Molecular phylogeny of caprines (Bovidae, Antilopinae): the question of their origin and diversification during the Miocene
Phylogénie moléculaire des caprinés (Bovidae, Antilopinae): origine et diversification au cours du Miocène
Abstract
enCaprines include all bovids related to sheep and goat. The composition of the group is controversial and inter-generic relationships have been widely debated. Here, we analysed 2469 characters draw from three distinct molecular markers, i.e. two mitochondrial genes (cytochrome b and 12S rRNA) and one nuclear fragment (exon 4 of the κ-casein gene). The taxonomic sampling includes all genera putatively described as caprines, as well as several other bovid genera in order to elucidate the position of caprines within the family Bovidae, and to determine the exact composition of the group. Phylogenetic analyses confirm firstly that Pseudoryx and Saiga do not belong to caprines, and secondly, that all tribes classically defined in the literature are not monophyletic, supporting the inclusion of all caprine species into a unique enlarged tribe Caprini sensu lato. Our results are in contradiction with previous investigations suggesting a sister-group relationship between Ovis (sheep and mouflons) and Budorcas (takins). By using a molecular calibration point at 18.5 Mya for the first appearance of bovids, we estimated divergence times with our molecular data. We also performed biogeographic inferences to better understand the origin and diversification of caprines during the Neogene. Our analyses suggest that caprines shared a common ancestor with Alcelaphini and Hippotragini in the middle-late Miocene (13.37 ± 0.70 Mya). Our results also indicate that the extant generic diversity of caprines resulted from a rapid adaptive radiation during the late Miocene, at 10.96 ± 0.73 Mya. We propose that this adaptive radiation resulted from the acquisition of reduced metacarpals, a key innovation which occurred during the late Miocene as a consequence of insularity isolation in the mountainous mega-archipelago between Mediterranean and Paratethys Seas.
Résumé
deLes caprinés incluent tous les bovidés apparentés aux chèvres et aux moutons. La composition du groupe et les relations inter-génériques sont très discutées dans la littérature. Dans cette étude, nous avons analysé 2469 caractères issus de trois marqueurs moléculaires: deux gènes mitochondriaux (ARNr 12S et cytochrome b) et un fragment nucléaire (exon 4 du gène de la κ-caséine). L’échantillonnage taxonomique comprend tous les genres supposés de caprinés et d'autres genres de bovidés afin d’élucider la position des caprinés au sein de la famille des Bovidae, et de déterminer la composition exacte de ce groupe. Les analyses phylogénétiques confirment, premièrement, que Pseudoryx et Saiga n'appartiennent pas aux caprinés, et deuxièmement, que les quatre tribus classiquement décrites dans la littérature ne sont pas monophylétiques, ce qui soutient le regroupement de toutes les espèces de caprinés dans une seule et même tribu appelée Caprini sensu lato. Par contre, nos données sont en contradiction avec des résultats antérieurs qui associaient les genres Ovis (moutons et mouflons) et Budorcas (takins). Nous avons estimé les temps de divergence entre les taxons sur la base de datations moléculaires en supposant une origine à 18,5 millions d'années pour les bovidés. Des inférences biogéographiques ont été réalisées afin de mieux comprendre l'origine et la diversification des caprinés au cours du Néogène. Nos analyses suggèrent que les caprinés partagent un ancêtre commun avec les Alcelaphini et les Hippotragini datéà la fin du Miocène moyen (13.37 ± 0.70 Ma). Nos résultats indiquent également que la diversité générique existant au sein des caprinés est la conséquence d'une rapide radiation adaptative au Miocène supérieur (10.96 ± 0.73 Ma). Nous supposons que cette radiation s'est produite suite à l'acquisition de métapodes réduits, et que cette innovation est apparue au cours du Miocène supérieur en raison d'un isolement insulaire dans l'archipel montagneux séparant, à cette époque, les mers Méditerranée et Paratéthys.
Introduction
The family Bovidae (Artiodactyla, Ruminantia) is composed of around 140 species, which are ranged into 45 genera (Grubb 1993). Bovids are distributed in all continents where they occupied diverse ecological niches, but they never reached Antarctica, Australia and South America. All bovids are clearly united by an unambiguous synapomorphy, i.e. the possession of typical horns in males and sometimes in females, which are composed of a bone core covered by a permanent unforked keratinous sheath. In the fossil record, the group emerged near 18.5 Mya (Vrba and Schaller 2000). From the taxonomic point of view, the family Bovidae is one of the most problematic groups within mammals (Simpson 1945). However, recent DNA analyses have somewhat clarified the situation by using partial 12S rRNA gene (Gatesy et al. 1997), complete cytochrome b gene (Hassanin and Douzery 1999a; Matthee and Robinson 1999), or a combination of mitochondrial and nuclear markers (Hassanin and Douzery 1999b; Matthee and Davis 2001; Hassanin and Douzery 2003). These analyses have evidenced a major dichotomy, which confirms the classification in two subfamilies previously proposed by Kingdon (1982, 1997): the subfamily Bovinae, which includes the three tribes Bovini (oxen), Boselaphini (nilgaut and four horned antelope) and Tragelaphini (spiralled-horned bovines), and the subfamily Antilopinae, which incorporates all other bovid species.
Within bovids, species of caprines are not united by an unambiguous morphological synapomorphy (Gentry 1992). The absence of diagnostic feature probably explains why the composition of this group is considerably variable in the literature. Simpson (1945) recognizes four distinct tribes within caprines: (1) the tribe Caprini, which includes goat, sheep and related species; (2) the tribe Rupicaprini, which groups the chamois, gorals, serows and Rocky mountains goat; (3) the tribe Ovibovini, which contains the muskox and takins; and (4) the tribe Saigini, which associates Saiga tatarica (saiga antelope) and Pantholops hodgsonii (chiru). However, Grubb (1993), as well as McKenna and Bell (1997), unite Pantholops and Saiga with gazelles and relatives, whereas Gentry (1992) ranges Pantholops with caprines, but Saiga with gazelles. Moreover, Thomas (1994) suggests that Pseudoryx (saola), the lately described bovid species of Vietnam (Dung et al. 1993), could be enclosed into the caprines. Molecular investigations based on multiple mitochondrial and nuclear markers (Hassanin and Douzery 1999b; Gatesy and Arctander 2000) have however shown that the saola is not a caprine as it belongs to the tribe Bovini within the subfamily Bovinae. In addition, molecular studies using sequences of the complete cytochrome b gene (Cyb) have also suggested that all caprine tribes defined by Simpson (1945) are not valid (Groves and Shields 1996, 1997; Hassanin et al. 1998a): the tribe Ovibovini was found polyphyletic, with, on the one hand, the muskox (Ovibos) allied with serow and gorals (Naemorhedus), which also implies the paraphyly of Rupicaprini, and on the other hand, the takin (Budorcas) allied with sheep (Ovis), which also implies the paraphyly of Caprini; and the tribe Saigini was also found polyphyletic as the saiga antelope (Saiga) was grouped with gazelles, while the chiru (Pantholops) was associated with other caprines. Because of the poly- or paraphyly of all caprine tribes described by Simpson (1945), all members of these tribes, but Saiga, have been ranged together in an enlarged tribe Caprini, named Caprini sensu lato (Hassanin and Douzery 1999a). According to the Cyb gene, this tribe is therefore supposed to include the following 11 genera: (1) Ammotragus (aoudad), (2) Budorcas (takins), (3) Capra (goats, ibexes, markhor and turs), (4) Hemitragus (tahrs), (5) Naemorhedus (gorals and serows), (6) Oreamnos (Rocky Mountain goat), (7) Ovibos (muskox), (8) Ovis (sheep, argalis and mouflons), (9) Pantholops (chiru), (10) Pseudois (bharals) and (11) Rupicapra (chamois and isards). Additional markers are however needed to definitively conclude on the taxonomic composition of the group.
Traditionally, palaeontologists consider that caprines emerged somewhere in Asia (e.g. Vrba 1985; Gentry 2000). However, this hypothesis is not really supported by the fossil record as the most ancient remains of caprines have been described in the middle Miocene of three different continents: Caprotragoides in Asia (India and Pakistan), Tethytragus in Europe (Spain and Turkey) and Gentrytragus in Africa (Kenya and Saudi Arabia) (Azanza and Morales 1994). By contrast, Alcalà and Morales (1997) do not consider these fossils as being closely related to caprines, and they suggest a more recent origin, during the late Miocene.
In the present study, we performed a molecular phylogeny of the family Bovidae with a special emphasis on the tribe Caprini sensu lato. Three different markers were analysed: two mitochondrial genes coding for cytochrome b protein (Cyb) and 12S rRNA (12S), and one nuclear gene fragment corresponding to the exon 4 of the κ-casein gene (κCas). The aims of this study are: (1) to evaluate the contribution of three markers (Cyb, 12S and κCas) and their combination (2469 characters) to decipher phylogenetic relationships between caprine genera; (2) to determine the time frame for caprine evolution, assuming an early Miocene (18.5 Mya) a priori divergence of bovids (Vrba and Schaller 2000); and (3) to build a biogeographic scenario explaining the current distribution of caprines and their extraordinary morphological diversification.
Materials and Methods
Taxonomic sample
The taxonomic sample used for this study contains 33 taxa with 30 bovid species and three outgroup genera belonging to the families Antilocapridae (Antilocapra), Cervidae (Cervus) and Giraffidae (Okapia) (Table 1). The ingroup incorporates seven species of the subfamily Bovinae, including representatives of the three different tribes (Bovini, Boselaphini and Tragelaphini) and 23 species of the subfamily Antilopinae sensuKingdon (1982, 1997, including representatives of nine different tribes, i.e. Aepycerotini, Alcelaphini, Antilopini, Caprini sensu lato, Cephalophini, Hippotragini, Oreotragini, Neotragini and Reduncini. Fourteen species of caprines are incorporated, which represents the totality of the 11 genera recognized into the tribe Caprini sensu lato (Hassanin et al. 1998a). In addition, this sample integrates two species described as possible caprine taxa in the literature: S. tatarica (saïga antelope) (Simpson 1945) and Pseudoryx nghetinhensis (saola) (Thomas 1994).
| Family | Tribe | Species (Grubb 1993) | Common name | Cytochrome b | 12S rDNA | κ-casein |
|---|---|---|---|---|---|---|
| Antilocapridae | Antilocaprini | Antilocapra americana | Pronghorn | AF091629 1 | M55540 9 | U37515 13/AF16567214 |
| Giraffidae | Okapini | Okapia johnstoni | Okapi | AY121993 2 | AY121987 2 | AY121996 2 |
| Cervidae | Cervini | Cervus elaphus | Deer | AJ000021 3 | AF091707 1 | U37505 13 |
| Bovidae | Bovini | Bos taurus | Domestic cow | NC_001567 4 | NC_001567 4 | X14908 15 |
| Bubalus bubalis | Asiatic buffalo | D88983 5 | AF231028 10 | AJ011387 16 | ||
| Syncerus caffer | African buffalo | AF036275 6 | AF091688 1 | AF030328 17/AF21015414 | ||
| Pseudoryx nghetinhensis | Saola | AF091635 1 | AF091705 1 | AY670667* | ||
| Boselaphini | Boselaphus tragocamelus | Nilgai | AJ222679 6 | M86494 11 | AF030331 17/AF16572814 | |
| Tetracerus quadricornis | Four-horned antelope | AF036274 6 | AF091690 1 | AY670668* | ||
| Tragelaphini | Tragelaphus imberbis | Lesser kudu | AF036279 6 | M86493 11 | AF030330 17/AF16573614 | |
| Neotragini | Neotragus moschatus | Suni | AJ222683 6 | AF091703 1 | AF210171 18 | |
| Oreotragini | Oreotragus oreotragus | Klipspringer | AF036288 6 | AF091702 1 | AF210172 18 | |
| Cephalophini | Cephalophus dorsalis | Bay duiker | AF091634 1 | AF091701 1 | AY122000 2 | |
| Antilopini | Gazella granti | Grant's gazelle | AF034723 7 | AY670652* | AY670669* | |
| Saiga tatarica | Saiga antilope | AF064487 7 | AF363777 12 | D32188 19 | ||
| Reduncini | Redunca fulvorufula | Mountain reedbuck | AF036284 6 | AF091704 1 | AY121999 2 | |
| Aepycerotini | Aepyceros melampus | Impala | AF036289 6 | M86496 11 | AY121998 2 | |
| Alcelaphini | Damaliscus pygargus | Blesbok | AF036287 6 | M86499 11 | AY122002 2 | |
| Hippotragini | Hippotragus niger | Sable antelope | AF036285 6 | AY670653* | AY122001 2 | |
| Caprini s.l. | Ammotragus lervia | Aoudad | AF034731 7 | AY670654* | AY670670* | |
| Budorcas taxicolor | Takin | AY669320* | AY670655* | AY670671* | ||
| Capra falconeri | Markhor | AF034736 7 | AY670656* | AY670672* | ||
| Capra nubiana | Nubian ibex | AF034740 7 | AY670657* | AY670673* | ||
| Capra sibirica | Asiatic ibex | AF034734 7 | AY670658* | AY670674* | ||
| Hemitragus jemlahicus | Tahr | AF034733 7 | AY670659* | AY670675* | ||
| Naemorhedus sumatraensis | Sumatran serow | AY669321* | AY670660* | AY670676* | ||
| Oreamnos americanus | Rocky mountains goat | AF190632 8 | AY670661* | AY670677* | ||
| Ovibos moschatus | Muskox | AY669322* | AY670662* | AY670678* | ||
| Ovis aries | Domestic sheep | AF034730 7 | AY670663* | AY670679* | ||
| Ovis dalli | Dall's sheep | AF034728 7 | AY670664* | AY670680* | ||
| Pantholops hodgsonii | Chiru | AF034724 7 | AF400659* | AY670681* | ||
| Pseudois nayaur | Bharal | AF034732 7 | AY670665* | AY670682* | ||
| Rupicapra rupicapra | Alpine chamois | AF034725 7 | AY670666* | D32182 19 |
- *This study; 1Hassanin and Douzery (1999b); 2Hassanin and Douzery (2003); 3Randi et al. (1998); 4Anderson et al. (1982); 5Kikkawa et al. (1997); 6Hassanin and Douzery (1999a); 7Hassanin et al. (1998a); 8Hassanin and Douzery (2000); 9Kraus and Miyamoto (1990); 10Kuznetsov et al. (2001); 11Allard et al. (1992); 12Kuznetsova and Kholodova (2002); 13Cronin et al. (1996); 14Matthee et al. (2001); 15Alexander et al. (1988); 16P. Das and L.C. Garg (unpubl. data); 17Ward et al. (1997); 18Matthee and Davis (2001); 19Chikuni et al. (1995).
DNA extraction
DNA was extracted from blood on Ammotragus lervia, Capra falconeri, Capra nubiana, Capra sibirica, Gazella granti, Hemitragus jemlahicus, Ovis aries, and Pseudois nayaur, from heart on Ovibos moschatus, from hair on Oreamnos americanus, and from cells for Hippotragus niger. The protocol of DNA extraction includes a digestion with CTAB (hexadecyltrimethylammonium bromide), followed by a deproteinisation with CIA (chloroform isoamyl alcohol) and thereafter a cold precipitation with propan-2-ol (Winnepenninckx et al. 1993). Bone fragment from specimens of the collections of the Museum National d'Histoire Naturelle of Paris were used for DNA extraction by applying the protocol detailed in (Hassanin et al. 1998a): Budorcas taxicolor (no. 1902-409), Naemorhedus sumatraensis (no. 1993-4240), Ovis dalli (no. 1938-124), P. hodgsonii (no. 1993-4237), Rupicapra rupicapra (no. 1998-146), and S. tatarica (no. 1962-393).
Amplification and sequencing
The sequence of 12S rRNA gene (955 bp, positions 431–1385 in the sequence of Bos taurus, accession number: NC_001567) was produced from several overlapping regions, which were generated by polymerase chain reaction (PCR) using the following five couples of primers: (1) U943: 5′-ATT GTA GCT GGA CTT AAC TGC-3′ with L1252: 5′-ARG AAG GGC TGG GAC CAA ACC T -3′, (2) U1230: 5′-CAC TGA AAA TGC CTA GAT GAG-3′ with L1505: 5′-TTA RCT TGG GTT AAT CGT ATG-3′, (3) U1425: 5′-ACC CCC ACG GGA AGA CAG CAG T-3′ with L1684: 5′-TGT TTA GGG CTA RGC ATA GTG-3′, (4) U1637: 5′-AAY GAC GAA AGT AAC CCT AC-3′ with L1946: 5′-CCC ATT TCT TCC CAY TCC AT-3′, (5) U1883: 5′-CCG CCA TCT TCA GCA AAC CCT-3′ with L2226: 5′-CTA GGT GTA AAC TAG RTG CTT-3′. The exon 4 of the κ-casein gene (402 bp, positions 84–481 in the sequence of B. taurus, accession number: X14908) was amplified with the two following primers: 5′-GTA TGT GCT GAG TAG GTA TCC TAG-3′ and 5′-GTC TTC TTT GAT GTC TCC TTA GAG T-3′. The complete cytochrome b gene was amplified with a set of primers previously published (Hassanin et al. 1998a; Hassanin and Douzery 1999a).
The standard PCR conditions were as follows: 3 min at 94°C; 30 cycles of denaturation/annealing/extension with 1 min at 94°C for denaturation, 1 min at 55°C for annealing and 1 min at 72°C for extension; and 7 min at 72°C. The PCR products were purified from Agarose gel using QIAquick PCR Purification kit (QIAGEN) to remove excess primers and unincorporated nucleotides. The purified PCR products were then used as starting templates for direct sequencing using the Thermo Sequenase cycle sequencing kit (Amersham, Uppsala, Sweden). Some PCR products were cloned into a Blue-script pKS plasmid (Stratagen, Kirkrand, WA, USA), modified according to Holton and Graham (1990). Three positive clones were sequenced on both strands by using the ‘Thermosequenase fluorescent-labelled primer cycle sequencing kit’ with 7-deaza-dGTP (Amersham, Pharmacia, Uppsala, Sweden), and a fluorescent primer labelled with CY5TM. The sequencing reactions were run on an automatic sequencer (Alf Express, Pharmacia, Uppsala, Sweden).
DNA alignment
The nucleotide sequences were aligned with Se-Al v1.0al (Sequence Alignment Editor Version 1.0 alpha 1; Rambaut 1996). The DNA sequences of the 12S rRNA gene were aligned using the models of secondary structure available in the SSU rRNA Database (http://rrna.uia.ac.be/ssu/; Wuyts et al. 2002) for O. aries (accession no. AF010406), Capra hircus (M55541), Damaliscus dorcas (M86499) and Oryx gazella (M86500). By this approach, we identify regions involved in helix, loop, or sites involved in non-standard pair (any pair other than G.C., A.U. or G.U.). The Cyb and κCas genes were aligned using the amino-acid sequences. All ambiguous regions for alignment were excluded from the analyses to avoid erroneous hypotheses of primary homology. Unambiguous insertions and deletions (indels) were coded as additional characters by using I and D symbols for insertion and deletion, respectively (Swofford 1993).
Phylogenetic analyses
The three markers were analysed separately and also combined to benefit from the maximum number of molecular characters. Bayesian and Maximum Parsimony (MP) methods were used for the tree reconstruction analyses.
Bayesian analyses were performed with Mr Bayes 3.0b4 (Huelsenbeck and Ronquist 2001). The Bayesian approach evaluates the posterior probability (PPB) of a tree given the character matrix, i.e. the probability that the tree is correct. The posterior probability is obtained after combining the prior probabilities of a tree and of the data with the likelihood of the data given that tree. The Bayesian approach combines the advantages of defining an explicit probability model of character evolution and of obtaining a rapid approximation of posterior probabilities of trees through the use of Markov chains Monte Carlo (MCMC). The likelihood model chosen is the General Time Reversible model (GTR, Yang 1994) with among-site substitution rate heterogeneity described by a gamma distribution with eight categories and a fraction of sites constrained to be invariable (GTR + I + Γ8). Different models were used for each of the eight partitions corresponding to the three codon-positions for Cyb and κCas, and helix and not helix for 12S. All analyses were conducted with five independent Markov chains – one cold chain and four incrementally heated chains – run for 1 000 000 Metropolis-coupled MCMC generations, with tree sampling every 100 generations, and burn-in after 1000 trees.
The MP analyses were conducted on paup 3.1.1 (Swofford 1993) with differential weighting of the character-state transformations as detailed in (Hassanin et al. 1998a,b): for each substitution type (i.e. A-G, C-T, A-C, A-T, C-G, G-T and I-D), the amount of homoplasy was measured through the consistency index excluding uninformative characters (CIex), and the saturation was assessed graphically by plotting the pairwise number of observed differences against the corresponding pairwise number of inferred substitutions calculated by paup [the slope of the linear regression (S) was used to evaluate the level of saturation]. The CIex and S values were calculated by distinguishing each of the three codon-positions separately for Cyb, and between the helix and not-helix regions of the secondary structure of 12S. The MP tree was found by heuristic search using 1000 replicates of random stepwise addition. Support for individual branches was assessed by Bootstrap Percentages (BP) (Felsenstein 1985) computed after 1000 replicates of the closest stepwise addition option.
Molecular datings
The divergence times were estimated using the ClockCombined model under BASEML in the paml 3.14beta software (Yang 2003). The first appearance of bovids in the fossil record was used as a calibration point for molecular datings (Vrba and Schaller 2000): the bovid divergence was fixed at 18.5 Mya with an interval between 18 and 20 Mya. The topology used for molecular datings is the Bayesian tree (Fig. 1), but Cervidae were considered as the sister-group of Bovidae, in agreement with the results published in Hassanin and Douzery (2003). The combined analysis was performed using a GTR model with a gamma distribution with eight categories for each of the eight following partitions: three codon positions separately for Cyb and κCas, and the helix and not-helix regions of the secondary structure of 12S. Independent analyses on each of the three markers were also conducted in the same way in order to evaluate the impact of each marker on the estimation of divergence times.
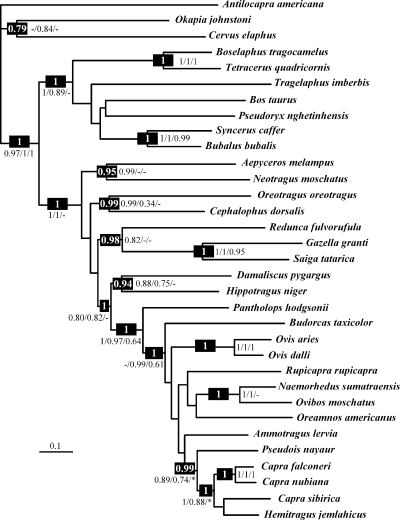
Bayesian tree. The tree was obtained from the combined analysis of the three markers (cytochrome b, 12S rDNA, and κ-casein). The values in the black spot are the posterior probabilities (PPB) greater than 0.75 obtained with the combination of the three markers. The other values are the PPB obtained with each of the three markers independently; from left to right: cytochrome b, 12S rDNA and κ-casein. Dash indicates that the node was not retrieved with the marker, but no alternative hypothesis was supported by PPB greater than 0.95. Asterisk indicates that an alternative hypothesis supported by a PPB greater than 0.95 was obtained with the marker
Saturation of nucleotide substitutions
The nucleotide substitution saturation of the three markers (12S, Cyb and κCas) under a GTR model was evaluated with a graphical method. For each of the three markers, the number of inferred substitutions between each pair of sequences was estimated from the ML tree. It corresponds to the sum of changes along branches linking these two sequences given in the output of BASEML. The saturation level was estimated by plotting the number of observed differences calculated using paup 4.0b10 (Swofford 2002) as a function of the number of inferred substitutions calculated using BASEML for all 528 pairwise comparisons for 33 sequences.
Biogeography analyses
Biogeography analyses were conducted from diva 1.1 (Ronquist 1996). diva is a program for reconstructing ancestral distributions in a phylogeny using dispersal–vicariance analysis (diva). This method, in which ancestral distributions are inferred, is based on a three-dimensional cost matrix derived from a simple biogeographic model (Ronquist 1997).
The biogeographic inferences were done using four distinct geographical areas, i.e. Africa, America, Asia and Europe. The extant distributions of wild species were determined by using the data available in Shackleton (1997). Different sets of outgroup taxa were tested for the analyses in order to know whether the number of outgroups may change the biogeographic results.
The following fully bifurcated tree was used for the analyses: (Suidae, (Hippopotamidae, (Tragulidae, (Antilocapridae, (Giraffidae, (((((Hydropotes, Capreolus), Alces), (Odocoileus, Mazama)), (Cervinae, Muntiacinae)), (Moschidae, ((((((Pantholops, other caprines), (Alcelaphini, Hippotragini)), (Redunca, (Gazella, Saiga))), (Cephalophus, Oreotragus)), (Aepyceros, Neotragus)), ((Bovini, Tragelaphus), (Boselaphus, Tetracerus)))))))))). The alternative grouping of Antilocapridae with Giraffidae was also tested. The relationships between outgroup taxa have been chosen according to the literature (Douzery and Randi 1997; Polziehn and Strobeck 1998; Hassanin and Douzery 2003).
Results
The Cytochrome b sequence of Budorcas
The complete Cyb sequence (1140 bp) produced during this study for B. taxicolor presents 104 differences (9.1%) with the sequence previously produced by Groves and Shields (1996) (accession no. U17868). We do not expect to find so much nucleotide differences between two specimens of the same species. Our sequence was obtained from a museum specimen (no. 1902-409) and its authenticity has been checked using two important criteria for ancient DNA studies: (1) the sequence was obtained from four different PCR products, which are overlapping on more than 100 bp and perfectly identical on the overlapping regions; and (2) the sequence was entirely reproduced twice at different periods of time, and in two different laboratories. The sequence generated by Groves and Shields (1996) was obtained for eight different specimens, and only nine differences were detected on the 1140 nucleotides, representing no more than 0.79% for divergence. However, the careful analysis of their sequences reveals that: (1) the fragment corresponding to the 5′ region of the Cyb gene (positions: 1–402) shows only 1.5% of difference with our sequence and 1–2% with three partial sequences of Budorcas available in the Genbank database (AY026509, AY026511 and AY026510). So, we can conclude that this region of the Cyb is authentic; (2) the fragment corresponding to the 3′ extremity of the Cyb gene (positions: 768–1140) is very different from our sequence of Budorcas (15%), but surprisingly it is very similar to the sequences of Ovis extracted from GenBank, with 99% of identity with four sequences of O. canadensis (U17859, AF112140, AF112139 and AF112138), and 98% of identity with O. dalli (AF034728). Therefore, this DNA fragment of the Cyb gene belongs to O. canadensis, and we believe that it was generated by exogenous contamination because Groves and Shields (1996) have also produced a Cyb sequence from O. canadensis in their phylogenetic study (U17859); (3) the fragment corresponding to the central region of the Cyb gene (positions: 403–767) is also very different from our sequence, with 11.5% of divergence, but the origin of this fragment is enigmatic as it does not present high similarities with any sequences of Cyb deposited in the databases.
Phylogenetic analyses
The three different markers (Cyb, 12S, κCas) were analysed separately and also combined (2469 sites including three sites with indels). In 1, 2 are presented the phylogenetic trees performed with Bayesian and MP methods, respectively. They appear very similar except a few topological differences, which are not strongly supported.
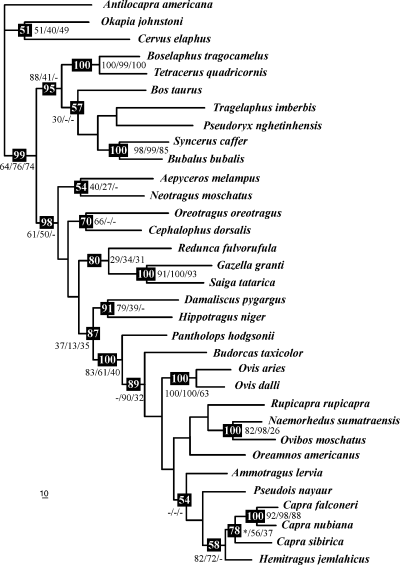
Most-parsimonious tree. The phylogram of 701053 steps was obtained from the combined analysis of the three markers (cytochrome b, 12S rDNA and κ-casein) and by using a differential weighting scheme based on the product of homoplasy and saturation estimators (see Materials and methods for details). The values in the black spot are the bootstrap percentages (BP) greater than 50 obtained with the combination of the three markers. The other values are the BP obtained with each of the three markers separately; from left to right: cytochrome b, 12S rDNA and κ-casein. Dash indicates that the node was not retrieved with the marker, but no alternative hypothesis was supported by BP greater than 75. Asterisk indicates that an alternative hypothesis supported by a BP greater than 75 was obtained with the marker
The family Bovidae was found monophyletic (PPB = 1; BP = 99), and this result was independently retrieved with each of the three markers (see values in 1, 2). The family Bovidae was divided into two subfamilies, i.e., Bovinae (PPB = 1; BP = 95) and Antilopinae (PPB = 1; BP = 98). The monophyly of these two subfamilies was independently found with both mitochondrial markers. Within the subfamily Bovinae, only two groupings were robust and supported by the three different markers: the monophyly of the tribe Boselaphini (PPB = 1; BP = 100), which includes the genera Boselaphus and Tetracerus, and the monophyly of the subtribe Bubalina (PPB = 1; BP = 100), which incorporates the genera Bubalus and Syncerus. Within the subfamily Antilopinae, Aepyceros was united with Neotragus, but their association was not robustly supported (PPB = 0.95; BP = 54). Oreotragus was allied with Cephalophus (PPB = 0.99; BP = 70). Saiga was grouped with Gazella into the tribe Antilopini (PPB = 1; BP = 100). This grouping was found independently with all the three markers and these two genera share an insertion of 12 codons in the κCas gene (position 379 in the sequence of B. taurus, X14908). The tribe Antilopini appeared closely related to Redunca (PPB = 0.98; BP = 80). The tribes Alcelaphini (represented by Damaliscus) and Hippotragini (represented by Hippotragus) were grouped together (PPB = 0.94; BP = 91), and they were associated with the tribe Caprini sensu lato (PPB = 1; BP = 87). All these results were independently found with at least two different markers. In addition, an insertion of two codons in the κCas (GWA CAC, position 367 in the sequence of B. taurus) is diagnostic of the clade uniting the tribes Alcelaphini, Caprini s.l. and Hippotragini.
The tribe Caprini s.l. was found monophyletic with the three markers analysed separately or in combination (PPB = 1; BP = 100). Moreover, this group is characterised by a diagnostic insertion of T in the 12S alignment (position 73 in the sequence of B. taurus, NC_001567). Within the tribe Caprini s.l., the inter-specific relationships were not highly supported in terms of PPB or BP values, but seven nodes were found independently with at least two markers: (1) Pantholops diverged first (PPB = 1; BP = 89), and all other caprines are diagnosed by a T at position 652 in the 12S of B. taurus; (2) Ovibos was grouped with Naemorhedus (PPB = 1; BP = 100); (3) Capra, Hemitragus, and Pseudois were enclosed into the same clade (PPB = 0.99; BP = 42); (4) Capra was united with Hemitragus (PPB = 1; BP = 58); (5) Capra was found monophyletic with 12S and κCas but not with the Bayesian analysis combining the three genes (Fig. 1); (6) C. falconeri was associated with C. nubiana (PPB = 1; BP = 100); and (7) Ovis was monophyletic (PPB = 1; BP = 100).
Molecular datings
Divergence times were estimated by analysing four distinct molecular data matrices: Cyb, 12S, κCas, and the whole data matrix combining all of the three markers (Table 2). By comparing the divergence times estimated with these four data matrices, it appeared that standard deviations (SD) on the molecular datings decrease when the number of informative sites increases (Table 2). This is particularly visible by comparing the results obtained from κCas sequences with the ones obtained from Cyb or the combined data matrix. When the divergence times calculated with each of the three markers were compared with the ages obtained with the combined data matrix (Fig. 3), it appeared that 12S ages are similar to those of the combined analysis. By contrast, nodes more ancient than the calibration point (18.5 Mya) tend to be older with κCas and younger with Cyb, and inversely, nodes more recent than the calibration point tend to be older with Cyb and younger with κCas.
| Inferred ancestral areas | Nodes in the tree of Fig. 1 | Molecular datings | |||
|---|---|---|---|---|---|
| Combined ± SD | Cyb ± SD | 12S ± SD | κCas ± SD | ||
| Antilocapra–Hemitragus | 22.99 ± 1.57 | 20.39 ± 0.97 | 24.49 ± 3.53 | 35.31 ± 13.34 | |
| Af | Bovidae | 18.50 | 18.50 | 18.50 | 18.50 |
| Af + As | Bovinae | 15.68 ± 0.63 | 15.88 ± 0.73 | 14.37 ± 1.39 | 17.b10,± 1.47 |
| As | Boselaphus–Tetracerus | 6.46 ± 0.64 | 7.56 ± 0.94 | 6.12 ± 1.15 | 0.70 ± 0.70 |
| Syncerus–Bubalus | 7.48 ± 0.74 | 8.70 ± 1.07 | 6.07 ± 1.16 | 5.25 ± 2.13 | |
| Af | Antilopinae | 15.20 ± 0.61 | 16.83 ± 0.58 | 14.36 ± 1.94 | 9.99 ± 1.95 |
| Af | Aepyceros–Neotragus | 13.14 ± 0.89 | 14.64 ± 1.05 | 11.45 ± 2.03 | 9.99 ± 1.95 |
| Af | Oreotragus–Cephalophus | 12.63 ± 0.88 | 13.76 ± 1.06 | 12.63 ± 2.13 | 8.19 ± 1.61 |
| Af/Af + As | Damaliscus–Hemitragus | 13.37 ± 0.70 | 15.29 ± 0.91 | 12.58 ± 1.86 | 8.19 ± 1.61 |
| Af | Damaliscus–Hippotragus | 11.98 ± 0.86 | 12.90 ± 1.19 | 11.71 ± 1.82 | 8.19 ± 1.61 |
| Af/Af + As | Redunca–Saiga | 13.11 ± 0.73 | 14.70 ± 0.95 | 12.73 ± 1.93 | 7.47 ± 1.60 |
| Gazella–Saiga | 8.24 ± 0.67 | 10.33 ± 0.96 | 6.71 ± 1.27 | 4.19 ± 1.28 | |
| As/Af + As/Am + Af + As/ Eu + Af + As/ Eu + Am + Af + As | Caprini sensu lato | 10.96 ± 0.73 | 12.71 ± 1.21 | 10.13 ± 1.72 | 7.14 ± 1.66 |
| Budorcas–Hemitragus | 9.87 ± 0.71 | 12.25 ± 1.19 | 8.15 ± 1.49 | 6.02 ± 1.51 | |
| Ovis aries–Ovis dalli | 3.81 ± 0.46 | 4.97 ± 0.80 | 2.26 ± 0.63 | 3.66 ± 1.22 | |
| Naemorhedus–Ovibos | 4.70 ± 0.55 | 6.46 ± 0.99 | 3.58 ± 0.86 | 1.46 ± 1.11 | |
| Pseudois–Hemitragus | 6.61 ± 0.61 | 8.85 ± 1.08 | 4.55 ± 0.98 | 3.85 ± 1.35 | |
| Capra falconeri–Hemitragus | 4.99 ± 0.50 | 6.56 ± 0.86 | 3.32 ± 0.77 | 3.85 ± 1.33 | |
| Capra falconeri–Capra nubiana | 2.31 ± 0.33 | 3.27 ± 0.57 | 1.61 ± 0.49 | 0.38 ± 0.38 | |
- Af, Africa; As, Asia; Eu, Europe; Am, America.
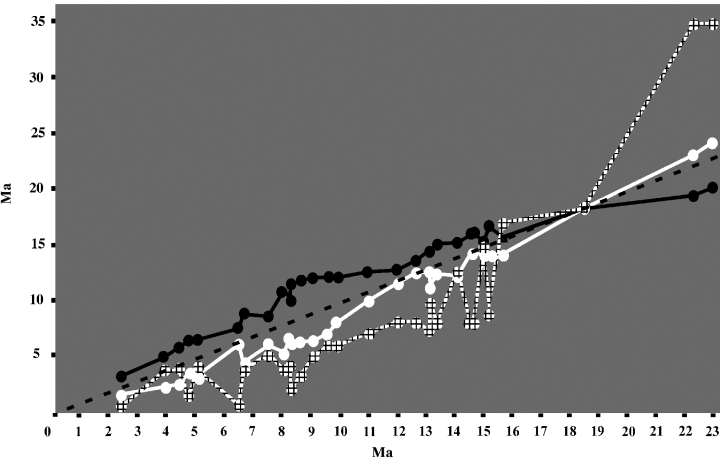
Comparison of molecular age estimations. The graphics were done by plotting for each node of the Bayesian tree (Fig. 1), the age obtained with the analysis combining the three markers (in abscissa) against the age obtained with each of the three markers, independently (in ordinate). The black line corresponds to the cytochrome b, the white line corresponds to the 12S rDNA, and the squared line corresponds to the κ-casein. The dotted line is used as a reference indicating the ages obtained with the combined analysis
Nucleotide saturation plots of the pairwise observed differences between the 33 sequences as a function of the pairwise number of substitutions inferred on the ML tree were presented in Fig. 4 for each of the three genes. The slope of the linear regression between the numbers of observed differences and inferred substitutions is an indication of the relative saturation level: the slope equals to one when no saturation is observed, and it is expected to decrease toward zero when the level of saturation increases. The comparison between the three markers revealed that the Cyb gene is more saturated (slope = 0.49) than the 12S (slope = 0.54), whereas the κCas is less saturated (slope = 0.84). These different levels of saturation correlated with the number of informative sites can explain the differences in age estimations: nodes close or more ancient than the calibration point tend to be younger with Cyb because of saturation at this level of the tree, whereas nodes more recent than the calibration point tend to be younger with κCas, simply because of lack of phylogenetic signal, i.e. 63 informative sites versus 218 and 428 for 12S and Cyb, respectively. According to this, we can conclude that ages of the most ancient nodes of the tree are probably underestimated in the combined analysis because of the saturation of mitochondrial markers.
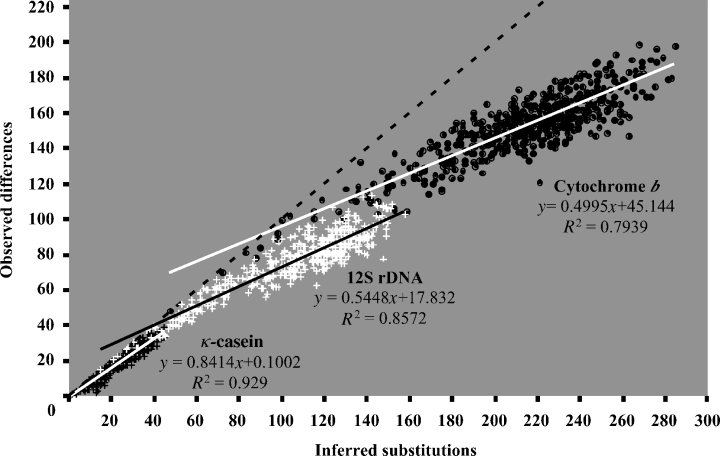
Graphics of saturation. Graphics were obtained by plotting the pairwise number of observed differences (in ordinate) against the corresponding pairwise number of inferred substitutions under the GTR model (in abscissa). Black points correspond to the cytochrome b; white crosses correspond to the 12S rDNA and black crosses correspond to the κ-casein. For each of the three markers, the slope of the linear regression was used to evaluate the level of saturation. The dotted line represents the area of equal numbers of observed and inferred changes, i.e. the theoretical situation for which no saturation is observed
Biogeography
diva was used for inferring the ancestral geographical distribution for different bovid taxa (Table 2). The analyses were done using a tree with multiple outgroup taxa in order to test whether the number of outgroups can modify the biogeography inferences. Despite some slight differences in the results, ancestral distributions remained unchanged when the more distant outgroups were successively removed (i.e. Suidae, Hippopotamidae, Tragulidae) or when additional taxa of Cervidae (Hydropotes, Capreolus, Alces, Odocoileus, Mazama, Muntiacinae) or Moschidae were excluded from the analyses (data not shown). Africa was found to be the ancestral area distribution for Bovidae, Antilopinae, Aepyceros + Neotragus, Oreotragus + Cephalophus, and Damaliscus + Hippotragus. Africa or Africa + Asia was found for the common ancestor of Antilopini + Redunca, and the one of Damaliscus + Hippotragus + Caprini s.l. The ancestral area distribution of Bovinae includes Africa and Asia. These analyses did not allow us to determine the ancestral area for the common ancestor of the tribe Caprini s.l. and five possibilities were found for this node: (1) Asia, (2) Africa + Asia, (3) Africa + America + Asia, (4) Africa + Asia + Europe or (5) Africa + America + Asia + Europe. This result was probably due to the lack of resolution for intergeneric relationships within caprines.
Discussion
Phylogenetic position of the tribe Caprini sensu lato
The present phylogenetic analyses (1, 2) based on three different markers (Cyb, 12S and κCas) show that the family Bovidae is separated into two distinct subfamilies: Antilopinae s.l. and Bovinae. The monophyly of these two subfamilies is supported by high posterior probability and Bootstrap values (PPB = 1; 95 < BP < 98). This result is in agreement with recent DNA analyses based on a combination of mitochondrial and nuclear markers (Hassanin and Douzery 1999a; Matthee and Davis 2001; Hassanin and Douzery 2003). So, the classification in two subfamilies previously proposed by Kingdon (1982, 1997) is confirmed by molecular data: the subfamily Bovinae includes the three tribes Bovini, Boselaphini and Tragelaphini, whereas the subfamily Antilopinae s.l. includes all other bovids.
All the analyses indicate that caprines belongs to the subfamily Antilopinae s.l. (PPB = 1, BP = 98), and that the tribe Caprini sensu lato is composed of 11 genera: Ammotragus, Budorcas, Capra, Hemitragus, Naemorhedus, Oreamnos, Ovibos, Ovis, Pantholops, Pseudois and Rupicapra. Our analyses therefore confirm that Pseudoryx and Saiga cannot be included in this group: Pseudoryx belongs to the subfamily Bovinae, in agreement with previous investigations concluding that the saola is a member of the tribe Bovini (Hassanin and Douzery 1999b; Gatesy and Arctander 2000); and Saiga is associated with Gazella (0.95 < PPB < 1; 91 < BP < 100), confirming that it should be considered as a member of the tribe Antilopini, as proposed by some morphologists (Gentry 1992; Vrba and Schaller 2000), and several previous molecular studies (Hassanin et al. 1998a; Hassanin and Douzery 1999a,b; Matthee and Robinson 1999).
Within Antilopinae s.l., the sister-group of Caprini s.l. is the clade grouping Alcelaphini (Damaliscus) and Hippotragini (Hippotragus). This result is in contradiction with the molecular studies based on partial sequences of 12S and 16S rRNA genes (Gatesy et al. 1997; Gatesy and Arctander 2000), in which caprines were found closely related to Hippotragini, with Alcelaphini at the outside. However, it agrees with other analyses based on molecular data (Hassanin and Douzery 1999a,b; Matthee and Davis 2001; Hassanin and Douzery 2003), cytogenetic data (Buckland and Evans 1978; Claro et al. 1995) and morphological data (Kingdon 1997; Vrba and Schaller 2000). This suggests therefore that classifications are not valid as Simpson (1945) grouped the tribes Alcelaphini, Hippotragini and Reduncini into the subfamily Hippotraginae, while Gentry (1992) and McKenna and Bell (1997) excluded the tribe Alcelaphini from the subfamily Hippotraginae.
Relationships between genera of the tribe Caprini s.l.
Previous investigations on the phylogeny of caprines have suggested a close association between the genera Budorcas and Ovis (Groves and Shields 1996, 1997; Hassanin et al. 1998a; Hassanin and Douzery 1999a; Lalueza-Fox et al. 2002). Some of them have also proposed that Ovis could be paraphyletic, because of the position of Budorcas as the sister-group of American sheep (O. canadensis and O. dalli), with the domestic sheep (O. aries) at the outside (Groves and Shields 1996, 1997). These results were considered with caution by the authors, firstly, because the monophyly of Ovis is supported by many morphological characters, and secondly, because sheep and takins do not share any phenotypic apomorphy. All these previous studies were based on the Cyb sequences of Budorcas produced by Groves and Shields (1996, 1997). In the present study, we have therefore sequenced three different markers: two mitochondrial genes, i.e. Cyb and 12S, and one nuclear gene, i.e. κCas. The genus Ovis is clearly found monophyletic (PPB = 1; 64 < BP < 100), and none of these markers gave any signal for grouping Budorcas with Ovis. By examining the Cyb sequence of Groves and Shields, we revealed that it is a chimeric sequence composed of three different fragments: (1) the 5′ region is an authentic sequence of Budorcas; but (2) the 3′ region is a contamination by O. canadensis; and (3) the central region is also doubtful, but its origin cannot be clearly determined (contamination by a nuclear pseudogene of Cyb or contamination by an undetermined bovid species). As exposed in Hassanin (2002), the use of chimeric sequence is particularly misleading for phylogenetic inferences. Here, the partial contamination by O. canadensis explains why Budorcas was found robustly enclosed with Ovis in all previous reports using the Cyb sequence of Groves and Shields (1996).
Our results show that none of the four Simpson’ tribes appear to be monophyletic. The tribe Ovibovini is not monophyletic because Ovibos is clearly associated with Naemorhedus (PPB = 1; BP = 100), while Budorcas cannot be positioned with our data. The grouping of Naemorhedus with Ovibos also involves that the tribe Rupicaprini is not monophyletic. From the morphological point of view, Ovibos and Naemorhedus can be united by unique shared specializations of horn cores and basioccipital region of the skull (Gentry 1992). The tribe Saigini is contradicted by our results, as Saiga is related to Antilopini, while Pantholops is the sister-genus of other caprine genera (PPB = 1; BP = 89). Pantholops shares with other caprines a series of behavioural and skeletal characters, and several morphological cladistic analyses have concluded that it must be separated from Saiga and Antilopini (e.g. Gentry 1992; Vrba and Schaller 2000). The only Simpson’ tribe that could be eventually monophyletic is the tribe Caprini: the four genera Ammotragus, Capra, Hemitragus and Pseudois are grouped together, but their possible association with the genus Ovis needs to be tested with additional markers.
To conclude, only four clades stand out from our analyses of inter-generic relationships within Caprini s.l.: (1) the clade enclosing all caprines, except Pantholops; (2) the association of Naemorhedus with Ovibos; (3) the clade including Capra, Hemitragus and Pseudois; and (4) the grouping of Capra with Hemitragus.
Adaptive radiation of caprines
According to Schluter (2000), adaptive radiation is the evolution of ecological and phenotypic diversity within a rapidly multiplying lineage. It occurs when a single ancestor diverges into an array of species that inhabit a variety of environments and that differ in phenotypic traits used to exploit those environments (i.e. morphological, physiological and behavioural traits). Several factors might facilitate adaptive radiation, including release from competition in an underutilized environment (e.g. new island and new lake), and key evolutionary innovations that open access to an entirely new range of resources (e.g. wings of birds).
We suggest that the extant diversity observed into the tribe Caprini s.l. resulted from an adaptive radiation during the late Miocene because the group presents all the three criteria used for defining an adaptive radiation: an explosive diversification, a phenotype–environment correlation and the appearance of a key innovation in their common ancestor.
Firstly, the explosive diversification during the late Miocene is supported by three arguments: (1) the fossil record indicates that caprines flourished rapidly during the late Miocene (Gentry and Heizmann 1996; Gentry 2000; Vrba and Schaller 2000); (2) the present analysis of 2469 nt characters did not give strong support for inter-generic relationships, suggesting rapid events of cladogenesis during a short interval of time; and (3) the molecular dating revealed that the common ancestor of extant caprines occurred during the late Miocene, at 10.96 ± 0.73 Mya.
Secondly, the fit between the diverse phenotypes and their divergent environments is particularly marked in this group. Extant genera are morphologically very distinct with great variations in body and horn size, colour patterns and horn shape depending of the habitats they occupy. Geist (1987) pointed out that caprine species present a gradient of evolutionary trends: from the tropics to alpine and arctic regions, species evolve increasingly larger horns or accentuated coat patterns, large rump patches, and shorter or bushier tails, terminating as‘grotesque giants’in climatically extreme, seasonal environments. In general, body size also increases with this progression.
Thirdly, the common ancestor of caprines was probably characterised by reduced limb length as all genera, except Ovis and Pantholops, share short metacarpals (Gentry 1992). The acquisition of reduced metacarpals constitutes a clear adaptation for climbing (Köhler 1993) that permitted the colonization of mountainous ecological niches that were not occupied by other competitor herbivores. Therefore, short metacarpals can be interpreted as the ‘key innovation’ (Simpson 1953) of caprines, i.e. a novel trait that influences ability to exploit resources hitherto little or not utilized, and that results in a wave of subsequent speciation.
Origin and diversification of caprines during the Miocene
Where did the caprines originate, and when and how did they come to occupy the place in which they live today? Most morphologists and palaeontologists believe that caprines are native to Asia (e.g. Schaller 1977; Kingdon 1982; Vrba 1995; Gentry 2000; Vrba and Schaller 2000), but to our knowledge, no argument really supports this hypothesis in the literature. One possible reason may be the fact that the greatest diversity of caprines is currently concentrated in Asia, where seven out of total 11 genera can be observed in this continent, i.e. Budorcas, Capra, Hemitragus, Naemorhedus, Ovis, Pantholops and Pseudois. By comparison, America and Europe contains three genera (Oreamnos, Ovis and Ovibos in America; Capra, Ovis and Rupicapra in Europe) while only two genera are present in Africa (Ammotragus and Capra). However, the distribution of caprines had been seriously modified during the Pleistocene epoch because of glaciations and climate variability. In addition, the fossil record of the Pliocene suggests that their distribution was very different at that time, with species closely related to Budorcas, Hemitragus, Naemorhedus and Ovibos in Europe (Guérin and Patou-Mathis 1996), and with species closely related to Budorcas and Ovibos in Africa (Gentry 1996). We therefore consider that the ‘centre of origin’ of caprines is an open question.
The fossil record of the Miocene is difficult to interpret because caprines are not diagnosed by an unambiguous skeletal synapomorphy. Some authors consider that caprines can be connected with three different genera of the middle Miocene, each found in one continent of the Old World: Caprotragoides in Asia (India and Pakistan), Tethytragus in Europe (Spain and Turkey) and Gentrytragus in Africa (Kenya and Saudi Arabia) (Azanza and Morales 1994). However, the taxonomic status of these three genera is controversial and many recent studies have concluded that they are not caprines (Azanza and Morales 1994; Alcalà and Morales 1997; Gentry 2000; Vrba and Schaller 2000). By contrast, all the famous late Miocene faunas of Pikermi, Samos (both Greece), and Maragheh (Iran) include several genera, such as Pachytragus, Palaeoryx, Protoryx, Pseudotragus, which have been related to either caprines or hippotragines (Gentry and Heizmann 1996; Gentry 2000). In addition, the first occurrence of caprines is well attested by the presence of short and robust metacarpals in Aragoral mudejar, a fossil species found in the Upper Vallesian (MN 10, approximately 9.7 Mya according to Agustí et al. 2001) of the Spanish locality of La Roma 2 (Alcalà and Morales 1997). The fossil record is therefore in perfect agreement with our molecular estimation suggesting that the tribe Caprini s.l. appeared during the late Miocene (10.96 ± 0.73 Mya). Unfortunately, the ancestral area of caprines cannot be clearly determined by our biogeographic inferences because of the lack of resolution for intergeneric relationships.
To understand the extraordinary diversification of caprines, we need to give answers to one major question concerning their evolution: What selection pressure was responsible for the acquisition of short and robust metacarpals in their common ancestor? An array of reasons suggests that this key innovation appeared as a consequence of insularity. First of all, caprines can be defined as dwarf stocky antelopes by comparison with their sister-group, i.e. Alcelaphini and Hippotragini. By using the data available in Nowak (1999), we show that caprines have an average shoulder height (923 mm) smaller than the one calculated for Alcelaphini (1228 mm) and Hippotragini (1167 mm), and that their average size/weight ratio (8.8) is higher than the one calculated for Alcelaphini (7.5) and Hippotragini (7.4), indicating that they are more thickset. Interestingly, these evolutionary trends are characteristic of insular faunas. By comparing continental and insular faunas of the Neogene, Caloi and Palombo (1994) have shown that large mammals colonizers of islands evolved towards forms with reduced size, and with shorter and more massive limbs. These processes affect all insular faunas, irrespective of the size of the island, of its climate and vegetation (Azzaroli and Guazzone 1979). By admitting that caprines developed a rock climbing adaptation because of insular endemism, the sole possible centre of origin is represented by Mediterranean islands of the Miocene. Four main arguments argue in favour of this hypothesis: (1) The orogenic movements which fused Africa and Eurasia also raised new mountains in the Taurides, the Hellenides, the Dinarides, and eventually, somewhat later in the middle or early late Miocene, the Helvetic Alps. This series of movements led to the emergence of an intercontinental mega-archipelago between the Mediterranean Sea and the Paratethys, an eastern European inland sea, which today encompasses the Black, Azov, Caspian and Aral Seas (Hsü et al. 1977; Rögl 1998). (2) During the middle and late Miocene, several insular faunas are known in the Western and Central Mediterranean islands (Moyà-Solà et al. 1999). (3) Our analyses indicated that the various tribes of Antilopinae s.l. flourished in Africa during the middle Miocene (15.2 ± 0.61 Mya), and that the common ancestor of the clade uniting Caprini s.l., Alcelaphini and Hippotragini appeared in the middle Miocene (13.37 ± 0.7 Mya) either in Africa or in a largest area including Africa + Asia. Two different biogeographic scenarios can be advanced for interpreting these results: either the common ancestor of alcelaphines, caprines and hippotragines dispersed out of Africa during the middle Miocene or the dispersal event occurred later during the late Miocene with the ancestor of caprines. (4) The faunal migration from Africa to Mediterranean is documented in the fossil record: numerous species related to either caprines or hippotragines are found in the late Miocene of the Sub-Paratethyan Province (Gentry and Heizmann 1996; Gentry 2000) and the faunas of this insular region contrast with the northern faunas of Asia and Central Europe. According to Moyà-Solà et al. (1999), these faunas may be of African origin. Interestingly, several post-skeletal remains indicate the presence of caprines with extremely short metapods, e.g. Aragoral from Spain and Tyrrhenotragus from Maremma (Italy) (Alcalà and Morales 1997; Moyà-Solà et al. 1999).
After acquisition of their climbing adaptation in the Miocene Mediterranean islands, caprines may have invaded Africa, Europe and Asia, probably just after the Tortonian salinity crisis, between 7.8 and 7.6 Mya (Krijgsman et al. 2000), or after the Messinian salinity crisis, between 6 and 5.3 Mya (Krijgsman et al. 1999). These two crises caused large falls in Mediterranean water level, and therefore permitted intense faunal migrations towards the continents. Such dispersions may explain the large distribution of caprines during the Plio-Pleistocene, their taxonomic diversity in Europe and Africa, as well as their intense diversification in the mountainous regions of Asia.
Acknowledgements
We thank Jean Deutsch for laboratory facilities. We are grateful to Jacques Cuisin, Jean-Marc Pons, Francis Renoult, Daniel Robineau, and Michel Tranier, who provided bone fragments from specimens of the MNHN collections. We thank Céline Canler and Vitaly Volobouev for frozen cells, Jean-Luc Berthier, Jacques Rigoulet, Claire Rejaud, Gérard Dousseau and Jean-François Marjarie for blood samples from specimens of the Ménagerie du Jardin des Plantes, Phillippe Chardonnet and Bertrand des Clers for tissues of Gazella granti, Jean-Claude Thibault for Ovis aries blood samples, Perry S. Barboza, Kevin Budsberg and Michel Perreau for tissues of Ovibos moschatus, Françoise Hergueta-Claro for Hippotragus niger cells, and Herbert Thomas for the skin sample from Pseudoryx nghetinhensis. We also acknowledge Damien Germain for help with the bibliography, Ziheng Yang for help with paml, and John Gatesy and one anonymous reviewer for their helpful comments on the first version of the manuscript. This work has been supported by funds from the International Foundation for the Conservation of Wildlife.




