110 | 1??10F1C15 | 842301C66C | 31310 |
| A. sp. jacui | 0110?01110 | 10010G0404 | GC3448806A | D8B80 |
| A. tembe | 0000100111 | 00001G4G4E | H30222920? | 4???0 |
| A. sp. pirapo | 1001010231 | 000307DAA7 | 35D8CCB963 | 24790 |
| A. sp. jacutinga | 1021010221 | 0003099B48 | 997C5D7772 | B2AB0 |
| A. sp. jacutinga Irai | 1021010221 | 00030E9F6A | DBA??????? | ????0 |
| A. sp. jacutinga Soberbio | 1021010221 | 00030D8E7E | FBGEBA6DC0 | E6CC0 |
| A. sp. jacutinga Iguaçu | ?0220????? | ?00305A84A | EDH??????? | ????0 |
| A. scitulus | 1011010221 | 000300FJCD | 52B7753A29 | 559A1 |
| A. scitulus quarai | 1011010221 | 000301EHDF | 63E98B9B98 | 492B1 |
| A. sp. uruguai | 1021201011 | 0000046156 | 6BF8671851 | CC5B0 |
| A. sp. paraguay | 1022101101 | 10000B4649 | BB8ADFD785 | DBD70 |
| A. facetus Argentina | 2022301001 | 0002086325 | 7B9DBE55BE | AA140 |
| A. facetus Uruguay | 2022301001 | 00020C3243 | 27CB9945AD | 77620 |
| A. cf. facetus Uruguay | 22021401001 | 000206C99C | A866C62336 | 9DD50 |
| A. cf. facetus Uruguay | 12021401001 | 000203B78B | CA51A40414 | 8E8?0 |
Appendix 6
Major morphometric and meristic differences between the putative species as summarized in a discriminant analysis
| facetus | facetoides | forquilha | jacui | jacutinga | uruguai | pirapo | paraguay | scitulus | tembe | |
|---|---|---|---|---|---|---|---|---|---|---|
| p = 0.0925 | p = 0.26432 | p = 0.13216 | p = 0.07489 | p = 0.07489 | p = 0.03965 | p = 0.03524 | p = 0.02203 | p = 0.23789 | p = 0.02643 | |
| HL/SL | 1.4295 | 3.34488 | −0.0095 | −3.8501 | −3.7253 | 0.2321 | −1.4753 | −0.2636 | −1.60236 | −0.6813 |
| Snout/HL | −5.1704 | −4.56985 | 6.3728 | 2.5658 | 1.2755 | 2.6012 | 2.0859 | −2.1373 | 1.55422 | 2.1572 |
| Bdep/SL | 2.5731 | −2.14628 | 1.0087 | 2.5425 | 3.3128 | −0.1336 | −1.9049 | 1.3215 | −1.0499 | 1.9113 |
| Orbit/HL | −6.3423 | −4.07887 | 7.9767 | 0.1332 | 3.8798 | 1.7378 | 5.3961 | −0.4356 | 0.45825 | −1.8294 |
| HW/HL | 1.1915 | 4.07271 | −1.7715 | −3.3657 | −4.8222 | −2.2698 | −1.9162 | −1.2795 | −0.53731 | −0.9796 |
| Intob/HL | 3.8198 | 1.74705 | −11.3921 | −0.8952 | 2.6089 | 7.2599 | 2.2763 | 5.3464 | 0.75264 | −3.889 |
| Preob/HL | −5.6135 | −2.80827 | 14.1836 | −1.2071 | −0.3688 | −4.6646 | 2.6313 | 0.1566 | −2.03844 | 2.9813 |
| Cpl/cpd | −0.4365 | −0.06442 | 3.7524 | 4.4211 | −2.3216 | −1.036 | −3.8678 | 0.1419 | −2.84472 | 9.6566 |
| Plen/SL | −1.6913 | −1.04804 | −0.4144 | −3.105 | 0.0378 | 0.6332 | 1.6837 | 1.0281 | 2.49084 | 0.6934 |
| Vlen/SL | 0.5834 | 0.16548 | 1.5923 | 2.1443 | 0.102 | −1.0629 | −0.6073 | −1.7758 | −1.42856 | −1.2821 |
| Dspl/SL | 0.7119 | 0.46975 | −1.0946 | −1.2214 | 0.3028 | −2.3498 | 0.7486 | 0.8744 | 0.43325 | −1.2151 |
| Prays | 0.8283 | 0.18996 | 0.3794 | 0.3492 | −2.3528 | −2.5755 | −1.4443 | −0.5488 | 0.46828 | 1.0133 |
| E0scale | 0.6395 | −0.06086 | 1.457 | −0.7865 | −1.268 | −0.8568 | 1.6323 | −2.029 | −0.3662 | 1.0019 |
| Cheek | −2.0506 | 0.39902 | 2.0732 | 1.2093 | −3.2412 | 1.8264 | −1.4474 | 1.2866 | −0.22439 | −1.2845 |
| C1raker | 2.4807 | 0.79502 | 4.1045 | −0.8269 | −3.1568 | −1.8101 | −1.0786 | 0.3852 | −2.36355 | −0.7636 |
| Vercaud | −2.3764 | −2.49419 | 4.9658 | −3.3731 | −1.6064 | −2.2929 | 5.096 | −3.4995 | 2.02261 | 3.8964 |
| Cpdvert | −2.5342 | 1.24744 | 2.4919 | 0.6924 | −0.8553 | −4.5523 | −4.4856 | −0.8036 | −0.77707 | 4.87 |
| Apterant | −1.4646 | 1.08383 | −1.5268 | −0.021 | −0.4545 | −0.9886 | −1.341 | −1.433 | 1.1026 | −2.1894 |
| Aspines | −2.7751 | 0.92035 | −2.7282 | −3.4513 | 0.3122 | −2.5944 | −0.7012 | −3.2662 | 3.63522 | −2.124 |
| Constant | −15.1303 | −7.10293 | −31.3893 | −14.689 | −14.8816 | −16.2619 | −15.0726 | −13.6168 | −8.16906 | −31.5437 |
- Highest and lowest values per character in bold. HL/SL, head length/SL; Snout/HL, snout length/HL; Bdep/SL, body depth/SL; orbit/HL, orbital diameter/HL; HW/HL, head width/HL; intob/HL, interorbital distance/HL; preob/HL, preorbital distance/HL; cpl/cpd, caudal peduncle length/caudal peduncle depth; plen/SL, pectoral fin length/SL; vlen/SL, ventral fin length/SL; dspl/SL, last dorsal fin spine length/SL; prays, pectoral fin ray count; E0scale, E0 scale count; cheek, cheek scale rows; c1raker, first lower ceratobranchial raker count; vercaud, caudal vertebrae count; cpdvert, caudal peduncle vertebrae count; apterant, anal pterygiophore count anteriorly from the first haemal spine; aspines, anal fin spine count.




