Comparative promoter analysis and its application to the identification of candidate regulatory factors of cartilage-expressed genes
Summary
Chondrocyte gene regulation is important for the generation and maintenance of cartilage tissues. Analysis of the transcriptional regulation of cartilage-specific genes, encoding both collagenous and noncollagenous proteins, provides a useful strategy to identify transcription factors (TFs) that control chondrocyte specification and differentiation. Our work aims at the identification of candidate TFs important for cartilage maintenance and development through an in silico approach. In order to better define the transcriptional regulatory networks that affect chondrogenesis in zebrafish we propose a combination of comparative promoter analysis and transcription factor binding site analysis using a TRANSFAC position weight matrix search to identify cis-regulatory transcription factor binding motifs in a set of cartilage characteristic genes. With this methodology we have successfully identified several transcription factors known to be important for chondrogenesis thus validating our in silico approach.
Introduction
Understanding the mechanisms of gene regulation is one of the major goals of comparative genomics as well as developmental biology. Even so, the functions of cis-acting regulatory sequences that are located in noncoding regions of DNA are still poorly understood (Clark, 2001). A large portion of the genome (up to 97%, Onyango et al., 2000) consists of noncoding DNA, of which an unknown proportion plays a role in regulating gene expression. Comparative DNA sequence analyses have become increasingly important in the study of regulatory gene expression since the high degree of conservation of regulatory elements was first recognized (e.g. Aparicio et al., 1995; Manzanares et al., 2000). However, the identification of functional elements in noncoding DNA sequences is often complicated by the fact that these elements are typically short (6–15 bp; e.g. Carroll et al., 2001) and reside at varying distances from their target gene. Functional elements tend to evolve at slower rates than nonfunctional regions, because they are subject to selection (Hardison et al., 1997; Hardison, 2000). Due to this slower rate of evolution, comparisons among evolutionarily distantly related genome sequences provide a tool to identify functional regions within noncoding DNA (Tompa, 2001; Blanchette and Tompa, 2002) – an approach that has been termed phylogenetic footprinting (Venkatesh et al., 2000). As an example, comparisons among closely related organisms, such as humans and mice (evolutionary distance of approximately 80 million years; Pough et al., 1999) revealed many of the functionally relevant binding sites (Onyango et al., 2000).
Regulation of transcription of eukaryotic genes is to a large extent combinatorial, i.e. the conditions under which a gene is expressed are determined by an intricate interplay of multiple positive and negative transcriptional regulators that recognize and bind to cis-regulatory elements within and beyond the gene’s promoter region. Therefore, a major goal in deciphering transcriptional regulation networks is to identify combinations of transcription factors (TFs) that functionally cooperate to form regulatory modules. Such modules can be recognized by the co-occurrence of the corresponding TF-binding sites in the same promoters (Sudarsanam et al., 2002; Elkon et al., 2003). Our aim in this work is to identify TFs important to cartilage maintenance and development. To this end, we computationally analyzed a set of four orthologous zebrafish and fugu gene promoters whose expression is documented to occur in cartilage. We report on nine TFs whose binding signatures are significantly over-represented in this set of promoters. The approach applied here is general and demonstrates the potential of computational promoter analysis for the identification of novel transcriptional modules.
Material and methods
Extraction of putative promoter sequences from zebrafish, fugu, tetraodon, and stickleback genomes
A set of four orthologous genes with documented expression in cartilage, i.e. aggrecan, dlx1, dlx2, and sox9, derived from zebrafish (Danio rerio), fugu (Takifugu rubripes), tetraodon (Tetraodon nigroviridis), and stickleback (Gasterosteus aculeatus), were selected for this analysis. An additional set of 14 genes expressed in liver with no documented expression in cartilage or bone (cholinergic receptor nicotinic alpha 5, coagulation factor II thrombin, complement component 9, deiodinase iodothyronine type II, fibrinogen alpha chain, glutamic pyruvate transaminase, GTP cyclohydrolase I feedback regulator, heparin cofactor II, methionine adenosyltransferase I, secreted immunoglobulin domain 4, solute carrier family 1, sterol carrier protein 2, transthyretin, fatty acid binding protein 10), derived from zebrafish, were also selected for testing specificity. The promoter sequences from these two gene sets were extracted for analysis by selecting 5000 bp upstream of the genes’ translation start sites. This length of sequence provided a reasonable assurance of containing the target gene’s transcription factor-binding sites (TFBSs). The promoter sequences were masked for repetitive elements by the program RepeatMasker (http://www.repeatmasker.org) with the slow and sensitive mode.
Comparative promoter TFBSs analysis
For each zebrafish–fugu orthologous promoter pair, the DNA Block Aligner (dba) software (http://www.ebi.ac.uk/Tools/Wise2) was used to extract conserved blocks of nucleotide sequences using the default parameter settings based on the assumption that conserved regulatory regions could be important for regulation of the gene. DBA aligns two sequences under the assumption that the sequences share a number of co-linear blocks of conservation separated by potentially large and varied lengths of DNA in the two sequences. DBA models four different types of conserved blocks, with corresponding sequence identities: type A, 60–70%; type B, 70–80%; type C, 80%–90%; and type D, 90–100%. The promoter sequences were then assessed for TFBSs by running MATCH against TRANSFAC 6.0 (http://www.gene-regulation.com) TF binding site position weight matrices (PWM). A matrix similarity score of 0.8 was considered to be significant. Only TFBSs that are located in these blocks in both zebrafish and fugu promoters were considered for further study.
TFBSs predictions, multiple sequence alignments and search for TFBSs across species
In the search of TFBSs we screened the TRANSFAC 6.2 database and the CORE_TF (Conserved and Over-REpresented Transcription Factor binding sites) (grenada.lumc.nl/HumaneGenetica/CORE_TF) that identifies common TFBSs in promoters of co-regulated genes (Hestand et al., 2008). We used this tool to search with PWMs from the TRANSFAC database for putative TFBSs that are over-represented in our zebrafish cartilage-expressed genes set when compared to a random set of liver-expressed promoter genes. The four zebrafish cartilage-expressed gene promoters extracted previously were used as the experimental promoter list. Fourteen liver-expressed promoter genes retrieved from zebrafish were used as the ‘control’ promoter list.
In the second phase of this study we used the ConTra tool (Hooghe et al., 2008) to identify TFBSs across four fish species (zebrafish, fugu, tetraodon, and stickleback). The ClustalW (Thompson et al., 1994) alignment program was used to create an alignment between sequences for each of the four orthologous genes (aggrecan, dlx1, dlx2, and sox9) and these aligned sequences of orthologous promoters were used with the ConTra web tool that allows the visualization of all predicted sites for selected TFs. Default stringency settings (core = 0.90, similarity matrix = 0.75) were used.
Results
Zebrafish and fugu promoter region comparison
For this study, a computational transcript-mapping approach was used to locate promoter sequences from zebrafish and fugu genes. Among the 5000 bp promoter sequences upstream of the translation start sites, an average of 3% of the total base-pairs were located in one of the four types of conserved blocks identified by the DBA software. The percentages of bases that belong to the above four categories in all the 5000 bp promoters were compared and plotted by their distance from the translation start sites. As demonstrated in Fig. 1, the overall conservation is higher in the most proximal 5′ promoter region (0–1000 bp upstream of translation start site) where 13% of this region is in blocks A, B, C, or D, in comparison to the most distal promoter region (4000–5000 bp upstream of translation start site) where only 2% of this region is in blocks A, B, C, or D. This observation is consistent with the experimental data already available which indicates that most of the regulatory modules are in the proximal region (Blanchette et al., 2006). Furthermore, the results show that the size of promoter regions in which the functional sites are likely to occur in zebrafish and fugu is substantially lower than 5000 bp. Reducing the size of the sampled promoter region would make the statistical computational analysis of the functional units more sensitive and specific.
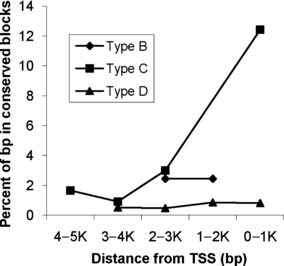
Distribution of percentage of base-pairs located in block B, C, or D along zebrafish and fugu promoters, for each of five 1000 bp segments from 0 to 5000 bp upstream of translation start signal
Analysis of regulatory elements using TRANSFAC PWM search
The promoter sequences of each of the four zebrafish and fugu genes, and the set of identified conserved blocks present within the promoters, were assessed for TFBSs by running MATCH program, and compared among them. A reduction from approximately 3000 TFBSs in the full promoter sequences to only forty TFBSs in the conserved blocks was observed, with the number of hits for the most frequently observed TFs being reduced over tenfold (Fig. 2). By identifying and using only conserved blocks, an average of 95% of all TFBSs in the full promoter regions was removed.
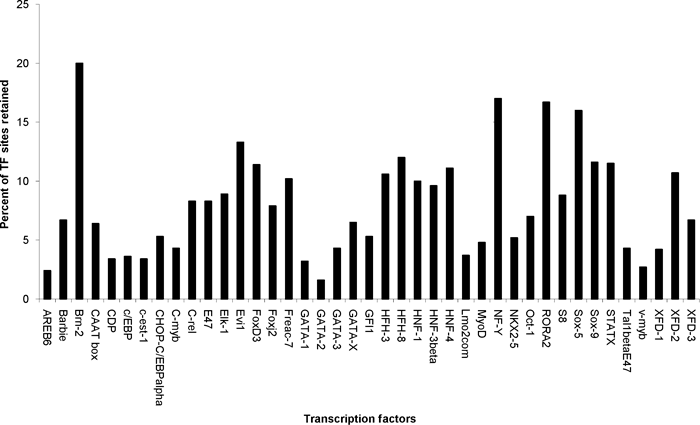
Retention of putative TF sites after comparative analysis. For each TF listed along the x-axis, respective bars represent the percentage (y-axis) of putative binding sites originally identified by TRANSFAC PWM that also survived comparative analysis
To determine whether our comparative screening of TFBSs resulted in the identification of TFs important to chondrogenesis, we searched for the presence of a variety of previously characterized TFs, known to regulate or potentially regulate cartilage genes, in the upstream sequences conserved in both zebrafish and fugu. These TF’s were C/ebp, Sox9, Sox5, c-Ets-1, Hnf-3beta, Hfh-3, Foxd3, and Areb6. Thus, by using the comparative approach combined with a TRANSFAC PWM based TF site search, more than 95% of the binding sites were removed, while the majority of TFs of known association with regulation of cartilage genes were retained. Several TFs important for chondrogenesis were identified.
The number of hits of the regulatory motifs in the four orthologous zebrafish and fugu selected cartilage genes were determined and compared with the number of hits obtained for the promoters of a ‘control’ set of 14 previously reported liver-expressed genes. The average number of hits of each motif and the standard deviation of samplings are presented in Table 1. These results show an over-representation of Caat box, Elk-1, Gata-1, Gata-2, Gata-X, Hnf-1, Lmo2 complex, RORalpha2, and Statx motifs in the promoters of the selected cartilage genes as compared to zebrafish liver- expressed genes.
| TF Name | Consensus sequence | Hits in cartilage genes | Hits in liver genes | P Value | ||
|---|---|---|---|---|---|---|
| Hits | SD | Hits | SD | |||
| Caat box | NNNRRCCAATSA | 46.63 | 18.77 | 32.36 | 12.18 | 0.042 |
| Elk-1 | NNNNCCGGAARTNN | 44.50 | 22.97 | 24.21 | 10.89 | 0.010 |
| Gata-1 | NNNNNGATANKGNN | 94.63 | 29.17 | 69.07 | 20.97 | 0.027 |
| Gata-2 | ASAGATAANA | 64.25 | 25.57 | 46.64 | 14.01 | 0.048 |
| Gata-X | NGATAAGNMNN | 31.38 | 10.28 | 21.21 | 7.07 | 0.012 |
| Hnf-1 | GGTTAATNWTTAMCN | 11.00 | 5.15 | 8.571 | 4.20 | 0.020 |
| Lmo2 complex | NMGATANSG | 27.13 | 8.64 | 19.14 | 5.91 | 0.018 |
| RORalpha2 | NWAWNTAGGTCAN | 5.75 | 3.45 | 3.21 | 1.93 | 0.038 |
| Statx | TTCCCGKAA | 25.75 | 5.12 | 18.21 | 7.50 | 0.021 |
Analysis of transcription factor binding sites in co-expressed genes using CORE_TF
Promoter analysis was performed on the four co-expressed genes to look for common TFBSs using CORE_TF, and 14 liver- expressed gene promoters as random set. In Table 2 we present the results indicating only the P-values below the defined threshold. The method resulted in 30 TFBSs over-expressed in the zebrafish cartilage-expressed genes. The top four over-represented TFBSs corresponded to the E2F-myc activator/cell cycle regulator (Table 2). E2F transcription factors (E2F1–E2F5) bind DNA in complex with DP proteins (DP1 and DP2). Studies have shown that constitutive E2F1 over-expression in transgenic mice and in the chondrogenic cell line ATDC5 inhibits chondrocyte differentiation, which results in delayed endochondral ossification in vivo, thus providing some indirect support to our in silico finding. In contrast, E2F4 over-expression has no effect on the differentiation program of chondrocytes (Scheijen et al., 2003).
| Name of matrix | P-value | Number of experimental promoters hit | Number of Random promoters hit | Random Frequencya | Hits in dlx1 | Hits in dlx2a | Hits in aggrecan | Hits in sox9 |
|---|---|---|---|---|---|---|---|---|
| AP4_01 | 0.033 | 2 | 3 | 0.21 | 1 | 1 | 0 | 0 |
| ATATA_B | 0.007 | 3 | 4 | 0.29 | 1 | 1 | 1 | 0 |
| BARBIE_01 | 0.034 | 3 | 6 | 0.43 | 1 | 1 | 1 | 0 |
| CHX10_01 | 0.028 | 1 | 1 | 0.07 | 1 | 0 | 0 | 0 |
| DEAF1_02 | 0.028 | 1 | 1 | 0.07 | 1 | 0 | 0 | 0 |
| E2F1DP1RB_01 | 0.002 | 3 | 3 | 0.21 | 2 | 1 | 0 | 1 |
| E2F1_Q3_01 | 0.007 | 3 | 4 | 0.29 | 1 | 2 | 1 | 0 |
| E2F1_Q4 | 0.002 | 3 | 3 | 0.21 | 2 | 1 | 0 | 1 |
| E2F1_Q6 | 0.010 | 2 | 2 | 0.14 | 2 | 0 | 0 | 1 |
| E2F1_Q6_01 | 0.016 | 3 | 5 | 0.36 | 2 | 2 | 0 | 1 |
| E2F4DP1_01 | 0.002 | 3 | 3 | 0.21 | 2 | 1 | 0 | 1 |
| E2F4DP2_01 | 0.007 | 3 | 4 | 0.29 | 2 | 1 | 0 | 1 |
| E2F_02 | 0.002 | 3 | 3 | 0.21 | 2 | 1 | 0 | 1 |
| E2F_Q4 | 0.034 | 3 | 6 | 0.43 | 2 | 2 | 0 | 1 |
| E2F_Q6 | 0.034 | 3 | 6 | 0.43 | 2 | 2 | 0 | 1 |
| E2F_Q6_01 | 0.028 | 1 | 1 | 0.07 | 1 | 0 | 0 | 0 |
| E2_Q6 | 0.002 | 3 | 3 | 0.21 | 1 | 1 | 0 | 1 |
| E47_01 | 0.016 | 3 | 5 | 0.36 | 1 | 2 | 1 | 0 |
| EVI1_03 | 0.028 | 1 | 1 | 0.07 | 0 | 0 | 1 | 0 |
| EVI1_05 | 0.028 | 1 | 1 | 0.07 | 0 | 0 | 1 | 0 |
| HFH4_01 | 0.033 | 2 | 3 | 0.21 | 0 | 1 | 0 | 2 |
| HIF1_Q3 | 0.033 | 2 | 3 | 0.21 | 1 | 0 | 0 | 1 |
| LPOLYA_B | 0.033 | 2 | 3 | 0.21 | 0 | 0 | 1 | 1 |
| NFKAPPAB50_01 | 0.007 | 3 | 4 | 0.29 | 1 | 0 | 3 | 1 |
| POU6F1_01 | 0.028 | 1 | 1 | 0.07 | 0 | 0 | 1 | 0 |
| PR_01 | 0.028 | 1 | 1 | 0.07 | 0 | 0 | 1 | 0 |
| RSRFC4_01 | 0.028 | 1 | 1 | 0.07 | 1 | 0 | 0 | 0 |
| STAT1_01 | 0.034 | 3 | 6 | 0.43 | 2 | 0 | 2 | 2 |
| SZF11_01 | 0.028 | 1 | 1 | 0.07 | 1 | 0 | 0 | 0 |
| WT1_Q6 | 0.016 | 3 | 5 | 0.36 | 1 | 0 | 2 | 1 |
| XPF1_Q6 | 0.034 | 3 | 6 | 0.43 | 1 | 1 | 0 | 3 |
- aFrequency of hits in the random data.
Analysis of transcription factor binding sites across species using ConTra
To analyze the functional implications of these TFBSs, we scanned the four cartilage-expressed gene promoter sequences from four fish species (zebrafish, fugu, tetraodon, and stickleback) for regulatory motifs using the ConTra tool (Hooghe et al., 2008). From the list of 30 TFBSs over-expressed in the cartilage-gene set obtained using CORE-TF, only Ap4, Chx10, Deaf1, E2F, Evi-1, Hif-1, Pou6F1, Pr, Stat1, Szf-1, and Wt-1 matrices are available at the ConTra website. In addition, we also searched for the TFBSs obtained using MATCH that were located in the blocks defined in both zebrafish and fugu promoters and that were available at the ConTra website: Areb6, C/ebp, Cdp, c-ets-1, Chop, Foxj2 (fork head box J 2), Evi-1, Gata-X, Gfi1, Hnf1, Hnf3 beta, MyoD, Lmo2 complex (complex of Lmo2 bound to Tal-1, E2A proteins, and Gata-1, half-site 1), Hnf4, Nf-Y, Nkx2-5, Oct-1, S8, Sox-9, and Tal1beta:E47 heterodimer. We were able to confirm the presence of all these TFBSs on the four cartilage-expressed promoter genes analysed. In Fig. 3 we show a part of the sequence alignment of the dlx1 promoter that reveals two Sox9 sites (PWM Sox9_B1) conserved between zebrafish and the other fishes analysed. Besides Sox9 sites, all the other TFBSs were conserved between zebrafish and at least one of the other fishes (results not shown). In addition, some of the TFBSs were shown to be overlapping in the alignment sequences. Given the premise that evolutionary conserved sites are more likely to be functionally relevant, those conserved sites that were found in all the fishes analysed should be tested for their ability to bind to TF in vivo.

Alignment of Sox9 binding sites in dlx1 promoter. The Sox9 sites are conserved between zebrafish, fugu, tetraodon, and stickleback
Discussion
Comparative analysis of genomic sequences is becoming increasingly important as a tool for identifying regulatory regions and functional elements. Despite the importance of promoters, the number of genes whose promoters have been identified and functionally analyzed experimentally is very limited compared to the growing number of known genes.
Accurate in silico detection of TFBSs is a difficult challenge that requires a combination of methods. Evidence used to identify sequences involved in the complex regulatory networks of eukaryotic genes is provided by the presence of the TFBSs, and the clustering of such binding sites in the promoter regions. Moreover, these features are usually conserved across species. For example, we have previously confirmed experimentally the cross-species transactivation between mouse and zebrafish: the mouse Runx2 was able to induce the zebrafish osteocalcin promoter, and vice-versa (Pinto et al., 2005). One drawback of using TRANSFAC PWM in identifying TFBSs is the large number of false positive hits identified by search programs such as MATCH. Performing statistical analysis can give informative results in some cases, but the high amount of false positives sometimes dilutes the existence of key TFBSs, making them statistically insignificant and therefore causing many false negative predictions (Qiu et al., 2002). However, by using a combination of methods such as those used in this work, the false negatives can also be substantially reduced.
In this study, we have taken a computational approach to annotate important cis-regulatory motifs in cartilage-expressed genes in fish. We performed promoter analysis and screened our fish cartilage-expressed gene set for previously characterized TFBSs from the TRANSFAC database.
Several TRANSFAC motifs were highly represented in our gene set for factors that play in zebrafish critical roles in (i) patterning process required for skeletal development (Nissen et al., 2003; Yokoi et al., 2009), (ii) cartilage differentiation (Yan et al., 2002), or (iii) cartilage gene regulation (Renn et al., 2006).
Areb6 (encoded by the gene zeb1 and also called delta Ef1), C/ebp, and possibly other transcription factor motifs highly represented in our data set, are negative regulators of cartilage-specific gene expression (Davies et al., 2002, 2007) (Fig. 2). Of the over-represented motifs (Table 1), the TFs, Stat, Gata-1 and -2, have previously been identified as being associated with common gene targets in osteoarthritis (Martensson et al., 2004; Millward-Sadler et al., 2006), Elk-1 function has been suggested to have a close link with matrix metalloproteinase-13 expression in human adult articular chondrocytes (Muddasani et al., 2007) and Lmo2 was predicted to be a TF involved in the bone response to mechanical loading by comparative model-based analysis (Chen et al., 2007).
Using data on evolutionary sequence conservation, as determined by both the multi-species sequence alignments and the in silico TFBSs predictions, it was possible to identify those sites most likely involved in regulating expression of target genes influencing growth and maintenance of cartilage. Regulatory regions with functional importance can be expected to exhibit sequence conservation due to selection. Thus, predicted TFBSs that are located in the orthologous positions in multiple genomes are likely to be functional. Indeed, our identification of evolutionary conserved sox9 binding sites in the dlx1 promoter supports this notion (Fig. 3). Conservation of TFBSs decreased as species became more evolutionarily divergent (Santini et al., 2003), so those TFBSs that are conserved between multiple species are more likely to be functionally important in the regulation of gene expression. This suggests that similar analyses extended to a larger pool of genes from a wider range of progressively more evolutionary distant species, could became increasingly informative.
Functional studies of gene regulatory processes involve a great deal of laboratory effort, and it is therefore critical to reduce the number of possible targets for function analysis prior to performing these studies. Our analyses confirm the value of comparative evolutionary genomic approaches in the identification and description of regulatory elements in genomes. Now that candidate TFBS have been identified, it would be particularly interesting to test some of the conserved regions (i.e. putative TFBS’s) that we have identified for a possible functional role in the activation and regulation of cartilage-expressed genes. With the increasing number of sequencing projects of whole genomes (e.g. pufferfish, zebrafish, and medaka) accomplished, new strategies for comparative genomic approaches can be followed to study distantly related organisms and thus uncover putative regulatory elements. Moreover, using distantly related genome comparisons between teleosts and, for example, mammals or amphioxus, highlights the divergence in gene regulation of paralogous genes that evolved subsequent to gene duplication. This type of analysis will help to increase the basic knowledge about our understanding of the characteristics, evolutionary conservation, and the position of functional elements, with respect to the genes that they control.
Acknowledgments
NC and BS are supported, respectively, by pos-doctoral and PhD grants from FCT (SFRH/BPD/48206/2008 and SFRH/BD/38083/2007).




