The systematic position of Aspidytidae, the diversification of Dytiscoidea (Coleoptera, Adephaga) and the phylogenetic signal of third codon positions
Die systematische Stellung der Aspidytidae, die Diversifikation der Dytiscoidea (Coleoptera, Adephaga) und das phylogenetische Signal der dritten Kodonpositionen
Abstract
enCharacters of the newly discovered larvae of the South African Cliff Water Beetle Aspidytes niobe were examined and integrated into a data matrix including all families of Dytiscoidea as well as Haliplidae. Fifty-three morphological characters of adults and larvae were analysed separately and combined with molecular data from six nuclear and mitochondrial genes. The phylogeny of the group is reconstructed for the study of the evolution of swimming behaviour and larval feeding habits, as well as the shift in diversification rates leading to the two most speciose lineages. The parsimony analysis of all equally weighted morphological and molecular characters combined resulted in a single well supported tree with the topology (Noteridae (Hygrobiidae ((Aspidytidae, Amphizoidae) Dytiscidae))), in agreement with the molecular data alone, but in contradiction to the morphological data, which favoured a topology in which Hygrobiidae is sister to Dytiscidae. The exclusion of third codon positions of the three protein coding genes resulted in a topology identical to that obtained with the morphological data alone, but the use of Bayesian probabilities or the amino acid sequence resulted in the same topology as that of the tree obtained with parsimony using all equally weighted characters. We concluded that interactions of third codon positions with the other data are complex, and their removal is not justified. There was a significant increase in the diversification rate at the base of the richest families (Noteridae and Dytiscidae), which could be associated with the development of simultaneous stroke and higher swimming performance, although data on the swimming behaviour of some basal groups of Noteridae are incomplete. The presence of larval mandibular sucking channels may have contributed to the diversification of Dytiscidae and the species-rich noterid genera Hydrocanthus and Canthydrus.
Zusammenfassung
deMorphologische Merkmale der neu entdeckten Larven der Aspidytidae wurden in einen Datensatz eingefügt, welcher alle Familien der Dytiscoidea sowie die Haliplidae umfasst. Insgesamt wurden dann 53 Merkmale von Imagines und Larven separat analysiert, sowie in Kombination mit DNA Sequenzdaten (vier mitochondriale und zwei kernkodierte Gene). Wir rekonstruieren damit die Phylogenie der Dytiscoidea, um die Evolution ihres Schwimmverhaltens, der larvalen Nahrungsaufnahmemechnismen sowie des Wechsels der Diversifikationsraten innerhalb der Gruppe zu beleuchten. Die Parsimonienanalyse aller morphologischen (‘‘equally weighted’’) und molekularen Merkmale kombiniert resultierte in einem einzigen Baum mit folgender Topologie: (Noteridae (Hygrobiidae ((Aspidytidae, Amphizoidae) Dytiscidae))). Diese stimmt mit dem Resultat der separaten Analyse von DNA-Sequenzdaten überein, nicht jedoch mit jener der morphologischen Merkmale alleine – hier waren Hygrobiidae die Schwestergruppe der Dytiscidae Der Ausschluss der dritten Kodonpositionen der drei proteinkodierenden Gene (CytB, COI, H3) führte zu einer Topologie, welche mit der separaten morphologischen Analyse kongruent ist. Allerdings führte eine Analyse inklusive der 3. Kodonpositionen mittels ‘‘Bayesian probabilities’’ oder der Aminosäuresequenzen zu der selben Topologie wie die kombinierte Parsimonieanalyse aller molekularen und morphologischen Daten. Darauf basierend folgern wir, dass die Interaktionen der 3. Kodonpositionen mit den anderen Merkmalen komplexer sind als erwartet – ihre Entfernung ist nicht zu rechtfertigen. Wir fanden einen signifikanten Anstieg der Diversifikationsraten an der Basis der artenreichsten Familien (Noteridae and Dytiscidae), welcher mit der Evolution des simultanen Ruderschlages der Schwimmbeine und damit verbesserter Schwimmfähigkeit bei diesen Familien zusammenhängen könnte. Das Auftreten von larvalen Mandibelsaugkanälen könnte die Diversifikation der Dytiscidae sowie der artenreichen Noteridengattungen Hydrocanthus und Canthydrus gefördert haben.
Introduction
A remarkable entomological discovery of the last years was the enigmfatic adephagan beetle family Aspidytidae (Ribera et al. 2002a; Balke et al. 2003) which has a highly disjunct range, with one species in China and one in South Africa. The recent finding of the larvae provides new important information for a more reliable placement of the taxon within Dytiscoidea, which form the major part of the aquatic representatives of adephagan beetles (including Noteridae, Amphizoidae, Hygrobiidae and Dytiscidae as defined by Bell 1966). The suborder Adephaga comprises c. 30 000 described species and most of them belong to the terrestrial Carabidae (ground and tiger beetles) (Shull et al. 2001). Some 5500 species of Adephaga are aquatic and have been assigned to seven families representing life forms as distinctive as the whirligig beetles (Gyrinidae), crawling water beetles (Haliplidae), burrowing water beetles (Noteridae) and predacious diving beetles (Dytiscidae), with 1000, 200, 250 and 4000 species respectively (Guignot 1933; Franciscolo 1979; Crowson 1981; Spangler 1981; Lawrence and Newton 1982; Ribera et al. 2002a; Balke et al. 2004a). They occur in a range of habitats from permanent and temporal ponds, streams and creeks, springs, underground rivers, high altitude pools, to water caught in epiphytic bromeliad plants, while a few of them are secondarily terrestrial (Wesenberg-Lund 1943; Spangler 1991; Balke et al. 2004a; Balke 2005).
In addition to the three large families of aquatic Adephaga, several isolated taxa have been recognized, which have apparently undergone very limited diversification. This includes the trout stream beetles (Amphizoidae) with five species in the genus Amphizoa; the squeak beetles (Hygrobiidae) including six species of Hygrobia; the recently described cliff water beetles (Aspidytidae) with two species in the genus Aspidytes; and Meruidae with a single species in Venezuela (Spangler and Steiner 2005, species not included here). These small families have widely disjunct distributions, posing a most interesting problem with respect to their geographical origin, age and phylogenetic relationships. Three species of Amphizoa occur on the west coast of North America and two in China (Kavanaugh 1986; Ji and Jäch 2003); Hygrobia occurs in four species in Australia, plus one species each in China and the Western Palaearctic (Hendrich 2001); and the two recently discovered species of Aspidytes are found in the Cape region of South Africa and in Central China (Balke et al. 2003).
These small families are also of interest because of their rather divergent ecology and behaviour. Only the Dytiscidae and Noteridae have developed advanced swimming abilities showing simultaneous stroke of all legs for propulsion, morphological modifications of the hind and middle legs and hydrodynamic body shape for increased swimming velocity and manoeuvrability (Nachtigall 1960; Ribera et al. 2002a). In contrast, the Aspidytidae and Amphizoidae instead crawl on wet rock faces or on logs, branches and wood debris in more or less fast flowing streams, respectively. Hygrobiids mostly crawl on the bottom of muddy ponds, although they are able to swim well. It is of great interest to explain the great discrepancies in apparent diversification rates of different lineages, and to correlate this to differences in the swimming and feeding behaviour, and possibly other factors.
Phylogenetic relationships of the small families are fundamental for understanding the diversification process in the group. Together with Noteridae and Dytiscidae, the small families have been grouped in the Dytiscoidea, a taxon introduced by Bell (1966) and confirmed as monophyletic by Beutel and Haas (1996) and recently by a combined cladistic analysis of morphology and DNA sequence data from three genes (18S rRNA, 16S rRNA and CO1) (Ribera et al. 2002a). The newly discovered family Aspidytidae was also shown to be part of this clade (Ribera et al. 2002a). However, the precise placement of the dytiscoid families relative to each other, and in particular the position of the small families, remained unsettled. For example, the Aspidytidae was found to be the sister to Hygrobia plus Dytiscidae, but alternative parameter settings in the tree searches placed it as sister to Amphizoa (Ribera et al. 2002a). Only an increased database can resolve the issues about dytiscoid relationships. We add three molecular markers to the existing data, with the aim of providing a robust hypothesis of the dytiscoid family relationships. In addition, we present an expanded morphological data set, which also includes the results of an anatomical study of the previously unknown larva of Aspidytes niobe (see also Alarie and Bilton 2005). We use the result of the cladistic analysis to analyse and discuss the causes and patterns of diversification within the Dytiscoidea.
Materials and Methods
Morphology
All described dytiscoid families were included in the study, with multiple representatives of the larger families (Appendix A). All trees were rooted in species of Haliplidae. Haliplids are an aquatic family clearly not part of Dytiscoidea, but possibly its sister group, as inferred from both morphology (e.g. Beutel 1995; Ribera et al. 2002a) and DNA sequence data (Ribera et al. 2002a,b). More distant groups were not included, as their relationships with Dytiscoidea are still controversial (Beutel 1995; Shull et al. 2001) and our focus was on intra-dytiscoid relationships.
Morphological studies of the newly discovered larvae of A. niobe were based on second and third instar larvae collected in the type locality (Ribera et al. 2002a), and their identity confirmed by sequencing a fragment of the CytB gene. Specimens were embedded in Historesin, sectioned at 3 μm, and stained with methylene-blue and acid fuchsine. Drawings were made using an ocular grid (Leica MZ8) or a camera lucida (cross-sections, Zeiss Axiolab). Kéler's (1963) muscular nomenclature was used in the text and the illustrations. Scanning electron microscopy was carried out with an FEI (Philips) XL 30 ESEM TMP after the specimens were critical point dried and sputter coated. The morphological data on larvae and adults of other groups are partly based on material in the collection of R.G. Beutel and partly extracted from the literature (e.g. Beutel 1993, 1995; Beutel and Haas 1996; Alarie et al. 2004). The data were not always available for the same species, but previous studies have shown that characters of the endo- and exoskeleton and of the musculature as used in the present analysis do not tend to vary at the generic level (e.g. Alarie et al. 2004). Morphological characters were summarized in a matrix provided in Appendix B.
DNA extraction, PCR, sequencing
Total DNA was extracted from single beetles either using the Qiagen DNeasy tissue kit (Qiagen, Hilden, Germany), or by standard phenol–chloroform extraction. Voucher specimens and DNA aliquots are kept in the Natural History Museum (London) and in the Museo Nacional de Ciencias Naturales (Madrid). Genes for molecular analysis were chosen to include different mtDNA regions (Otto et al. 1996) and several nuclear genes. We sequenced six gene regions, including the 3’ ends of the mitochondrial 16S rRNA (16S), the 3’ portion of cytochrome c oxidase I (COI) genes, a central fragment of cytochrome b (Cytb), a central fragment of 12S rRNA (12S), the full-length nuclear 18S rRNA gene (18S), and a fragment of histone H3 (H3) (Table 1). Gene fragments were amplified using standard PCR procedures (Ribera et al. 2002a; Balke et al. 2004b). Amplification products were purified with Qiagen Qiaquick PCR purification columns or Millipore Multiscreen 96-well plates (Millipore, Billerica, MA, USA). Sequencing reactions were purified by ethanol precipitation and electrophoresed on an ABI3700 sequencer. Sequences were edited using the Sequencher 4.1 software package (GeneCodes Corp., Ann Arbor, MI, USA). New sequences have been submitted to GenBank under accession numbers indicated in Appendix A.
| Gen | Primername | F/R | Primer sequence (fragment number) | Reference |
|---|---|---|---|---|
| CO1 | Jerry | F | CAA CAT TTA TTT TGA TTT TTT GG | Simon et al. (1994) |
| Pat | R | TCC AAT GCA CTA ATC TGC CAT ATT A | ||
| CytB | CB3 | F | GAG GAG CAA CTG TAA TTA CTA A | Barraclough et al. (1999) |
| CB4 | R | AAA AGA AA(AG) TAT CAT TCA GGT TGA AT | ||
| 12S | 12S ai | F | AAA CTA GGA TTA GAT ACC CTA TTA T | Simon et al. (1994) |
| 12S bi | R | AAG AGC GAC GGG CGA TGT GT | ||
| 16S | 16s aR (M14) | F | CGC CTG TTT AAC AAA AAC AT | Simon et al. (1994) |
| ND1 A (M223) | R | GGT CCC TTA CGA ATT TGA ATA TAT CCT | ||
| H3 | H3aF | F | ATG GCT CGT ACC AAG CAG AC(AG) CGC | Colgan et al. (1998) |
| H3aR | R | ATA TCC TT(AG) GGC AT(AG) AT(AG) GTG AC | ||
| 18S | 18S 5′ | F | GAC AAC CTG GTT GAT CCT GCC AGT (1) | Shull et al. (2001) |
| 18S b5.0 | R | TAA CCG CAA CAA CTT TAA T (1) | ||
| 18S ai | F | CCT GAG AAA CGG CTA CCA CAT C (2) | ||
| 18S b2.5 | R | TCT TTG GCA AAT GCT TTC GC (2) | ||
| 18S a1.0 | F | GGT GAA ATT CTT GGA CCG TC (3) | ||
| 18S bi | R | GAG TCT CGT TCG TTA TCG GA (3) | ||
| 18S a2.0 | F | ATG GTT GCA AAG CTG AAA C (4) | ||
| 18S 3′I | R | CAC CTA CGG AAA CCT TGT TAC GAC (4) |
Phylogenetic analysis
We opted for a two-step procedure for the alignment of length variable regions, with an initial hypothesis of character homology (i.e. an alignment) and a subsequent, independent step of tree search (Phillips et al. 2000; Simmons 2004).
Different alignments were constructed with ClustalX (Thompson et al. 1994) using various parameter settings for gap opening and gap extension cost (Table 2). Starting from a ClustalX default (gap opening/extension 15/6.66), we build a manual alignment based on the visual identification of conserved motives flanking length variable portions, as alignments constructed in a similar way were shown to perform better than alignments directly obtained by ClustalX as measured with the ILD (e.g. Balke et al. 2004b; Ribera et al. 2004) or taxonomic congruence (Ouvrard et al. 2000; Xia et al. 2003; Kjer 2004). The hypervariable 18S gene regions V4 and V6 (Tautz et al. 1988) were excluded from the analysis, as homology statements among nucleotide positions were difficult to hypothesize.
| Alignment/part weights | Length of individual partitions | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 18S | 16S | 12S | CO1 | Cytb | H3 | Morph | Sum part. | Length comb. | ri | ILD | WILD | Chars | Trees | Topology | |
| Mol 1 | 569 | 665 | 469 | 1252 | 720 | 523 | – | 4198 | 4331 | 0.415 | 133 | 0.03 | 4155 | 1 | ((d(as + am))h) |
| Mol 2 | 601 | 707 | 496 | 1252 | 720 | 523 | – | 4299 | 4441 | 0.398 | 142 | 0.032 | 4159 | 3 | ((d(as + am))h) |
| Mol 3 | 581 | 688 | 495 | 1292 | 725 | 523 | – | 4304 | 4444 | 0.394 | 140 | 0.031 | 4218 | 2 | ((d(as + am))h) |
| Manual | 569 | 665 | 469 | 1252 | 720 | 523 | – | 4198 | 4331 | 0.415 | 133 | 0.03 | 4155 | 1 | ((d(as + am))h) |
| Combined analysis, equal | 569 | 665 | 469 | 1252 | 720 | 523 | 90 | 4287 | 4426 | 0.439 | 138 | 0.031 | 4208 | 1 | ((d(as + am))h) |
| Morph = 4 | 569 | 665 | 469 | 1252 | 720 | 523 | 360 | 4558 | 4706 | 0.5 | 148 | 0.031 | 4208 | 2 | ((d + h)(as + am)) |
| Morph = 30 | 569 | 665 | 469 | 1252 | 720 | 523 | 2700 | 6898 | 7046 | 0.726 | 148 | 0.021 | 4208 | 2 | ((d + h)(as + am)) |
| Morph = 1, no 3rd | 569 | 665 | 469 | 286 | 220 | 41 | 90 | 2340 | 2402 | 0.555 | 62 | 0.025 | 3734 | 1 | ((d + h)(as + am)) |
| Morph = 1, no 3rd CO1 | 569 | 665 | 469 | 286 | 720 | 523 | 90 | 3322 | 3450 | 0.474 | 128 | 0.037 | 3955 | 3 | (d + as + am + h) |
| Morph = 1, no 3rd CO1 and CytB | 569 | 665 | 469 | 286 | 220 | 523 | 90 | 2822 | 2929 | 0.512 | 107 | 0.036 | 3837 | 2 | (d + h(as + am)) |
| Morph = 4, no 3rd | 569 | 665 | 469 | 286 | 220 | 41 | 360 | 2610 | 2672 | 0.632 | 62 | 0.023 | 3734 | 1 | ((d + h)(as + am)) |
| Morph = 1, 18S = 5 | 2845 | 665 | 469 | 1252 | 720 | 523 | 90 | 6564 | 6723 | 0.503 | 159 | 0.023 | 4208 | 1 | (((d + h)as)am) |
| Morph = 1, 18S = 5, no 3rd | 2845 | 665 | 469 | 286 | 220 | 41 | 90 | 4616 | 4687 | 0.599 | 71 | 0.015 | 3955 | 1 | (((d + h)as)am) |
| Morph = 1, 18S = 10 | 5690 | 665 | 469 | 1252 | 720 | 523 | 90 | 9409 | 9578 | 0.546 | 169 | 0.018 | 4208 | 1 | (((d + h)as)am) |
| Morph = 1, CO1 = 5 | 569 | 665 | 469 | 6260 | 720 | 523 | 90 | 9296 | 9483 | 0.4 | 187 | 0.019 | 4208 | 1 | ((((as + am)d)d)d)h) |
| Morph = 1, 16S = 5 | 569 | 3325 | 469 | 1252 | 720 | 523 | 90 | 6948 | 7100 | 0.475 | 152 | 0.021 | 4208 | 1 | ((d(as + am))h) |
| 18S, 16S, CO1 | 569 | 665 | – | 1252 | – | – | – | 2486 | 2514 | 0.468 | 28 | 0.011 | 3116 | 1 | ((d(as + am))h) |
| 18S, 16S, CO1 no 3rd | 569 | 665 | – | 286 | – | – | – | 1520 | 1533 | 0.566 | 13 | 0.008 | 2863 | 1 | (((d + h)as)am) |
- Mol 1, 2, 3 – Clustal X alignments of the molecular data (mol 1, gap opening/gap extension was 15/6.66; mol 2, 8/4; mol 3, 4/2); sum part. – sum length individual partitions; length comb. – tree length in combined analysis; ILD: incongruence-length difference test (Farris et al. 1994; ILD = length combined – sum length individual partitions); WILD: Wheeler's ILD (Wheeler and Hayashi 1998; WILD = ILD/length combined); Chars – number of characters. Topologies of Dytiscoid families: d, Dytiscidae; as, Aspidytidae; Am, Amphizoidae; h, Hygrobiidae.
Results obtained with alternative alignments were compared using the incongruence length difference test to evaluate our ‘manual’ alignment (ILD; Mickevich and Farris 1981; Farris et al. 1994; Yoder et al. 2001) and Wheeler's normalized ILD (Wheeler and Hayashi 1998; ‘WILD’: Dowton and Austin 2002).
The data matrix was partitioned in 10 character sets, corresponding to each of the six genes, plus the morphological character set, and further separating the third codon positions for each of the three protein coding genes (for Bayesian inference the third positions were pooled). Parsimony analysis was conducted under equal weights in PAUP* version 4.0b10 (Swofford 2002), using TBR heuristic searches with 1000 random addition sequences. Gaps were coded as a fifth character state (Giribet and Wheeler 1999). Molecular and morphological data were analysed separately and in combined analysis. Node support was measured with Partitioned Bremer Support values (PBS) (Baker and DeSalle 1997), searching on constraint trees generated with TreeRot (Sorenson 1996). Non-parametric bootstrapping was conducted with 1000 pseudoreplicates and 100 random additions each (Felsenstein 1985). Parsimony jackknife replications (Farris et al. 1996) were based on 1000 replicates with 100 random addition sequences and deletion of 37% of the data. Following Grant and Kluge (2003), these analyses were designed to test which clades remain weakly supported and thus constitute the object of future concern.
Preliminary analyses of the data revealed that differences with previous results (Ribera et al. 2002a) and between the results obtained with morphological and molecular data where apparently because of the inclusion or exclusion of third codon positions of the protein-coding genes. There is a considerable body of literature concerning the adequacy of including or excluding third codon positions (which are in general assumed to be more homoplastic, introducing more ‘noise’ and less ‘signal’ than first or second codon positions) in phylogenetic studies (e.g. Baker and DeSalle 1997; Källersjö et al. 1999; Baker et al. 2001; Cognato and Vogler 2001; Damgaard and Cognato 2003; Simmons et al. 2004b). We opted for a combined analysis, weighting all characters equally as suggested in the aforementioned works, but evaluated our results in two ways. One possible way of partially by-passing the possible analytical artefacts introduced by third codon positions is to use the amino acid instead of the nucleotide sequence of the genes (but see Simmons 2000; Felsenstein 2004) which was obtained using MacClade 4.0 (Maddison and Maddison 2000). It must be noted that transversions in the third codon positions do have an effect in the coded amino acid, so by the use of the amino acid sequence not all third position changes are ignored. Secondly, the use of model-based phylogenetic methods should in principle be less sensitive than parsimony to the biases introduced by highly saturated or homoplasious data (e.g. Swofford et al. 1996). We performed exploratory analyses using Bayesian probabilities (Rannala and Yang 1996) as implemented in the computer program MrBayes 3.0b4 (Huelsenbeck and Ronquist 2001), as it allows the estimation of different evolutionary models for the user-defined data partitions. We used a GTR model (Tavaré 1986) with gamma distributed among site rate variation and estimating the proportion of invariable sites (Yang 1993). The parameters of all partitions were estimated independently. Despite some risk of bias because of over-parameterization (Lemmon and Moriarty 2004), we opted for a complex model to favour the expression of possible differences in the evolutionary models among partitions, maximizing their effect in the resulting tree topology. Searches were conducted using the default priors (uniform probabilities) starting with random trees, with three heated and one cold Markov chains for 1 000 000 generations, sampled at intervals of 100 generations. To determine the point at which the Markov chains reached stationarity, the log-likelihood scores were plotted against generation time, and visually determined when the log-likelihood values reached a stable equilibrium.
Posterior probabilities were used to assess node support. Although generally higher when compared with bootstrap support, posterior probabilities higher than 95% can be taken as indicative of strong node support (Suzuki et al. 2002; Alfaro et al. 2003; Douady et al. 2003; Erixon et al. 2003; Simmons et al. 2004a). In fact, Huelsenbeck and Rannala (2004) suggest that, assuming the correct model has been chosen, the posterior probability of a tree is the probability that the tree is correct.
Rate of lineage diversification
We evaluated possible differences in the diversification rates of sister clades within Dytiscoidea, trying to establish the origin of the highly diverse lineages within the group. The probability of observing a significant difference of species richness in sister clades can be estimated under the equal-rates Markov random branching model (Yule 1924; Nee et al. 1994). All possible partitions of N species into the two clades are equi-probable (Farris 1976), and thus the (two-tailed) probability of an equal or greater magnitude of split under the null model is given by p = 2n/(n + N−1), where n is the number of species of the less diverse clade, and N the number of species in the richest clade (Slowinski and Guyer 1989a,b; Mayhew 2002). The rejection of the null model of cladogenesis suggests that the sister pair has experienced non-random rates of diversification, i.e. speciation or extinction rates were significantly different between the two lineages.
Results
Morphological characters
Adults
1. Head shape: (0) not shortened and laterally rounded, eyes protruding (1) shortened, laterally rounded and streamlined, eyes not protruding. The head is shortened and laterally rounded (1.1) in adults of Aspidytes, Noteridae, and Dytiscidae. The eyes are not protruding in the adults of these taxa.
2. Shape of scapus: (0) parallel-sided, longer than wide; (1) shortened, large globular basal part; (2) shortened, basal and distal part globular; (3) shortened, without globular basal part. A shortened scapus with a large globular basal part and a moderately elongate distal part is present in Noteridae (Beutel and Roughley 1987; Belkaceme 1991) (2.1). The scapus is divided into a globular proximal part and an equally globular distal portion in Aspidytes (2.2). The scapus of Haliplidae is also strongly shortened, but without enlarged globular part (2.3).
3. Pedicellus: (0) elongate, not enclosed by distal part of scapus; (1) strongly shortened and partly enclosed by distal part of scapus. A strongly shortened pedicellus partly enclosed by the globular distal part of the scapus (3.1) is characteristic for adults of Aspidytes.
4. Flagellomeres of males: (0) elongate, not broadened; (1) antennomeres 3–9 slightly broadened; (2) antennomeres 5 and more than one of the following segments distinctly broadened. Expansion of antennomeres in males (e.g. 5, 7, 9; Belkaceme 1991) is a characteristic feature of Noteridae (4.2). Antennomeres 3–9 are very slightly dilated in Aspidytes (4.1). Expansion of antennomeres of males or females also occurs in few derived representatives of Dytiscidae (not included in present study).
5. Galea: (0) palp-like, 2-segmented; (1) palp-like, 1-segmented. The galea is one-segmented (5.1) in Amphizoa (Beutel 1988). It is 2-segmented (5.0) in Aspidytes and adults of other groups of Adephaga.
6. Sensorial field of distal labial palpomere: (0) absent or very small and not transverse; (1) present, elongate; (2) present, large covering whole apical part of palpomere. A transverse, elongate sensorial field is present on the apex of the distal labial palpomere (6.1) in adults of most noterid genera (absent in Notomicrus; Belkaceme 1991). A conspicuous sensorial field covers the whole apical part of the apical palpomere in Aspidytes (6.2). A very small sensorial field is present on each of the subapices in adults of Copelatus (6.0).
7. Shape of prosternal process: (0) rather elongate, converging towards apex; (1) posteriorly rounded; (2) strongly broadened and apically truncate. The prosternal process is rounded postero-laterally or posteriorly (7.1) in adults of Aspidytes, Phreatodytes, Mesonoterus, Noterus (Belkaceme 1991: Fig. 66), Amphizoa, and Trachypachidae. It is strongly broadened postero-laterally and with truncate hind margin (7.2) in Haliplidae and most genera of Noterinae (Siolius, Renotus, Suphis, Hydrocanthus, Canthydrus, Suphisellus; Belkaceme 1991). The apical part is acuminate or at least converging towards the apex (7.0) in adults of Dytiscidae and Hygrobia, and also in basal representatives of Noteridae (Notomicrus, Hydrocoptus; Belkaceme 1991: Figs 63–65).
8. Ventral procoxal joint: (0) with distinct coxal condyle; (1) coxal condyle strongly reduced. The ventral procoxal joint is largely reduced (8.1) in adults of Dytiscidae (Baehr 1979). It is distinct in all other groups of Adephaga (8.0).
9. Profemoral cleaning device: (0) absent; (1) present. A profemoral cleaning device is present (9.1) in Aspidytes and adults of all noterid genera (Belkaceme 1991), and also in some representatives of Dytiscidae (e.g. Copelatus, Laccophilus). It is absent in all other groups of Adephaga (9.0).
10. Protibial burrowing spur: (0) absent; (1) one large spur; (2) two large spurs. The presence of a strong, curved protibial spur (10.1) is a characteristic feature of Noterinae excluding Notomicrus, Hydrocoptus and Pronoterus (Beutel and Roughley 1987). Two large, flattened burrowing spurs (10.2) are present in adults of Hygrobia (Beutel 1986).
11. Protibial row of flattened thorns on apical part of protibia: (0) absent; (1) present. A regular, short row of flattened, ribbed spines is present (11.1) on the outer apical edge of the protibia of Noterinae excluding Notomicrus, Hydrocoptus and Pronoterus (Belkaceme 1991). It is extended towards the proximal part of the protibia in Hydrocanthus, Canthydrus and Suphisellus (Belkaceme 1991). The row of thorns is absent in other adephagan and non-adephagan beetles.
12. Outer edge of apical part of the protibia: (0) not rounded; (1) rounded. The outer edge of the protibial apex is rounded (12.1) in adults of Noterinae excluding Notomicrus, Hydrocoptus and Pronoterus (Belkaceme 1991).
13. Tibial groove or concavity for reception of burrowing spur: (0) absent; (1) present. A tibial concavity for reception of the burrowing spur is present (13.1) in adults of Noterinae excluding Notomicrus, Hydrocoptus, Pronoterus and Noterus (Belkaceme 1991). It is a furrow in adults of Synchortus and Mesonoterus, but a deep pit in adults of the remaining genera (Belkaceme 1991).
14. Curved spurs on ventral side of protarsomeres 1–3: (0) absent; (1) present. Curved spurs are present (14.1) on the protarsomeres 1–3 of adults of Noterinae excl. Notomicrus (Belkaceme 1991).
15. Prothoracic defense gland: (0) absent; (1) present. Prothoracic glands are present (15.1) in Hygrobia and Dytiscidae, but absent in other groups of Adephaga or non-adephagan beetles. Minor structural differences were pointed out by Forsyth (1968, 1969). For a discussion of the possible convergent evolution in Hygrobiidae and Dytiscidae see Miller (2001).
16. Mesocoxal cavity: (0) laterally bordered by mesepimeron; (1) laterally bordered by mesepimeron and metathoracic anepisternum. In Amphizoa and most representatives of Dytiscidae (e.g. Copelatus, Laccophilus), the mesocoxal cavities are laterally closed by the mesepimeron and the metathoracic anepisternum (complex-type sensuBell 1967) (16.1). The same condition is found in Aspidytes. The mesocoxal cavities are only closed by the mesepimeron (disjunct-type sensuBell 1967) (16.0) in adults of the other aquatic groups, in extant members of Trachypachidae, and in basal groups of Carabidae (e.g., Bell 1967).
17. Swimming hairs on meso- and metathoracic legs: (0) absent; (1) sparse fringe of very thin and fine hairs; (2) dense row of longer hairs. Dense rows of hairs are present in adults of Haliplidae, Hygrobiidae, and Dytiscidae, and also in most representatives of Noteridae (17.2). Only a sparse fringe of fine hairs (17.1) is present in Amphizoa and Notomicrus.
18. Noterid platform of metaventrite: (0) absent; (1) present. A flattened, platform-like median part of the metaventrite is characteristic for Noterinae excl. Notomicrus and Phreatodytes (Beutel and Roughley 1987) (18.1). It is absent in all other adephagan beetles.
19. Transverse ridge of metaventrite: (0) complete; (1) partly reduced; (2) absent. The transverse ridge of the metaventrite is fully present (19.0) in Haliplidae, and partly reduced (19.1) in adults of Aspidytes, Trachypachidae, Hygrobia and Amphizoa. It is absent in Noteridae, and Dytiscidae (25.2).
20. Articulation between pro- and metasternal process: (0) absent; (1) present. The prosternal process articulates with the metasternal process (20.1) in Haliplidae, Noterinae, Hygrobiidae and Dytiscidae (articulation lost in few specialized species) (Baehr 1979). This is not the case in Aspidytes, Amphizoa and other groups of Adephaga, even thought the apex of the prosternal process may reach the metasternal process when the ventral intersegmental muscles are contracted.
21. Metafurca: (0) originates from katepisternum; (1) originates from intercoxal septum. The metafurca originates from the intercoxal septum (and not from the katepisternum) in all Dytiscoidea examined (21.1).
22. Size of metafurca: (0) large, with well developed lateral arms; (1) strongly reduced in size, lateral arms present. The metafurca of Amphizoidae is strongly reduced in size (22.1), but short lateral arms are present (Beutel 1988). The metafurca is well developed with large lateral arms in most adephagan beetles.
23. Mesal walls of metacoxa: (0) free; (1) connected, contact area extensive (forming an intercoxal septum). A broad mesal attachment area and an extensive intercoxal septum (23.1) is characteristic for all Dytiscoidea (Beutel 1995). The mesal metacoxal walls are free (23.0) in adults of Haliplidae (Belkaceme 1986) and Carabidae.
24. Anterior margin of the metacoxa: (0) almost straight or slightly extended anterolaterally; (1) with distinct angle; (2) rounded, strongly extended anteriorly. A characteristic anteromesal angle (24.1) of the metacoxa is present in all adults of Noterinae (Beutel and Roughley 1987). The metacoxae are strongly extended anteriorly (24.2) in adults of Dytiscidae, and Phreatodytinae (Uéno 1957). The anterior margin is rounded in Dytiscidae, but oblique in the latter taxa, which are also characterized by a strongly reduced metaventrite (Uéno 1957). The metacoxae are slightly extended anteriorly (24.0) in Amphizoa, Hygrobia and Aspidytes, and the anterior margin is almost straight like in adults of Trachypachidae and Carabidae.
25. Metacoxal plates: (0) well developed, moderately sized; (1) largely reduced; (2) strongly enlarged. Metacoxal plates are largely reduced (25.1) in adults of Dytiscidae and Hygrobiidae (and †Charonoscaphinae; Ponomarenko 1977), but very strongly enlarged (25.2) in Haliplidae (and †Triaplidae; Ponomarenko 1977). Moderately sized metacoxal plates (25.0) are present in Aspidytes, Amphizoa, Noteridae, and Geadephaga.
26. Lateral edge of metacoxal plates: (0) diverging anteriorly; (1) converging anteriorly; (2) equally rounded, reaching anterior margin of metacoxae laterally. The lateral edges of the metacoxal plates are strongly diverging anteriorly and reach the anterior metacoxal margin laterally in Trachypachidae and Carabidae. They are at least slightly diverging (26.0) in Aspidytes, Amphizoa, Hygrobia, and Dytiscidae. A clearly delimited internal lamina of the metacoxal plates with anteriorly converging external edges (26.1) is characteristic for Noterinae. The external edge of the plates is equally rounded and reaches the anterior coxal margin laterally in Haliplidae (26.2).
27. Mm. furca-coxalis anterior: (0) present; (1) absent. The muscle is absent (27.1) in adults of Dytiscoidea and Gyrinidae excl. Spanglerogyrus (Larsén 1966; Beutel 1990), but present (27.0) in Haliplidae (Belkaceme 1986) and geadephagan adults.
28. Mm. furca-coxalis posterior: (0) present; (1) absent. The muscle is present (28.0) in Geadephaga and Haliplidae (Belkaceme 1986), but absent (28.1) in adults of Dytiscoidea and Orectochilus (Larsén 1966; Baehr 1975; Beutel and Haas 2000).
29. Abdominal segments III and IV: (0) clearly separated; (1) largely or completely fused. The line separating the abdominal sternites III and IV is extremely indistinct or absent (29.1) in Noteridae (Uéno 1957; Belkaceme 1991: Figs 63 and 65–69). It is distinct (29.0) in other Adephaga.
30. Laterotergite: (0) orientation strictly caudal, laterotergite articulates with cranial portion of gonocoxa; (1) strictly cranial orientation of laterotergite; (2) short apodeme extended cranially, rest of laterotergite with caudal orientation. In females of Aspidytes, Amphizoa, Hygrobia and Dytiscidae the laterotergite articulates with the cranial portion of the gonocoxa and has usually a strict caudal orientation in the resting position (37.1). In Noteridae the laterotergite has either a short apodeme, which is extended cranially while the rest has a more or less caudal orientation (Notomicrus) (30.1), or the laterotergite is much longer than the gonocoxa and completely orientated cranially (see Burmeister 1976) (37.0). A similar condition is present in Haliplidae and Gyrinidae (Burmeister 1976). However, the laterotergite is only slightly longer than the gonocoxa (37.0).
Larvae
31. Egg bursters in first instar larvae: (0) present; (1) absent (Alarie and Bilton 2005). Egg bursters are absent (38.1) in larvae of Gyrinidae and Haliplidae (Ruhnau 1985; Beutel 1993), but are present (38.0) in first instar larvae of A. niobe and in first instar larvae of the other groups of Adephaga. The presence in second instar larvae of Aspidytidae (Alarie and Bilton 2005) is an autapomorphy of that family.
32. Shape of anterior margin of clypeolabrum: (0) with distinct nasale; (1) with strongly reduced nasale in median concavity (2) evenly convex. A prominent, completely immobilized nasale with four distinct teeth as it is found in most larvae of Gyrininae, in basal groups of Carabidae (e.g. Metrius, Carabus, Nebria; Thompson 1979; Arndt 1993), and in first instar larvae of Hydrotrupes (Dytiscidae; Beutel 1994) is arguably a groundplan feature of Adephaga. A distinct but modified nasale without teeth is present in larvae of A. niobe (Fig. 1a,c,d). It is indented medially and densely covered with short spines and sensilla (32.0). Six to eight small teeth are present in larvae of Trachypachidae (Arndt and Beutel 1995). The nasale is represented by a small, complex structure with a pair of paramedian triangular teeth or strongly modified sensorial pegs in larvae of Noterus and Canthydrus (32.1) (R.G. Beutel, personal observations; Dettner 2004: Fig. 6.3.1.F,H), but is apparently absent (32.2) in larvae of Suphis (Spangler and Folkerts 1973; Spangler 1991: ‘anterior margin arcuate’). It is also absent (32.2) in larvae of Haliplidae (Jaboulet 1960; Beutel 1986), Amphizoa (Beutel 1991), Hygrobia (Alarie et al. 2004) and almost all larvae of Dytiscidae (excl. Hydrotrupes). In these cases, the anterior clypeolabral margin is evenly rounded.
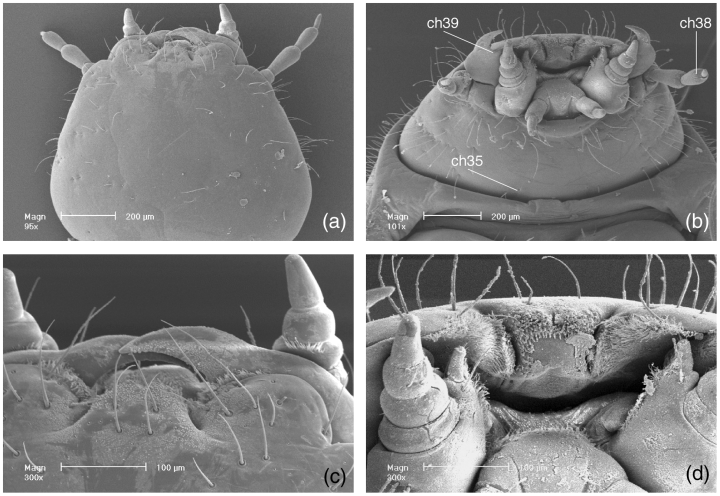
Larva of Aspidytes niobe, SEM micrographs (ch35, 38, 39 = character number, see Appendix B and Results). (a) head, dorsal view; (b) head, ventral view; (c) clypeolabral region, dorsal view; (d) clypeolabrum and mouthparts, frontal view; (e) abdominal tergite I, with spiracle; (f) abdominal tergite IV, with spiracle
33. Short sensorial setae of anterior clypeolabral margin of third instar larvae: (0) 6 or less; (1) 24 or more. The presence of four short sensilla on the nasale or anterior frontoclypeolabral margin is probably a groundplan feature of Adephaga (33.0). This condition is found in larvae of Gyrinidae, Trachypachidae (Bousquet and Goulet 1984: coded as FR10−11), in most larvae of Carabidae (Bousquet and Goulet 1984; Arndt 1993) and in first instar larvae of Dytiscidae: Hydrotrupes (Beutel 1994; Alarie et al. 1998). Only one pair (33.0) is present in larvae of Haliplidae and Noteridae according to Ruhnau (1986), even though two pairs of strongly modified sensorial structures seem to be present in larvae of Canthydrus (Dettner 2004: Fig. 6.3.1.F,H). Six sensilla (33.0) are present in larvae of A. niobe, but difficult to distinguish from the surrounding spines (Fig. 1d). An increased number (33.1) of 24 pointed sensorial setae is present in third instar larvae of Amphizoa (Ruhnau 1986; Beutel 1991), and 24 or more spatulate setae (lamellae clypeales; Bertrand 1972) are present in third instar larvae of Hygrobia (Alarie et al. 2004) and Dytiscidae (e.g. Bertrand 1972; De Marzo 1976a,b; Ruhnau 1986; Alarie et al. 1998).
In contrast to other interpretations (e.g. Alarie et al. 1998, 2002) we assume that the spatulate setae or ‘lamellae clypeales’ are homologous with the short spine-like sensilla (sensorial pegs) of the anterior clypeolabrum. The alternative would imply that the spatulate setae would have evolved de novo (e.g. the four spatulate setae described for first instar larvae of Hydrotrupes by Alarie et al. 1998), which appears much less likely than to assume that some or all setae have modified their shape.
34. Frontal arms (frontal suture): (0) straight or evenly curved; (1) with indistinct indentation; (2) sinuate. The frontal arms are sinuate in all carabids larvae (incl. Rhysodidae), in all known larvae of Noterinae (suture obsolete in Phreatodytes), and in most larvae of Hydroporinae (Bertrand 1972; Alarie 1991; Beutel 1993). They form a V posteriorly and are almost straight anteriorly (34.1) in larvae of A. niobe (Fig. 1a). An indistinct indentation is present at the level of the attachment of the dorsal tentorial arm. The frontal sutures form a V or U (Copelatus) and are straight or evenly curved (41.0) in larvae of Amphizoa and Hygrobia, and in most larvae of Dytiscidae (Bertrand 1972; Alarie 1991; Alarie et al. 1998).
35. Origin of posterior tentorial arms: (0) central region of ventral wall of head capsule or slightly posterior to it; (1) adjacent to foramen occipitale. The posterior tentorial arms of most larvae of Adephaga originate in the central region of the ventral wall of head capsule (42.0). They are slightly shifted posteriorly in larvae of A. niobe (35.0) (Fig. 1b, head capsule retracted ventrally), but adjacent with the foramen occipitale (35.1) in noterine larvae (Bertrand 1972; Ruhnau 1985; Beutel 1993; condition in Phreatodytes unknown).
36. Caudal tentorial arm: (0) absent or very short; (1) present elongate and slender. A short caudal tentorial arm (36.0) is present in larvae of Trachypachus, Noterus (Beutel 1993) and Hydrocanthus. It is strongly elongated and slender and attached to the head capsule posteriorly in larvae of Amphizoa (Beutel 1991: Fig. 4), Hygrobia (Alarie et al. 2004), Dytiscidae (De Marzo 1979; Ruhnau 1986), and A. niobe (36.1) (Fig. 2).
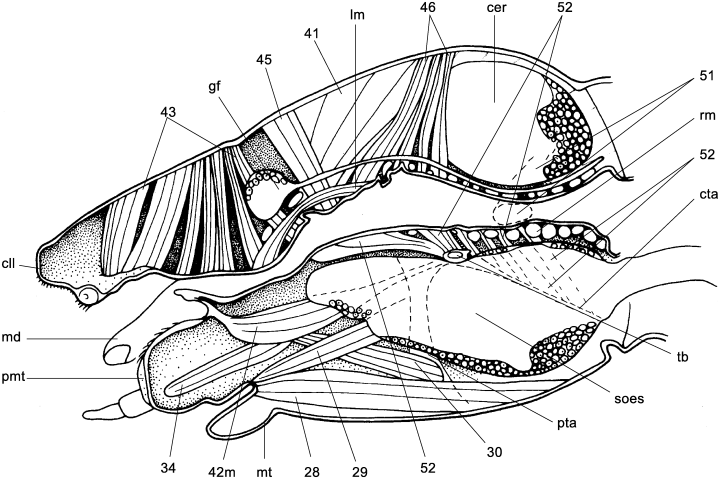
Larva of Aspidytes niobe. (A) head, sagittal section; abbreviations: cer, cerebrum; cta, caudal tentorial arms; gf, frontal ganglion; hy, anterior hypopharynx; lm, longitudinal muscle; md, mandible; mt, mentum; nrec, nervus recurrens; ph, pharynx; pl, palpus labialis; pmt, prementum; pph, prepharynx; pta, posterior tentorial arms; soes, suboesophageal ganglion; 11, M. craniomandibularis externus; 28, M. submentopraementalis; 29, M. tentoriopraementalis inferior; 30, M. tentoriopraementalis superior; 43, M. clypeopalatalis; 41, M. frontohypopharyngalis; 41, M. tentoriohypopharyngalis; 45, M. frontobuccalis anterior; 46, M. frontobuccalis posterior; 51, M. verticopharyngalis; 52, M. tentoriopharyngalis
37. Tentorial bridge: (0) connecting posterior arms; (1) connecting proximal part of caudal arms; (2) distinctly shifted posteriorly on caudal arms; (3) reduced, with posteriorly shifted origin of M. tentoriohypopharyngalis. The tentorial bridge connects the posterior tentorial arms (37.0) in most adephagan larvae. It arises from the proximal part of the caudal arms (37.1) in larvae of A. niobe (Fig. 2) and Amphizoa, but is distinctly shifted posteriorly on the slender caudal arms (37.2) in larvae of Dytiscidae. The tentorial bridge is absent or extremely reduced (37.3) in larvae of Hygrobia (Alarie et al. 2004). The origin of M. tentoriohypopharyngalis is distinctly shifted posterioroly, which suggests a caudal position of the bridge before its reduction.
38. Sensorial appendage of penultimate antennomere: (0) absent; (1) present. A distinct sensorial appendage is present (38.1) on the subapical antennomere of larvae of Trachypachidae (strongly modified; Arndt and Beutel 1995), Carabidae (Thompson 1979; Arndt 1993), Haliplidae, Hygrobia, and Dytiscidae, and also in larvae of A. niobe (Fig. 1b). It is strongly shortened in larvae of Amphizoa (38.1), and absent (38.0) in larvae of Noteridae.
39. Retinaculum: (0) present; (1) strongly reduced in size; (2) absent. The retinaculum is distinct (39.0) in most larvae of Adephaga, but absent (39.2) in Dytiscidae and Hygrobia. It is very small or vestigial (39.1) in larvae of Haliplidae and Amphizoa, but well developed in almost all geadephagan larvae and in larvae of Noteridae and A. niobe (Alarie and Bilton 2005) (Fig. 1b).
40. Mandibular groove in mature larvae: (0) completely absent, one mesal cutting edge; (1) two distinct cutting edges delimiting a distinct groove; (2) mandibular sucking channel. One mesal edge of the mandible (40.0) is present in mature larvae of Carabidae and Hygrobiidae (Ruhnau 1986). An upper and a lower cutting edge (40.1) is present in larvae of Trachypachidae (Arndt and Beutel 1995), Noterus (Ruhnau 1985; R. Beutel, personal observation), Amphizoa (Beutel 1991: Fig. 6), Copelatus, Hydrotrupes (Beutel 1994), and A. niobe. Mandibular sucking channels (40.2) are present in larvae of Gyrininae, Haliplidae, Hydrocanthus and Canthydrus (Ruhnau 1986), and in larvae of Dytiscidae (excl. Copelatus and Hydrotrupes; Beutel 1994). They are short and restricted to the proximal part of the mandible in the noterid genera.
41. Adductor tendon: (0) undivided; (1) divided into an upper and lower portion. The mandibular adductor tendon is divided into an upper and a lower portion (41.1) in larvae of Amphizoa (Beutel 1991; Fig. 6a), Hygrobia (Alarie et al. 2004), Dytiscidae (Ruhnau 1986), and A. niobe. In Hygrobiidae and Dytiscidae the two branches of the tendon are separated by a strong lateral component of M. verticopharyngalis.
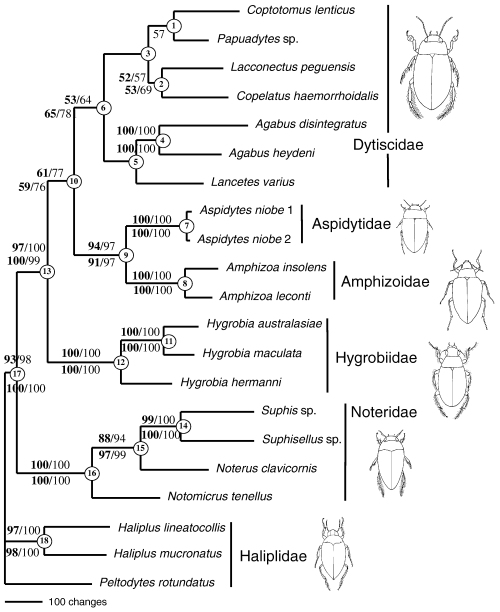
Phylogram of the single most parsimonious tree obtained with the analysis of the molecular data (all characters equally weighted, gaps as fifth character state). The topology is the same as that of the single most parsimonious tree obtained with the analysis of the combined morphological and molecular data. Above branches, bootstrap/jackknife support values for the molecular data alone; below branches, bootstrap/jackknife support values for the combined data. See Table 4 for the partition Bremer support values of the different nodes
42. Maxillary groove: (0) present; (1) absent. The maxillary groove is distinct in larvae of Gyrinidae, partly reduced but still present (49.0) in larvae of Haliplidae (Beutel 1993: Fig. 7), and completely absent (42.1) in all other adephagan larvae including A. niobe (Fig. 1b). The complete reduction results in a strongly increased moveability of the maxilla in all directions.
43. Articulation of cardo or maxillary base: (0) head capsule without elongate articulatory process articulating with socket on cardo base; (1) elongate articulatory process of head capsule articulates with socket of cardo base; (2) maxillary base retracted. An elongate, slender process of the head capsule articulates with a socket of the cardo (43.1) in larvae of Noteridae (Beutel 1993: Fig. 9), A. niobe, and Dytiscidae (Ruhnau 1986). This process is very short or absent in larvae of Haliplidae (Beutel 1993: Fig. 7) and Trachypachidae (Beutel 1993: Fig. 8). A condyle of the cardo articulates with a socket of the head capsule in carabid larvae (Ruhnau 1986; Beutel 1991: Fig. 2). The maxillary base is moderately retracted (43.2) in larvae of Amphizoidae (Beutel 1991: Fig. 2) and inserted into a deep pouch (43.2) in larvae of Hygrobiidae (Alarie et al. 2004).
44. Cardo: (0) as broad as stipital base; (1) strongly narrowed or reduced. A distinctly reduced cardo, which is about half as broad as the stipes (44.1) is characteristic for larvae of Noteridae (Beutel 1993: Fig. 9), Amphizoa (Beutel 1991: Fig. 2), Dytiscidae (Bertrand 1972), and A. niobe. The cardo is scarcely recognizable in some specimens of A. niobe because of its almost vertical position when the maxilla is retracted (Fig. 1b). The cardo is absent or fused with the stipes (44.1) in larvae of Hygrobia (Alarie et al. 2004). A broad cardo (44.0) is present in larvae of Gyrinidae, Haliplidae and Trachypachidae (Beutel 1993: Figs 6–8). It is represented by a mesal and a lateral sclerite in most larvae of Carabidae (Beutel 1993: Fig. 10).
45. Position of cerebrum: (0) posterior part of head (Fig. 2); (1) anterior part of head. The cerebrum is strongly shifted anteriorly (45.1) in larvae of Dytiscidae (De Marzo 1979) and Hygrobia (Alarie et al. 2004). A similar condition is not found in other larvae under consideration.
46. Trochanteral annulus: (0) absent; (1) present. A trochanteral annulus is present (46.1) in larvae of Hygrobia and Dytiscidae, but absent in other taxa under consideration (this and the following three characters are from Alarie and Bilton 2005).
47. Primary setae FE7-FE10: (0) absent; (1) present. These setae are present (47.1) in larvae of A. niobe, Hygrobia, Amphizoa and Dytiscidae, but absent in larvae of the other groups of Adephaga.
48. Additional pore on tibia: (0) absent; (1) present. An additional pore is present on the tibia of all legs (48.1) in larvae of A. niobe and Amphizoa, but absent in larvae of the other groups of Adephaga.
49. Segment IX: (0) well developed; (1) distinctly reduced (3, 4); (2) vestigial or absent. The abdominal segment IX is distinctly reduced but still visible in dorsal view (49.1) in larvae of A. niobe. It is vestigial or absent (49.2) in larvae of the other groups of Dytiscoidea.
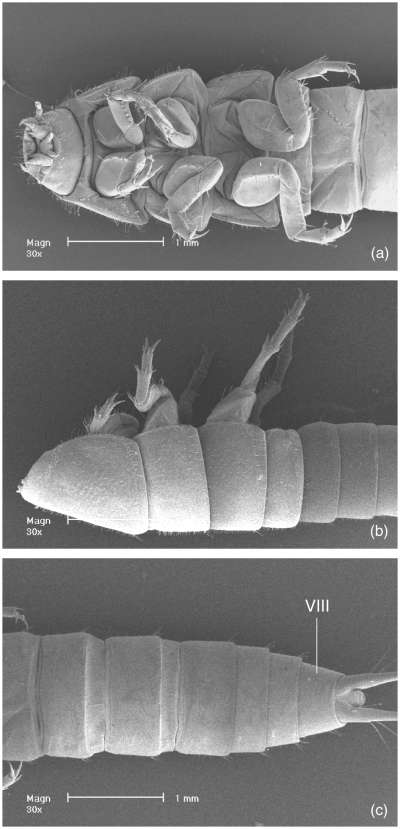
Larva of Aspidytes niobe, SEM micrographs. (a) head, thorax and anterior abdomen, ventral view; (b) thorax and anterior abdomen, lateral view (head removed); (c) posterior abdomen, ventral view (VIII = segment VIII)
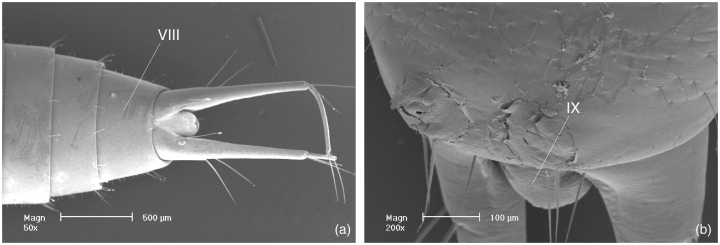
Larva of Aspidytes niobe, SEM micrographs (VIII, IX = segment number). (a) abdominal apex with urogomphi, ventral view; (b) segments VIII and IX, dorsal view
50. Segment X: (0) present; (1) absent. The abdominal segment X is absent (50.1) in larvae of all groups of Dytiscoidea including A. niobe (2, 4).
51. Spiracle VIII: (0) normally developed; (1) enlarged, terminal; (2) reduced; (3) small, shifted to dorsal side of segment VIII. Enlarged terminal spiracles VIII are present (51.1) in larvae of Amphizoidae and Dytiscidae. They are reduced (51.2) in larvae of Hygrobia, Haliplidae, and Gyrinidae, and very small and shifted to the dorsal side of tergite VIII (51.3) in A. niobe (Fig. 7b). They are terminal and large in larvae of Noteridae, but non-functional in 3rd instar larvae (Spangler 1991; 51.1).
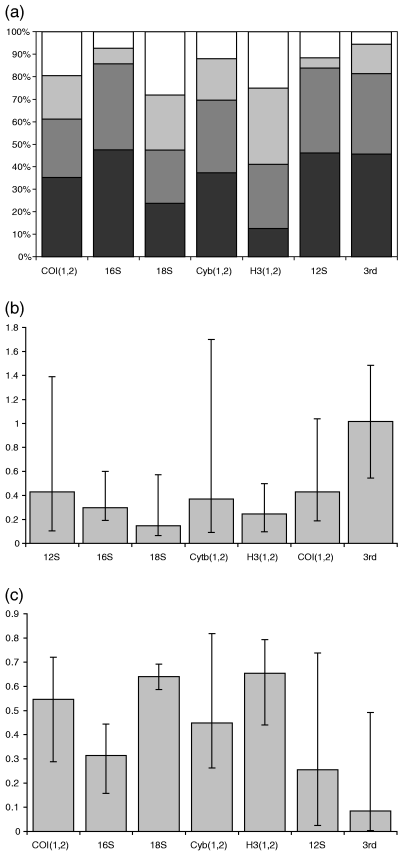
Parameters of the Bayesian analysis of the nucleotide data, with third codon positions of the protein coding genes (COI, Cytb, H3) grouped as a separate partition. A GTR + I + G model was optimized for each partition separately (see text for details). (a) Proportion of nucleotides (from top down: G, C, A, T); (b) parameter alfa of the gamma distribution; (c) proportion of invariable sites. Error bars in (b) and (c), 95% confidence intervals
52. Collar-like semi membranous connections between sclerites: (0) absent; (1) present. Longitudinally striated, collar-like semi membranous connections between body segments (52.0) are characteristic for larvae of A. niobe (Fig. 3b), Noterus and Canthydrus (Dettner 2004). This condition is apparently absent from larvae of Suphis (Spangler and Folkerts 1973). However, it is possible that these structures are only visible if the body segments are distinctly extended. The presence or absence in larvae of Phreatodytes is unknown.
53. Size of urogomphi: (0) longer than maximum width of head capsule; (1) shorter. The urogomphi are very elongate and slender (53.0) in larvae of A. niobe (Fig. 4a), Hygrobia (Alarie et al. 2004), and many larvae of Dytiscidae (Bertrand 1972; Alarie et al. 2002). They are comparatively short in larvae of Amphizoa and Copelatus (53.1) and very short (53.1) in larvae of Noteridae (Uéno 1957; Spangler 1991). The urogomphi are also distinctly or completely reduced in larvae of Trachypachidae (Arndt and Beutel 1995).
Larval feeding apparatus of Aspidytes niobe
The head is slightly inclined but clearly prognathous and the labrum is fused to the clypeus (1, 3). The functional mouth is wide open. A preoral or prepharyngeal filter apparatus is absent (Fig. 1d). The lateral margins of the posterior hypo- and epipharynx are fused, thus forming a closed prepharyngeal tube (Fig. 2). The prepharyngeal dilators (M. clypeopalatalis, M. 43 v. Kéler 1963) are only moderately developed (Fig. 2). The anterior hypopharynx is distinctly separated from the upper side of the prementum, and retractile (Fig. 2). The dorsal postcerebral dilator (Fig. 2; M. verticopharyngalis, ‘M. 51’: v. Kéler 1963) is very large. Together with the very strong ventral pharyngeal dilator (Fig. 2; M. tentoriopharyngalis, ‘M. 52’: v. Kéler 1963), which arises from the strongly elongated posterior tentorial arms, ‘M. 51’ forms a very strong postcerebral, pharyngeal pumping apparatus.
The structural features suggest that larvae seize their prey with the sickle-shaped mandibles, squeeze it against the clypeolabral margin (Fig. 1b,d), process it with the well developed retinaculum and the cutting edges (Fig. 1b), and mingle it with digestive fluid in the space between the mesal mandibular furrow and the epi- and hypopharynx. The maxillae, which are moveable in all directions, are also likely involved in the handling of food particles (see Spence and Sutcliffe 1982). The more or less liquified parts of the prey are sucked back through the comparatively wide prepharynx and pharynx (Fig. 2).
Phylogenetic analysis of morphological and molecular data
Parsimony analysis of the 53 morphological characters under equal weights resulted in five shortest trees of 90 steps, CI (consistency index) = 0.83 and RI (retention index) = 0.93. In the strict consensus tree of all most parsimonious trees (Fig. 5) Noteridae are the sister of all other Dytiscoidea, comprising four lineages, i.e.: (Aspidytidae + Amphizoidae) and (Hygrobiidae + Dytiscidae). Nodes defining family relationships are supported with bootstrap and jackknife values over 75%, but are lower for the node grouping Aspidytes and Amphizoa (Fig. 5). In the following, the family relationships as inferred from the morphological data will be referred to as ‘topology 1’.

Strict consensus of the five most parsimonious trees obtained with the analysis of the morphological data. Numbers above branches are bootstrap/jackknife support values, below branches Bremer support values
The combined molecular data set of our preferred manual alignment included 4155 characters, of which 989 were parsimony informative. They were contributed to about equal proportions by the three rRNA and three protein coding genes, with about twofold lower character consistency in the latter as measured with the ILD (Table 3). A heuristic search of the manually aligned matrix including all six molecular markers resulted in a single tree of 4331 steps with CI = 0.44 and RI = 0.42. This tree showed Noteridae as sister to the remaining Dytiscoidea, Hygrobiidae sister to (Dytiscidae + Amphizoidae + Aspidytidae), and Aspidytidae sister to Amphizoidae (Fig. 6; hereafter referred to as topology 2). Support for most deep nodes was high, but lower for the node defining the monophyly of Dytiscidae (Fig. 6, node 6) and the monophyly of (Dytiscidae + Aspidytidae + Amphizoidae) (Fig. 6, node 10). The COI gene contributed the largest proportion of the total PBS, followed by H3 and 18S, whereas the total PBS for CytB was negative (Table 4). In each of the three protein coding genes, the first and second positions showed several negative nodal support values, in contrast to the third positions which played a key role in driving PBS in the simultaneous analysis tree (Table 4). Alternative alignments constructed with ClustalX, or removing all 130 character columns containing indels from the matrix, yielded the same family relationships. ILDs for alternative ClustalX alignments were the same (default) or slightly higher than for the manual alignment (Table 2).
| Partition | Unaligned | Aligned | Inf | Const | ri | ci | Trees |
|---|---|---|---|---|---|---|---|
| 18S | 1797–1812 | 1834 | 174 | 1513 | 0.6585 | 0.7065 | 55 |
| 16S | 506–512 | 524 | 175 | 296 | 0.5326 | 0.4722 | 2 |
| 12S | 354–364 | 374 | 133 | 202 | 0.4925 | 0.4968 | 1297 |
| CO1 | – | 758 | 258 | 433 | 0.3764 | 0.3794 | 2 |
| Cytb | – | 354 | 152 | 170 | 0.3436 | 0.3764 | 373 |
| H3 | – | 311 | 97 | 187 | 0.3887 | 0.4015 | 39 |
| Sum mol | – | 4155 | 989 | 2801 | 0.415 | 0.4399 | 1 |
| Morph | – | 53 | 53 | 0 | 0.9296 | 0.8333 | 5 |
| Combined | – | 8363 | 2031 | 5602 | 0.439 | 0.4474 | 1 |
| Node | CO1, 1 & 2 | CO1, 3 | 16S | 18S | CB, 1 & 2 | CB, 3 | H3, 1 & 2 | H3, 3 | 12S | Morph | Sum |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 6 | 7 | 5.5 | 0 | −3.5 | 1 | 0.5 | 10 | 3 | −0.5 | 29 |
| 2 | −3 | 5 | −3 | −2 | −2 | 1 | 1 | 6 | 0 | −1 | 2 |
| 3 | −3 | 5 | −3 | −2 | −2 | 1 | 1 | 6 | 0 | −1 | 2 |
| 4 | 3 | 3.7 | 3.3 | −1 | −3.3 | −1 | 0.3 | 3.7 | −1.7 | 0 | 7 |
| 5 | −3 | 5 | −3 | −2 | −2 | 1 | 1 | 6 | 0 | −1 | 2 |
| 6 | −1 | −1 | 2 | 2 | 2 | −1 | 0 | 0 | 0 | 4 | 7 |
| 7 | 7 | 16 | 13 | 23 | 0 | 0 | 0 | 0 | 18 | 11 | 88 |
| 8 | 2 | 13 | 7 | 24 | 1 | 2 | 3 | 15 | 12 | 2 | 81 |
| 9 | −1 | 16 | 0 | −10 | 0 | −1 | 2 | 9 | 1 | −3 | 13 |
| 10 | 2 | 3 | 1 | 0 | 0 | 2 | 0 | 0 | −3 | 0 | 5 |
| 11 | 1 | 19.5 | 13.5 | 28 | −2.5 | 0.5 | 0.5 | 6 | 7.5 | −1 | 73 |
| 12 | 6 | 1 | 5 | −2 | 2 | 5 | 0 | 0 | 9 | 3 | 29 |
| 13 | −0.5 | 0.5 | 4 | 9.5 | −4 | −0.5 | 0.5 | 4 | 1 | 4.5 | 19 |
| 14 | 1.5 | 1 | 10.5 | 5 | −1 | 2 | 0.5 | 6 | 3 | 0.5 | 29 |
| 15 | 4 | 0 | 5 | 6 | −3 | −2 | 0 | −2 | 0 | 6 | 14 |
| 16 | 7 | −1 | 8 | 9 | 3 | 0 | −1 | −3 | 0 | 4 | 26 |
| 17 | −1 | 2.5 | −3 | 11 | −1 | 0.5 | 1.5 | 4.5 | 1 | 9 | 25 |
| 18 | 4 | 5.5 | 4 | −1 | 0 | 4 | −0.5 | 2.5 | 3 | 0.5 | 22 |
| Sum PBS | 31 | 101.7 | 69.8 | 97.5 | −16.3 | 14.5 | 10.3 | 73.7 | 53.8 | 37 | 473 |
| Lgth const | 292 | 973 | 671 | 581 | 246 | 520 | 47 | 519 | 482 | 95 | 4426 |
| PBS/lgth const | 0.106 | 0.104 | 0.104 | 0.168 | −0.066 | 0.028 | 0.219 | 0.142 | 0.112 | 0.389 | 0.107 |
The simultaneous analysis of all molecular and morphological data (4208 characters, 1042 informative) resulted in a single tree with length 4426, RI = 0.44 and CI = 0.45, and a topology identical to the tree from molecular data alone (i.e. topology 2, Fig. 6). Support was again high for most nodes, with a slight increase in support levels over the molecular based tree for the two weakest nodes (Fig. 6, nodes 6 and 10). The morphological data contributed less than one-tenth of the total PBS, but the signal per character was almost four times greater than the average of the molecular characters (Table 4).
Investigating the role of third codon positions
The current data set contains three protein coding regions, compared with only one in Ribera et al. (2002a), with nearly half of the total character changes and nodal support contributed by fast evolving third positions. The latter show incongruence with the morphological characters in all three protein coding genes. In pairwise comparisons of PBS values of the morphological partitions and the COI and H3 partitions, Spearman rank correlations were significantly negative (−0.5, p < 0.1, 16 d.f. and −0.6, p < 0.05), respectively, whereas the correlation with the Cytb third positions was −0.2 (n.s.). There was only one other case of negative correlation with the morphological characters, the first and second positions of H3 (−0.4, n.s.), but this pertained to a very small partition with little statistical power. The third positions of the COI gene were also found to produce incongruence in the previous analysis (Ribera et al. 2002a) and were removed from the preferred set of parameters that was chosen based on character congruence. We tested the role of third codon positions in the simultaneous analysis in more detail, in particular to assess the possibility of spurious phylogenetic signal arising from the presumed higher level of homoplasy in these characters.
First, the analysis was conducted with the exclusion of the third positions of the three protein coding genes (3734 characters, 672 parsimony informative) which resulted in a single tree of 2402 steps, with CI = 0.54; RI = 0.56, and a topology identical to that obtained with the morphological data only (topology 1), contrary to that obtained when all nucleotides were included (topology 2).
Second, the three protein coding genes were translated into amino acids (AA) and their AA sequences analysed together with the nucleotide sequences for the three ribosomal genes, producing a matrix of 3205 characters, of which 569 are informative (87 of them amino acid characters). Under equal weighting two shortest trees of 2153 steps and CI = 0.56 and RI = 0.55 were obtained, the consensus of which had the same family relationships as the tree obtained based on the nucleotide sequence (topology 2). Node support for the sister relationship between Aspidytes and Amphizoa was high (bootstrap support 80%), but was lower for their sister relationship with Dytiscidae. The node defining the Noteridae as sister to all other dytiscoids had a bootstrap support of 99%, and that defining Dytiscoidea 87%.
We also tested the effect of potential bias in evolutionary rates of third position characters by applying a model based method. Bayesian analysis on the full set of nucleotide data under a GTR + I + G model (estimated for each gene separately) resulted in a topology identical to that obtained using parsimony (topology 2). All nodes had a high posterior probability (Table 5), and the family relationships also remained fully resolved in the strict consensus of the top three topologies (with a joint probability of 0.98), further indicating the high levels of stability for the basal nodes. Only when the fourth most likely topology was included (with a pooled probability of 0.985) the position of Hygrobia became unresolved. Bayesian analysis was also conducted with the estimation of a separate evolutionary model for the third codon positions of all protein coding genes pooled as a seventh partition. This analysis demonstrated differences in the estimated parameters in the third versus all other positions, showing more extreme AT bias, a much lower proportion of invariable sites, and a higher alpha parameter (shape parameter of the gamma distribution), suggesting a more homogeneous between-character rate variation (Fig. 7) (although note the large confidence intervals of the estimations, rendering most differences among partitions not statistically significant). Under these settings topology 2 was recovered again, indicating that a model allowing a separate estimation of the variation in the third positions resulted in the same topology, despite apparent differences in tempo and mode of their variation. Posterior probabilities were also high, with all nodes defining family relationships at 100%, and only the node including (Dytiscidae (Amphizoidae, Aspidytidae)) at 98% (Table 5).
| Bayesian analysis | Node | ||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
| All nucleotides, partitions = genes (6) | 100 | 99 | 100 | 100 | – |
| All nucleotides, partitions = genes + third positions (7) | 100 | 98 | 100 | 100 | – |
| Nucleotides, excluding third positions, partitions = genes (6) | 97 | – | 100 | 100 | 74 |
| Nucleotides and amino acids, partitions = ribosomal, protein coding genes (2) | 100 | 98 | 100 | 100 | – |
- Nodes: 1, (Aspidytidae, Amphizoidae); 2, (Dytiscidae (Aspidytidae, Amphizoidae)); 3, Dytiscoidea excluding Noteridae; 4, Dytiscoidea; 5, (Hygrobiidae, Dytiscidae). See text for details of the analyses.
When third positions of all protein coding genes were excluded in the bayesian analysis, the optimal topology placed Hygrobia as sister to Dytiscidae (with posterior probability 74%), and Aspidytidae sister to Amphizoidae (posterior probability 97%), i.e. recovering topology 1 and in accordance with the parsimony analysis on the same data set (Table 5). Similarly, the results from the bayesian and parsimony analysis were consistent when translated amino acid sequences were analysed in combination with the nucleotide data of the ribosomal genes. Using two partitions, the AAs with a MTREV (Adachi and Hasegawa 1996) + I + G model, and the pooled ribosomal genes with a GTR + I + G model, topology 2 was obtained. The posterior probabilities were also high, with all nodes defining family relationships having 100%, and only the node including Dytiscidae plus (Amphizoidae, Aspidytidae) having 98% (Table 5). The results of bayesian and parsimony analyses were thus identical: when the third codon positions of the protein coding genes are excluded Hygrobiidae is sister to Dytiscidae, and when they are included Hygrobiidae is sister to Dytiscidae plus (Amphizoidae + Aspidytidae). The special role of the third codon positions cannot thus be readily attributed to a different model of evolution that could be accounted for by a model-based phylogenetic approach.
Discussion
New data or different method?
The two major differences in the preferred topology of Dytiscoidea compared with an earlier study (Ribera et al. 2002a) were the sister relationship of Aspidytidae and Amphizoidae, and of both families with Dytiscidae, leaving Hygrobia in a more basal position within Dytiscoidea. The larval characters of Aspidytes were previously unknown, and the molecular data included the 18S, COI and 16S genes only. The procedures for data analysis also differed in the earlier study, using a direct optimization method for nucleotide homology assignment, as implemented in the computer program POY (Wheeler et al. 2002). Searching a large parameter space that included different weights for the insertion of indels and the conserved and variable portions of each marker, the optimal parameters selected based on the congruence criterion (Wheeler 1995) were a gap cost = 2, with third positions of COI excluded, and the weight of the 18S partition set to five times the weight of the other data sets. Applying the same molecular data (18S, 16S, COI) and parameter settings of Ribera et al. (2002a) to the current alignment in a parsimony search using PAUP*, we obtained a single tree of 4292 steps, CI = 0.66 and RI = 0.65, with the same topology as in the previous study [i.e. (((Dytiscidae, Hygrobiidae) Aspidytidae) Amphizoidae) Noteridae)]. Only using 18S, 16S, COI, and removing third positions of COI, also led to the lowest ILD/WILD among a number of differential weighting schemes explored here (Table 2). Setting all characters (including indels) to equal weight did not change that topology (a single tree of 1625 steps, CI = 0.57, RI = 0.61). However, including the third positions of COI resulted in a single tree of 2608 steps with CI = 0.49, RI = 0.50 that did change the topology with respect to that of the simultaneous analysis (topology 2). Finally, when all molecular partitions were included, but without the third positions, a heuristic search resulted in a single tree (2402 steps, CI = 0.54, RI = 0.56) with the same topology as that obtained with the morphological data alone (topology 1).
Hence the discrepancies with the previous study are not because of the different analytical procedures, but are mostly affected by the inclusion of additional molecular data. However, with the expanded data set only the sister relationship between Aspidytidae and Amphizoidae is widely supported (topologies 1 and 2), whereas the basal position of Hygrobiidae (toplogy 2) apparently depends on the signal from third codon positions. This finding was independent of the method of tree construction or the evolutionary model applied to the third positions, indicating complex interactions among the phylogenetic information of the three codon positions in the protein coding genes. Interestingly, the tree obtained with the analysis of the morphological data alone produced the same topology (topology 1) as the molecular data excluding the third positions. The role of the third positions in the incongruence of molecular and morphological data would be supported by the fact that third positions also showed significantly conflicting signal in tests of character congruence (Table 4).
The conflicting role of third codon positions could lead to the option of excluding them from the analyses. However, data removal should be considered with caution, in particular when it affects nearly half of the total number of character changes. First, even if incongruence of the third positions with some other data (morphology in our case) is clearly apparent, its level of significance is rather low. For example, constraining the analysis of all data to the topology obtained with morphological characters alone (i.e. Hygrobiidae sister to Dytiscidae, topology 1) as a backbone constraint in a heuristic search resulted in shortest trees of 4436 steps, compared with a shortest unconstrained tree of 4426 steps (topology 2), a difference which was only marginally significant (p = 0.099) according to the non-parametric test of Templeton (1983). Hence the argument for data removal based on incongruence would depend on a rather low level of conflict.
Second, the sister relationship of Amphizoidae and Aspidytidae was only recognized in both the morphological and the molecular data after increasing the size of the character matrix beyond that available in Ribera et al. (2002a), turning what was initially recovered as a paraphyletic series into a strongly supported clade. The addition of further data, even if highly homoplastic, could add further phylogenetic signal, and hence removal of third positions would lead to a less desirable tree. This view is, perhaps, supported by the fact that the translated sequence also supports the topology of the combined analysis (topology 2), although the bias resulting from homoplasy or nucleotide composition should be less apparent under this type of character coding. As such, it would be unjustified to discard a priori a data partition. Likelihood models that estimate separate parameters for the third positions do not result in a topology different from that obtained with parsimony (i.e. they also recover topology 2), even if they are supposed to account for the higher homoplasy of these data. If the third position indeed produce spurious results, these findings indicate that model based analyses of the kind used here would not overcome biases of character evolution.
Implications for the phylogeny of Dytiscoidea
Our analyses confirmed the position of Noteridae as sister to the remaining Dytiscoidea (Beutel 1993; Ribera et al. 2002a,b), but contrary to traditional taxonomic arrangements which considered noterids as a subfamily of Dytiscidae (e.g. Sharp 1882; Guignot 1933; Franciscolo 1979) and morphological analyses considering them as sister to Dytiscidae (Kavanaugh 1986; Miller 2001).
The placement of Aspidytidae within Dytiscoidea was here also supported by larval characters: the small size of the cardo (character state 44.1), and the absence of segment X (50.1). Larval segment IX (character 49) is distinctly reduced in size in Aspidytes (49.1), but still clearly visible in dorsal view in contrast to larvae of Noteridae, Amphizoidae, Hygrobiidae and Dytiscidae (49.2) (4, 7; Y. Alarie, personal communication). The spiracles VIII of Aspidytes differ strongly from the typical dytiscoid condition (enlarged and terminal, 51.1): their small size and dorsal position on tergite VIII (51.3.) is an autapomorphy of Aspidytes, likely to be related to their hygropetric habitat.
The inclusion of Aspidytes within a clade comprising Dytiscoidea exclusive of Noteridae is supported by a longitudinally divided larval mandibular adductor tendon (41.1), and the presence of very long and thin caudal tentorial arms (36.1). This placement of Aspidytidae within Dytiscoidea excl. Noteridae implies that the enlarged tergite IX in Aspidytes is secondarily derived, or that a strong degree of reduction has taken place at least two times independently within Dytiscoidea. It also implies that the collar-like semi-membranous connections between the body segments have evolved independently in Noteridae and Aspidytidae (52.1).
The position of Hygrobiidae outside of (Dytiscidae + Aspidytidae + Amphizoidae) in the analysis containing all molecular data (topology 2), contradicts most previous phylogenetic studies of the group. Most recent studies suggested a sister relationship of Dytiscidae and Hygrobiidae (Ruhnau 1986; Beutel and Haas 1996; Shull et al. 2001; Ribera et al. 2002a). However, this hypothesis was weakly supported by morphological evidence. Larval synapomorphies suggested by Ruhnau (1986) are problematic due to insufficient taxon sampling (Y. Alarie, personal communication, 2000). The only potential synapomorphy of Dytiscidae + Hygrobiidae found by Beutel and Haas (1996) was the presence of prothoracic defensive glands. However, Miller (2001), based on structural differences pointed out by Forsyth (1968, 1969), suggested independent gain of such glands. The hypothesis presented here implies either that, or secondary loss in (Aspidytidae + Amphizoidae).
Additional potential synapomorphies of Dytiscidae and Hygrobiidae found in this study are the strongly reduced condition of the metacoxal plates (25.1), the cranial shift of the larval cerebrum (45.1), the absence of the larval retinaculum (39.1), and the presence of a trochanteral annulus in the larvae (46.1). The alternative hypothesis of Hygrobiidae outside of the clade (Dytiscidae (Aspidytidae Amphizoidae)) implies homoplasy in these cases, and also other characters such as the strongly increased number of labral pegs or spines (33.1), which is found in larvae of Amphizoa, Hygrobia and Dytiscidae, but not in Aspidytes. It would also imply that the comparatively large abdominal tergite IX in the larvae of A. niobe (49.1) is because of reversal (see above).
Evolution of adult swimming behaviour
The evolution of the swimming behaviour and related morphological traits were reconstructed by Ribera et al. (2002a). The only change introduced by the family-level topology presented here (Fig. 6) is the single loss of swimming abilities in the branch leading to (Aspidytidae + Amphizoidae), instead of the alternative of two independent loses postulated in Ribera et al. (2002a). This, along with observations suggesting alternate leg movement in basal Noteridae now reveal that the alternate locomotory mode is indeed a groundplan character of the Dytiscoidea + Haliplidae.
Adults of Aspidytidae are stout beetles lacking the swimming hairs that are characteristic of most hydradephagan species. This morphology is common to most hygropetric hydradephagans (see Ribera et al. 2003 for a review).
Evolution of larval feeding in Dytiscoidea
The results of the analysis show that a feeding apparatus without larval mandibular sucking channels (40.0) is ancestral for Dytiscoidea, and that advanced feeding habits have evolved independently in Dytiscidae (absent in Copelatinae and Hydrotrupes) and the species-rich noterid genera Hydrocanthus (c. 50 spp.) and Canthydrus (c. 60 spp.; not included in the analysis). Whether the presence may be ancestral for Adephaga has to be clarified in a broader phylogenetic context. Sucking channels are also present in Gyrininae (very short in Gyrinini, possibly absent in the unknown larvae of Spanglerogyrus) and Haliplidae (tubular, with prominent apex; Beutel 1986). The prepharyngeal structures (including musculature) associated with the mandibular sucking channels differ considerably. The prepharynx is transverse and short in Dytiscidae, but elongated in Gyrinini, Orectochilini, Haliplidae, and the noterid genera.
The larval feeding apparatus of A. niobe is probably very close to the ancestral condition of Dytiscoidea. The larvae possess several apomorphic groundplan features of the suborder Adephaga, which are correlated with carnivorous feeding habits: prognathous head (2, 3), labrum fused to the clypeus (Fig. 1a,b), mandibles with well developed retinaculum (39.0), but lacking a mola and a prostheca, maxilla inserted at or close to the anteroventral margin of head capsule (Fig. 1b), and lateral margins of posterior hypo- and epipharynx fused (prepharyngeal tube, Fig. 2). The open functional mouth is clearly plesiomorphic (Fig. 1d). More or less elaborate closing mechanisms have evolved in Dytiscidae (De Marzo 1979), apparently in correlation with the presence of mandibular sucking channels (absent in Aspidytes, see above). Other plesiomorphic features of larvae of A. niobe are the absence of a strong and complex prepharyngeal pumping apparatus (present in Dytiscidae; De Marzo 1979; Beutel 1986, 1993), the absence of a dense pre-oral filter with long hairs (Fig. 1d) (present in Carabidae; Beutel 1993), the absence of a prepharyngeal filter apparatus (present in Gyrininae; Beutel and Roughley 1994), and the presence of a well developed and retractile anterior hypopharynx (Fig. 2). The latter condition is also found in Amphizoidae and basal noterids, and also in larvae of Trachypachidae (Beutel 1993). A completely flattened anterior hypopharynx is characteristic for ‘higher’ Carabidae (see Beutel 1993).
A derived condition which has evolved within Dytiscoidea is the very large size of the dorsal postcerebral dilators (Fig. 2; M. verticopharyngalis, ‘M. 51’: v. Kéler 1963) and the presence of a very strong postcerebral pumping apparatus. This is a likely synapomorphy of the dytiscoid families with the exception of Noteridae (Beutel 1993; Alarie et al. 2004). Another derived feature within Dytiscoidea is the complete loss of the retinaculum (character state 39.2). This is either a synapomorphy of Hygrobiidae and Dytiscidae (topology 1) or has evolved independently in these two taxa (topology 2) (as in enhydrine and orectochiline Gyrindae).
Whereas the larval feeding apparatus of Amphizoa and Noteridae (excluding Hydrocanthus and Canthydrus) is similar to that of A. niobe, the structures involved with the feeding process are highly derived in Hygrobia. Hygrobiid larvae possess mandibles with only one cutting edge (second and third instar larvae) and without retinaculum (see above), strongly retracted and simplified maxillae, an enlarged ligula, a highly modified epi- and hypopharynx, and an exceptionally strong pharyngeal musculature (Alarie et al. 2004). The preferred prey of larvae of Aspidytes is unknown. However, it is noteworthy that at least the larvae of the other two groups comprising only few species are restricted to a very specialized diet, i.e. tubificid worms in Hygrobiidae and plecopteran larvae in Amphizoidae.
The diversification of Dytiscoidea
Numbers of species differ dramatically among the families of Dytiscoidea, with most lineages being species poor and only two relatively rich clades: Dytiscidae (with 3810 species, Nilsson 2001, 2003) and the subfamily Noterinae within the Noteridae, with 235 species (Nilsson, unpubl.) (the uncertainties of the internal phylogeny of Dytiscidae do not allow more precision in the characterization of the species-rich clades). Using the equal-rates Markov random branching model, and irrespective of the position of Hygrobiidae, the two species-rich clades show a clear asymmetry with respect to their closest sisters, although in the case of the Noterinae this is only marginally significant (Table 6).
| Species poor clade | Species rich clade | n | N | p | |
|---|---|---|---|---|---|
| 1 | Aspidytidae + Amphizoidae | Dytiscidae | 7 | 3810 | 0.004 |
| 2 | Notomicrinae | Noterinae | 9 | 235 | 0.07 |
| 3 | Hygrobiidae | Dytiscidae | 6 | 3810 | 0.003 |
| 4 | Noteridae | Dytiscidae | 250 | 3810 | 0.12 |
- N, species number of the species rich clade; n, species number of the species poor clade; p, probability that the species richness of the two clades is the product of the same underlying random cladogenetic process (computed with the statistic 2n/(n + N−1), see text for details). No correction for multiple tests was applied, as each of them is testing a different null hypotheses. Contrasts 1 and 2 correspond to the optimal phylogenetic hypothesis of the combined dataset (Fig. 2). Contrast 3 correspond to the results of the morphological data alone, and of most previous phylogenetic hypotheses (see text for details). Contrast 4 correspond to traditional taxonomy and the phylogenetic hypothesis of Miller (2001).
There is an apparent association between mode of swimming and species diversification: both species-rich clades (Dytiscidae and Noterinae) swim with simultaneous movements of the hind and medium legs, while the rest of Dytiscoidea lineages and their putative sister group, Haliplidae, either do not swim at all, or use alternate movements of the legs (Ribera et al. 2002a and references therein). Haliplidae swim with alternate leg movements and possess slender legs, and a moderately developed swimming setation (Holmen 1987; Vondel 1997). A similar morphological condition is found in species-poor subfamilies of Noteridae: Phreatodytinae (with six cave, non-swimming species in a single genus, Phreatodytes, Uéno 1996) and Notomicrinae (with nine species in a single genus, Notomicrus) (Belkaceme 1991: Figs 63 and 65). Recent observations suggest that adults of Notomicrus swim with alternate leg movements (M. Balke in French Guyana, 2002, and C.H.S. Watts in Australia, 2003, which remain to be confirmed by high speed photography due to length of beetle <2 mm).
If the preliminary observations of the swimming mode of Notomicrus are confirmed, a parsimony reconstruction on the optimal phylogenetic hypothesis illustrated in Fig. 6 shows that alternate swimming behaviour with developed swimming hairs is likely to be the groundplan feature of Dytiscoidea. This locomotory pattern with alternate leg movement is preserved in Hygrobiidae, which possess dense fringes of natatorial setae on all legs and exhibit a good swimming performance. We assume that swimming abilities were lost once in the common ancestor of the clade comprising Amphizoa and Aspidytes, which exhibit the plesiomorphic behaviour, i.e. alternate leg movement. Poorly developed fringes of thin and short hairs are still present in Amphizoa, whereas they are completely absent in Aspidytes. The change to simultaneous leg movements took place independently in Dytiscidae (Nachtigall 1960) and in ‘higher’ Noteridae (subfamily Noterinae) (Belkaceme 1991).
This interpretation of the simultaneous stroke as a ‘key innovation’ triggering an increase in the diversification rate would be supported by the fact that a third lineage of aquatic Adephaga, the family Gyrinidae, may show a similar trend. The members of the species-rich subfamily Gyrininae (with c. 1000 species, Balke et al. 2004a) are all excellent swimmers, with simultaneous stroke of the middle and hind legs, which are highly modified (Nachtigall 1961). Their sister group, Spanglerogyrininae (Beutel 1995; Ribera et al. 2002b), includes a single species in the genus Spanglerogyrus, with less modified legs. Field observations (R.G. Beutel, personal observations, 1987) suggest alternate movements, but a closer examination with a suitable experimental set up is clearly required.
It must be noted that by pooling all species in each of the two sister clades of a node the effect of any shift in the rate of diversification further up in the tree is also taken into account. It is, therefore, essential to try to precise as much as possible the exact node in which the increase in the number of species is produced. At present there are no available robust complete phylogenies of Noterinae or Dytiscidae, and it is thus not possible to precise the exact node in which the diversification shift was produced. Accepting the hypotheses of Belkaceme (1991) and Miller (2001) (based both on morphological data alone), the basal node of Noterinae will be symmetrical (Neohydrocoptus, with 28 species, vs. the rest of Noterinae, with 207 species, p = 0.2, Table 6), but the basal node of Dytiscidae would be highly asymmetrical (Matinae, with eight species, vs. the rest of Dytiscidae, with 3802 species, p < 0.005, Table 6). The species of Matinae already exhibit the simultaneous stroke (W. Steiner and C.H.S. Watts, observations in Australia and the USA respectively, 2003). This would contradict the hypothesis of the simultaneous stroke as the factor triggering a higher diversification rate, and call for the search of alternative explanations for the high asymmetry in the species richness of the different lineages of Dytiscoidea.
As pointed out above, a reliable interpretation of the evolution of the feeding apparatus has to be clarified in an analysis which includes representatives of all adephagan families. However, it may be noted that sucking channels are present in the larvae of the species-rich Dytiscidae (with the exception of Copelatus and Hydrotrupes), in the relatively diverse families Gyrinidae and Haliplidae, and also in the two most species-rich genera of Noteridae. It is likely that none of these groups are closely related to each other (Beutel and Haas 1996 and present analyses), which indicates that sucking channels must have evolved several times independently. This suggests that a specialized feeding apparatus, which prevents dilution of digestive fluids in the aquatic medium, might also have contributed to the diversification of some families of Dytiscoidea.
Acknowledgements
We particularly thank D.T. Bilton for providing larvae of Aspidytes niobe; Y. Alarie for unpublished information on larval chaetotaxy; W. Steiner and C.H.S. Watts for observations of the swimming behaviour of Matinae and Notomicrus; and all persons mentioned in Appendix A for providing specimens for study. We also thank Alried P. Vogler, Joan Pons, Jesús Gómez-Zurita, Yves Alarie and Gonzalo Giribet for comments on earlier versions of the manuscript. MB was supported by a Marie Curie Postdoctoral Fellowship of the European Community programme IHP, with additional funds from the Linnean Society of London and the Deutsche Forschungsgemeinschaft. Financial support of the European Community's programme ‘Structuring the European Research Area’ under SYNTHESYS at the Museo Nacional de Ciencias Naturales (CSIC), contract number ES-TAF 193 is also greatly acknowledged.
IR was supported by the Ramón y Cajal programme. Work on the molecular systematics of Coleoptera in A. Vogler's laboratory was supported by The Leverhulme Trust (F696/H) and NERC.
Appendix
Appendices
| Origin | Collector | CO1 | CytB | 12S | 16S | H3 | 18S | |
|---|---|---|---|---|---|---|---|---|
| Amphizoidae | ||||||||
| Amphizoa insolens LeConte, 1853 | USA | Cognato | AY071796 | AY334160 | AY745638 | AY071770 | AY745672 | AJ3186757 |
| Amphizoa lecontei Matthews, 1872 | USA | ? | AY071797 | AY745656 | AY745639 | AY071771 | AY745673 | AJ318678 |
| Aspidytidae | ||||||||
| Aspidytes niobe Ribera et al., 2002 | RSA | Ribera & Cieslak | AY0718089 | AY745657 | AY745640 | AY0717823 | AY745674 | AY071794 |
| Dytiscidae | ||||||||
| Agabus disintegratus Crotch, 1873 | CA, USA | Shepard | AY071795 | AY745658 | AY745641 | AY071769 | AY745675 | AY071793 |
| Agabus heydeni Wehncke, 1872 | Spain | Fery | AY039274 | AY745659 | AY745642 | AY039263 | AY745676 | AJ318689 |
| Coptotomus lenticus Hilsenhoff, 1980 | USA, NY | Miller | AY071802 | AY745660 | n/a | AY071776 | AY745677 | AJ318686 |
| Copelatus haemorrhoidalis (F., 1787) | Germany & UK | Balke, Ribera | AY071800 | AY334182 | AY745643 | AY071774 | AY745678 | AJ318679 |
| Papuadytes sp. IR73 | New Guinea | Balke | AY071801 | AY334215 | AY745644 | AY071775 | AY745679 | AJ318682 |
| Lancetes varius (F., 1775) | Chile | Ribera | AY071810 | AY745661 | AY745645 | AY071784 | AY745680 | AJ318684 |
| Lacconectus peguensis Brancucci, 1986 | Myanmar | Schillhammer | AY071811 | AY745662 | AY745646 | AY071785 | AY745681 | AJ318680 |
| Haliplidae | ||||||||
| Haliplus lineatocollis (Marsham, 1802) | Spain | Ribera | AY071803 | AY745663 | AY745647 | AY071777 | AY745682 | AJ318666 |
| Haliplus mucronatus Stephens, 1829 | Spain | Ribera | AY071804 | AY745664 | AY745648 | AY071778 | AY745683 | AJ318667 |
| Peltodytes rotundatus (Aube, 1836) | Spain | Ribera | AY071816 | AY745665 | AY745649 | AY071790 | AY745684 | AJ318668 |
| Hygrobiidae | ||||||||
| Hygrobia australasiae (Clark, 1862) | Australia | Watts | AY071805 | AY745666 | AY745650 | AY071779 | AY745685 | AJ318672 |
| H. hermanni (F., 1775) | Spain | Ribera | AY071806 | AY334185 | AY745651 | AY071780 | AY745686 | AJ318673 |
| H. maculata Britton, 1981 | Australia | Norton | AY071807 | AY745667 | AY745652 | AY071781 | AY745687 | AJ318674 |
| Noteridae | ||||||||
| Notomicrus tenellus (Clark, 1863) | Australia | Watts | AY071813 | AY745668 | n/a | AY071787 | AY745688 | AJ318671 |
| Noterus clavicornis (De Geer, 1774) | UK | Ribera | AY071814 | AY745669 | AY745653 | AY071788 | AY745689 | AF201416 |
| Suphis sp. IR-2002 | Venezuela | Bilton | AY071817 | AY745670 | AY745654 | AY071791 | AY745690 | AF012523 |
| Suphisellus sp. | Venezuela | Bilton | AY071818 | AY745671 | AY745655 | AY071792 | AY745691 | AJ318669 |
| 11111111112222222222233333333334444444445555 | |
| 12345678901234567890123456789012345678901234567890123 | |
| Agabus | 10000001000000112021101212110102100121221111111021100 |
| Lancetes | 10000001?00000??2021101212110?02100??1221111?110211?0 |
| Coptotomus | 10000001?00000??2021101212110?02100??1221111?110211?0 |
| Lacconectus | 10000001?000001?2021101212110???????????????????????? |
| Copelatus | 10000001100000112021101212110102100121211111111021100 |
| Papuadytes | 10000001100000112021101212110???????????????????????? |
| Hygrobia | 00000000020000102011101011110102100131201121111021200 |
| Aspidytes | 12110210100000010010101001110100010111011111001111310 |
| Amphizoa | 00001010000000011010111001110102100111111121001121100 |
| Notomicrus | 110200001000000010211011031112??????????????????????? |
| Noterus | 11020110111101002121101103111001021000010111000021111 |
| Suphis | 110201201111110021211011031110?2021??001?111000021101 |
| Suphisellus | 110201201111110021211011031110??????????????????????? |
| Haliplus | 03000020000000002001000024000012000001120000000000200 |
| Peltodytes | 03000020000000002001000024000012000001120000000001200 |




