Detection of quantitative trait loci affecting non-return rate in French dairy cattle
Summary
The purpose of this study was to map quantitative trait loci (QTL) influencing female fertility estimated by non-return rate (NRR) in the French dairy cattle breeds Prim’Holstein, Normande and Montbeliarde. The first step was a QTL detection study on NRR at 281 days after artificial insemination on 78 half-sib families including 4993 progeny tested bulls. In Prim’Holstein, three QTL were identified on Bos taurus chromosomes BTA01, BTA02 and BTA03 (p < 0.01), whereas one QTL was identified in Normande on BTA01 (p < 0.05). The second step aimed at confirming these three QTL and refining their location by selecting and genotyping additional microsatellite markers on a sub-sample of 41 families from the three breeds using NRR within 56, 90 and 281 days after AI. Only the three QTL initially detected in Prim’Holstein were confirmed. Moreover, the analysis of NRR within 56, 90 and 281 days after AI allowed us to distinguish two FF QTL on BTA02 in Prim’Holstein, one for NRR56 and one for NRR90. Estimated QTL variance was 18%, 14%, 11.5% and 14% of the total genetic variance, respectively, for QTL mapping to BTA01, BTA02 (NRR90 and NRR56) and BTA03.
Introduction
During the last years, the fertility of dairy cows has continuously deteriorated: Royal et al. (2002) have reported that calving rates to first service have fallen by as much as 0.45% in the US and 1% per year in the UK. In France, a 1% decrease in conception rate has been observed over the last 7–12 years (Grimard et al. 2006). Reduced fertility results in higher costs due to supplementary artificial inseminations (AI), curative interventions and longer calving intervals.
Female fertility (FF) in cattle is a complex trait that can be divided into at least two components: interval traits such as interval from calving to first insemination and success traits such as non-return rates (NRR). However, these two measures of fertility do not have the same significance: the interval from calving to first insemination describes the ability of a cow to show oestrus while the NRRs is related to the capacity of a cow to conceive when inseminated (Andersen-Ranberg et al. 2005).
Female fertility is influenced by several physiological factors (post partum return to cyclicity, occurrence of oestrus, AI success, pregnancy success) and also largely by environmental factors leading to high phenotypic variability. Hence, FF traits have a low heritability but show high genetic variation (Boichard & Manfredi 1994; Wall et al. 2003). Identification of quantitative trait loci (QTL) controlling FF variability can be used in marker assisted selection (MAS) programmes to increase the genetic progress of this trait and better describe molecular mechanisms involved in its variability. Several studies have already reported QTL underlying traits related to FF such as ovulation rate (Gonda et al. 2004), pregnancy rate (Ashwell et al. 2004; Schnabel et al. 2005; Muncie et al. 2006) or NRR (Schrooten et al. 2000, 2004; Holmberg & Andersson-Eklund 2006).
In France, a first QTL detection programme was carried out in a grand-daughter design (GDD) composed of 14 paternal half-sib families belonging to Prim’Holstein (PH), Normande (NO) and Montbeliarde (MO) breeds (Boichard et al. 2003). Twenty-four different traits were analysed including FF as estimated by the rate of success for each insemination of the daughters of a bull. Three QTL mapping to Bos taurus chromosomes (BTA) BTA01, BTA07 and BTA21 were detected (Boichard et al. 2003). In addition, this QTL programme made it possible to implement a first-generation MAS programme for which 45 markers were chosen to track 14 genomic regions containing QTL underlying different traits of interest across the dairy cattle population (Druet et al. 2006).
In this paper, we report the detection of QTL underlying NRR within 281 days (NRR281) after AI in 12 genomic regions covered by the MAS programme on 78 paternal half-sib families originating from the three breeds (PH, MO and NO). The most significant genomic regions were further investigated by increasing marker information on a sub-sample of 41 half-sib families and considering, in addition to NRR281, NRR within 56 (NRR56) and 90 (NRR90) days after AI.
Material and methods
Animals
In the QTL detection study, we used the extended mapping GDD available in PH, NO and MO breeds (Gautier et al. 2006b). They were composed of 78 half-sib families with more than 25 sons: 47 in PH, 18 in NO and 13 in MO. A total of 4993 progeny tested sires were available with an average of 70 sires per family (from 25 to 240). As shown in Table 1, for the QTL confirmation study, which required additional genotyping, a sub-sample of 41 families was chosen. As described previously (Gautier et al. 2006b), we tried to maximize the number of informative families for the QTL underlying the different traits initially analysed (FF but also production and disease resistance traits) while avoiding the overrepresentation of some closely related families. The 41 families included: 26 PH, 9 NO and 6 MO families. All 14 families (9 PH, 3 NO and 2 MO) from the first QTL programme were included to provide a link with the previous results.
| Breed | Number of families | Average family size | Number of individuals considered in the QTL analysis | |
|---|---|---|---|---|
| QTL detection | PH | 47 | 70 | 3275 |
| NO | 18 | 56 | 956 | |
| MO | 13 | 56 | 762 | |
| Total | 78 | – | 4993 | |
| QTL confirmation | PH | 26 | 83 (35–236) | 2042 |
| NO | 9 | 63 (39–76) | 527 | |
| MO | 6 | 67 (41–99) | 356 | |
| Total | 41 | – | 2925 |
Phenotypic data
For each individual, daughter yield deviation (DYD) and reliabilities were estimated (VanRaden & Wiggans 1991) and used as phenotypes. When reliability was lower than 0.3, the corresponding DYD was removed. DYD for NRR281 was used in the QTL detection study. To improve QTL characterization and detection, two additional NRR were analysed: NRR56 and NRR90. Measuring these two phenotypes should make it easier to distinguish between reproduction events that occur early in gestation and those, which continue to occur during the whole gestation period. The estimated correlations between NRR56 and NRR90 NRR56 and NRR281 and NRR90 and NRR281 were 0.91, 0.81 and 0.94, respectively (Guillaume, personal communication). The average DYD reliabilities for NRR56, NRR90 and NRR281 were 0.67, 0.66 and 0.70, respectively.
Genotyping data
For the QTL detection study, we used genotyping data from the 78 half-sib families already available from the French MAS programme (Boichard et al. 2006). As shown in Table 2, the data set consisted of genotypes for two to five microsatellites within 12 genomic regions (39 markers in total), which covered approximately 17% of the bovine genome. Based on QTL detection results, marker density on BTA01, BTA02 and BTA03 was further increased (see Table 3 for details on the different markers). For BTA01 and BTA02, respectively, nine and 11 microsatellites were chosen from the USDA database (http://www.marc.usda.gov/genome) for genotyping, in addition to the four and two markers already available for these two chromosomes from the MAS programme (Table 2). For BTA03, genotypes for four microsatellites were already available from the previous MAS programme (Table 2) and also for 14 microsatellites in PH (Guillaume et al. 2007). These latter markers were genotyped in the nine NO and six MO families. Since the region spanning the first 30 cM on BTA03, corresponding to the most likely QTL position (Guillaume et al. 2007), contained too few markers, seven additional microsatellite markers were selected for genotyping (BMS2904, DIK4193, INRA3001, INRA3012, INRA3013, INRA3015 and URB006).
| Bovine chromosome | Number of markers | PH | NO | MO |
|---|---|---|---|---|
| BTA01 | 4 | ** | ** | N |
| BTA02 | 2 | ** | N | N |
| BTA03 | 4 | ** | N | N |
| BTA06 | 3 | * | N | N |
| BTA07 | 3 | N | N | N |
| BTA14 | 2 | N | N | N |
| BTA15 | 3 | N | N | N |
| BTA19 | 4 | N | N | N |
| BTA20 | 5 | * | N | N |
| BTA21 | 3 | N | * | N |
| BTA23 | 2 | * | N | N |
| BTA26 | 4 | N | N | N |
- N: no QTL detected. *p < 0.05; **p < 0.01.
| Marker name | BTA | Position (cM) | Number of genotyped animals | Alleles | Number of informative meiosis | Forward primer | Reverse primer | ||
|---|---|---|---|---|---|---|---|---|---|
| PH | NO | MO | |||||||
| BM4307 | 1 | 38.0 | 1908 | 479 | 361 | 8 | 1240 | ATAACACAAAAAGTGGAAAACACTC | ATTTTATCTCAGGTCCCTTTTTATC |
| BMS4000 | 1 | 51.6 | 1946 | 504 | 235 | 13 | 1499 | TCCCTCCTTGATGGATTCAG | TTGAAGTAGCCTGAGAAAAGGG |
| BM1312 | 1 | 58.5 | 2046 | 525 | 369 | 9 | 1133 | CCATGTGCTGCAACTCTGAC | GGAATGTTACTGAACCTCTCCG |
| BMS4030 | 1 | 62.6 | 1890 | 500 | 285 | 9 | 1548 | TGTACCCAACACAGGAGCAC | TGACAGAGGGACCCATATCC |
| DIK2886 | 1 | 65.4 | 1988 | 496 | 243 | 7 | 969 | TTCATGTTTTTGGCCCAAGT | GAGAATGCCCAAGGACAGAG |
| INRA073 | 1 | 73.6 | 1988 | 545 | 368 | 4 | 1031 | ACTGAGGAACTAAGCACCGC | GAAAAGCAAGGCTGTCCGAC |
| BM8246 | 1 | 79.3 | 2025 | 529 | 366 | 12 | 1321 | AATGACAAATTGAGGGAGACG | AGAGCCCAGTATCAATTCTTCC |
| DIK4587 | 1 | 88.7 | 2044 | 522 | 364 | 8 | 866 | CAGAGGAAGGCTGATGGAG | GGAGTGGGTGTTCCCTTCTC |
| BM864 | 1 | 97.5 | 1952 | 522 | 354 | 15 | 1530 | TGGTAGAGCAATATGAAGGCC | GGAAATCCAAGAAAGAGGGG |
| BMS4028 | 1 | 106.5 | 2020 | 536 | 370 | 10 | 1523 | TGGGGTTGAAAAAGAACTGG | ATAAAGCAGATGTGGTGTGTGC |
| DVEPC32 | 1 | 111.9 | 1964 | 485 | 325 | 8 | 1172 | TGGGGTTGAAAAAGAACTGG | ATAAAGCAGATGTGGTGTGTGC |
| BMS4041 | 1 | 116.8 | 1846 | 529 | 358 | 4 | 900 | TAACAGAGGAGCCTGGCG | TTTGGATCATTTTGCAGTGTG |
| BM1824 | 1 | 122.3 | 957 | 297 | 221 | 5 | 922 | GAGCAAGGTGTTTTTCCAATC | CATTCTCCAACTGCTTCCTTG |
| DIK2496 | 2 | 49.0 | 2049 | 529 | 392 | 2 | 963 | GGAGGAATTGGCTATGGTCTC | CGACTTCCGTCTGTCTCACA |
| DIK4673 | 2 | 56.3 | 2025 | 516 | 254 | 6 | 1408 | GTACTTGGGGAAGCCCTCTC | ACTGCGCTTGAGGGAAAATA |
| BM4440 | 2 | 56.8 | 1988 | 526 | 372 | 8 | 1181 | CCCTGGCATTCAACAAGTGT | CACCCTGTTAGGAATCACTGG |
| DIK2719 | 2 | 58.9 | 2028 | 495 | 302 | 7 | 1888 | TGATCCCATTTGAGACAGCA | CCTTACTGTCCCCACTCTGC |
| DIK4972 | 2 | 65.6 | 1902 | 474 | 266 | 3 | 659 | GCCTCCCAGAATCCTGACTA | GAATTTCCTGGCAGTTCCTG |
| BMS778 | 2 | 69.3 | 1996 | 533 | 295 | 9 | 1604 | CTTGGGGAGGCAGAATTTA | CCTCTCCACATACTCTTCTCCA |
| RM041 | 2 | 70.0 | 1936 | 528 | 357 | 6 | 1613 | GATCACAGAAACATGCA | TTCCTAACATCACAGCACCTCC |
| DIK4208 | 2 | 75.0 | 2015 | 532 | 378 | 4 | 1597 | GTCCGTGTTGCTGCAAATAG | AGCAGCACTAGTTGCCAAGG |
| BMS1866 | 2 | 79.1 | 1949 | 502 | 353 | 9 | 1272 | CAGGGACTGAAAAATAATGCC | TTCCATGTTGATTGTTTCTTCC |
| BM1223 | 2 | 96.0 | 1997 | 474 | 359 | 5 | 613 | AGGCAAATTGTGTTTCCAGC | TCATAAGGGTTTGGAGGCTG |
| BMS2519 | 2 | 108.1 | 1983 | 504 | 351 | 10 | 1744 | CATGGTTCTCATCTGGTGTG | AGTGAAGACCTACTGCAGCC |
| BM2113 | 2 | 115.2 | 1029 | 357 | 240 | 7 | 1047 | GCTGCCTTCTACCAAATACCC | CTTCCTGAGAGAAGCAACACC |
| IDVGA-2 | 2 | 136.3 | 1715 | 475 | 264 | 9 | 1337 | CCTTGGATTTCCTTCCCATT | TATGGATACCCACCAAGCTG |
| BMS871 | 3 | 0.0 | 2091 | 535 | 366 | 6 | 863 | GAACATGAGGTTGACAAAGGA | TCCAGTGACTCTTTTCTGCC |
| URB006a | 3 | 12.3 | 2037 | 472 | 323 | 5 | 513 | GTTTCTACCCTTGCCTGTTGCCT | CCTTCAATGCTCTTCCTCCCATC |
| INRA3001a | 3 | 19.8 | 1903 | 435 | 285 | 7 | 962 | CATGTTATACCAAGTAGATGTGCAAT GAGACAGTTTTGCTGAGTCAAT | TCCATGAGGACTTCTCTGACC |
| INRA3015a | 3 | 21.4 | 2006 | – | – | 5 | 762 | AATGGCAATAGCTTGGACAGA | GTGGCTGTGTTCCACAAAGTT |
| INRA3012a | 3 | 21.7 | 2014 | – | – | 6 | 1113 | GGCTTGATCCCTGTGTTAGG | TCTCCTTAGCTCCAGAGCACA |
| INRA3013a | 3 | 22.5 | 2012 | – | – | 4 | 711 | AGGAATATCTGTATCAACCTCAGTC | TGGAGCAACCTGGATAGCA |
| MB101 | 3 | 22.6 | 2093 | 541 | 377 | 3 | 188 | GTGCTGTGGTACCTGGGACT | CTGAGCTGGGGTGGGAGCTATAAATA |
| DIK4193a | 3 | 23.6 | 2043 | – | – | 3 | 646 | GTGACCTGGAGAAGTTTTCC | TGCTCCAGTTTTCTTGTCTGAA |
| ILSTS096 | 3 | 25.6 | 1999 | 408 | 355 | 8 | 1534 | TGGCAGTGAGAGAGGGGAC | ACCACGCTCTGACTTGTAGC |
| BMS2904a | 3 | 26.5 | 2016 | – | – | 4 | 412 | CCCAGCACCAATGTGACAA | TCCACTTGAGAGCGTGTCTG |
| DIK2101 | 3 | 27.7 | 2079 | 542 | 379 | 6 | 1634 | CCAGGGTGCTGATATCTGGAGG | TGCGTGTGTGTTTGGGTA |
| RM019 | 3 | 28.8 | 1998 | 499 | 346 | 4 | 369 | AGGCAGGTGTTTTGGTTCTG | TTCAATGAACAAGATGGAGACTT |
| BMS2522 | 3 | 28.8 | 2067 | 539 | 270 | 9 | 801 | AATCTGATTTTCTCCGTAGCTG | AGGCCATGTAGAGGTAAAGTGC |
| MNB65 | 3 | 30.4 | 2032 | 526 | 365 | 6 | 1216 | TGCAAGATCCAGTGAACGTC | GGTTGAGTTAAAACTGTTTAAGAGA |
| DIK4196 | 3 | 32.0 | 2000 | 533 | 365 | 8 | 998 | GGAGGATGAAGGAGTCTTTGG | CACTTGGGGAAACCATAATGA |
| BMS963 | 3 | 32.0 | 2044 | 542 | 376 | 6 | 1156 | ACTTCCCCAGTCTTCCCAGT | AATTTACCACAGTCCACCGC |
| BMS482 | 3 | 32.9 | 2065 | 543 | 378 | 8 | 1335 | TCCAAAGCATATGCCAGAAA | TGGTGGACAGTCCCATACAG |
| DIK2434 | 3 | 35.3 | 2043 | 543 | 370 | 11 | 1752 | TCTAGTCCAGACATAAACAAATGC | AACTGAAGGCACCCTTCTCC |
| MNB-86 | 3 | 37.2 | 1908 | 478 | 351 | 12 | 1300 | CCTCTGCCATCTTTATTCCG | GTTCTGTATGCACGTGCAGAGA |
| BL41 | 3 | 42.2 | 1994 | 510 | 366 | 6 | 1022 | CAATGGGAGACGACTTTCCT | AAGATCAACTTATTCCTCACAGTGG |
| DIK2609 | 3 | 45.3 | 2112 | 542 | 377 | 2 | 374 | AACCTTTATTAGGGGAGTTCGG | AACCCCAGCCTCGTCTACAT |
| BM4129 | 3 | 45.3 | 1941 | 543 | 312 | 3 | 476 | GAGTAGAGCTACAAGATAAACTTC | TAAGCTGTGGAGTGCAGCAA |
| INRA023 | 3 | 50.4 | 923 | 327 | 206 | 8 | 985 | CTGGAGGTGTGTGAGCCCCATTTA | TAACTACAGGGTGTTAGATGAACTC |
| INRA003 | 3 | 58.4 | 1831 | 482 | 333 | 11 | 1239 | TGTTTTGATGGAACACAGCCTCCATCA | GGTACGAGCGTTGGGAAAGGT |
| ILSTS029 | 3 | 63.8 | 2030 | 530 | 369 | 7 | 1271 | AAGTATTTGAGTGCAA | TGGATTTAGACCAGGGTTGG |
| HUJI177 | 3 | 93.8 | 1237 | 307 | 147 | 7 | 973 | ATAGCCCTACCCACTGTTTCTG | |
- aMarkers used in the QTL confirmation study.
Among these seven additional new markers, only INRA3001 and URB006 were genotyped in the three breeds as part of a first round of genotyping, the five other markers were subsequently developed and genotyped only in PH because no NRR QTL segregated in the two other breeds (see Table 3 and Results). The markers INRA3001, INRA3012, INRA3013 and INRA3015 were selected and developed from available bovine genome sequence data to target the region of interest according to the Btau_3.1 assembly (see ftp://ftp.hgsc.bcm.tmc.edu/pub/data/Btaurus/fasta/Btau20060815-freeze/ReadMeBovine.3.1.txt). Their expected positions on BTA03 were confirmed by radiation hybrid mapping using standard procedures (Gautier et al. 2003). Selected markers were then genotyped as previously described (Gautier et al. 2006a). Genotypes for 13, 13 and 26 (21 in NO and MO) markers, respectively, were available for BTA01, BTA02 and BTA03. Finally, for each of the three chromosomes, only individuals genotyped for at least 60% of the markers were considered in the analysis. A total of 2925 sons from the three breeds were included in the analysis (Table 1).
Linkage map construction
Linkage maps were constructed using crimap 2.4 software (Green et al. 1990). For each linkage group, the marker order obtained from known linkage maps and the bovine genome assembly (see above) was challenged using the flips option with a four-marker window. The Chrompic option was used to identify unlikely double crossing-overs. Final map distances were estimated using the Haldane mapping function.
QTL analysis
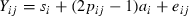
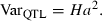
Results
QTL detection study
In the extended PH design (47 half-sib families), we detected three significant (p < 0.01) QTL underlying NRR281 and mapping respectively to BTA01, BTA02 and BTA03 chromosomes and three suggestive (p < 0.05) QTL mapping to BTA06, BTA20 and BTA23. In the NO extended pedigree (18 half-sib families), one significant QTL mapping to BTA01 and one suggestive QTL mapping to the BTA21 chromosome were detected. No QTL was identified in the extended MO pedigree (Table 2). Among the three QTL initially detected in the French QTL programme, which was based on 14 families (Boichard et al. 2003), the QTL on BTA01 was confirmed in the NO and PH breeds. The QTL mapping to BTA21 was only suggestive in NO while the QTL mapping to BTA07 could not be confirmed in any of the three breeds considered. Since only a few markers were genotyped for each chromosome (Table 2), we were not able to provide precise mapping intervals on the whole chromosome.
QTL confirmation study
As detailed in Table 3, on average 1152 informative meioses (from 188 for MB101 to 1888 for DIK2719) were available in PH to construct the linkage maps for BTA01 (13 markers), BTA02 (13 markers) and BTA03 (26 markers). Marker order and distances were in agreement with published data for markers mapping to BTA01 and BTA02 (http://www.marc.usda.gov/genome/). However, for BTA03, the marker order between RM019 and DIK2101 was different from published data (http://www.marc.usda.gov/genome/) but in agreement with Btau_4.0 assembly (ftp://ftp.hgsc.bcm.tmc.edu/pub/data/Btaurus/fasta/Btau20070913-freeze/README.Btau20070913.txt) (Table 3).
In the PH breed, at least one QTL underlying one of the three NRR traits was detected at a 5% significance threshold. No significant QTL was identified for the three different NRR traits in NO and MO. Results for each chromosome are detailed in the following paragraphs.
Chromosome BTA01
One significant QTL underlying NRR90 was detected in PH (p < 0.009). The peak position (at 97 cM on our genetic map) was located near marker BM864 (Figure 1). When considering NRR281, the QTL was confirmed at a slightly lower significance level (p < 0.019) and the peak position was approximately the same as that for NRR90. Based on the T-test value at the maximum peak position, four sires (3442, 3481, 3498 and 7210) were heterozygous (p < 0.05) for both NRR281 and NRR90 while sire 6865 was heterozygous (p < 0.05) only for the QTL underlying NRR90. The average QTL allele substitution effect was estimated as 4.86% NRR90 and the QTL explained 18% of the total genetic variance (Table 4).
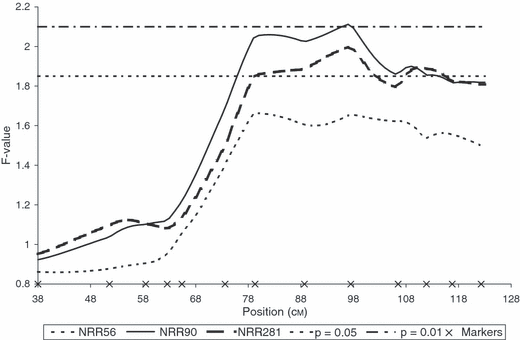
F-statistic profiles for NRR56, NRR90 and NRR281 on BTA01 in PH. Marker positions are indicated with crosses on the x-axis. Significance thresholds at 5 and 1% are valid for all NRR data.
| Chromosome | Marker informativity ± SD | Trait | Familya | Marker bracketb | pchromosome | a (%) | VarQTL (%) |
|---|---|---|---|---|---|---|---|
| BTA01 | 0.77 ± 0.05 | NRR90 | 3442, 3481, 3498, 6865, 7210 | [INRA073-BM1824] | ** | 4.86 | 18.00 |
| NRR281 | 3442, 3481, 3498, 7210 | [DIK4587-BMS4028] | * | – | – | ||
| BTA02 | 0.73 ± 0.10 | NRR56 | 3468, 3481, 3502, 5245, 6109, 6865 | [DIK2496-BM2113] | * | 3.80 | 11.50 |
| NRR90 | 3487, 3480, 3514, 6865 | [DIK4972-IDVGA-2] | * | 4.84 | 14.00 | ||
| NRR281 | 3468, 3480, 3514, 6865 | [DIK4972-IDVGA-2] | * | – | – | ||
| BTA03 | 0.71 ± 0.02 | NRR90 | 3514, 4442, 4729, 6145, 6151 | [BMS871-BL41] | * | 4.72 | 14 |
- aCode number for segregating (p < 0.05) families.
- b90% CI computed for only QTL that have a type I error <0.05.
- *p < 0.05; **p < 0.01.
Chromosome BTA02
One suggestive QTL (p < 0.047) underlying NRR56 was detected in PH. The peak position was located near BMS778 and RM041 at 70 cM on our genetic map. Although they showed a similar trend, Linkage Analysis (LA) curves obtained for NRR90 and NRR281 did not reach the 5% significance threshold (Figure 2). Six sires were heterozygous (p < 0.05) for the QTL: 3468, 3481, 3502, 5245, 6109 and 6865 (Table 4). The average QTL allele substitution effect was estimated as 3.8% NRR90 and the QTL variance explained 11.5% of the total genetic variance.
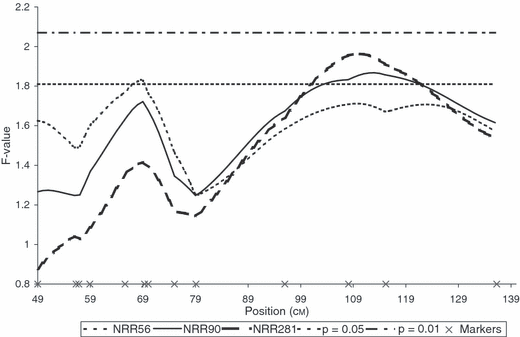
F-statistic profiles for NRR56, NRR90 and NRR281 on BTA02 in PH. Marker positions are indicated with crosses on the x-axis. Significance thresholds at 5 and 1% are valid for all NRR data.
A QTL underlying both NRR90 (p < 0.048) and NRR281 (p < 0.034) was detected on BTA02 at a more telomeric position. The peak position was situated near BM2113 at position 115 cM according to our genetic map (Figure 2). Sires 3480, 3514 and 6865 were heterozygous (p < 0.05) for both NRR90 and NRR281 data while sire 3487 heterozygous (p < 0.05) only for NRR90. The average QTL allele substitution effect was estimated as 4.84% NRR90 and the QTL explained 14% of the total genetic variance (Table 4).
Chromosome BTA03
A QTL underlying NRR90 (p < 0.015) was detected on BTA03 in the PH breed. The peak position (25 cM on our linkage map) was near ILSTS096 (Figure 3). When considering NRR281 as a trait, the LA curve displayed a similar trend but did not reach the 5% significance threshold (p < 0.057). Sires 3514, 4442, 4729, 6145 and 6151 were heterozygous (p < 0.05) for this QTL. The 90% bootstrap CI spanned 32 cM between markers BMS871 and BMS482 which is half the size of that obtained using only the markers included in Guillaume et al. (2007). The average QTL allele substitution effect of the identified QTL was estimated as 4.72% NRR90 and the QTL explained 14% of the total genetic variance (Table 4).

F-statistic profiles for NRR56, NRR90 and NRR281 on BTA03 in PH. Marker positions used in the previous study (Guillaume et al. 2007) are indicated with crosses on the x-axis. Positions for the new markers are indicated with triangles. Significance thresholds at 5 and 1% are valid for all NRR data.
Discussion
Among the QTL detected in the 14 families of the first French QTL programme (Boichard et al. 2003), the QTL mapping to BTA01 was confirmed in NO and PH while the QTL on BTA21 was confirmed only in NO. It is more likely that this QTL is segregating only in NO as the first French QTL programme analysis included all three breeds together while these latter, were analysed separately in the QTL detection study. The QTL mapping to BTA07, which was further investigated (Gautier et al. 2002; Guillaume et al. 2007), could not be confirmed in the QTL detection study. This is due to the difference between the phenotypes used in the two studies. Indeed, in the first French QTL programme, FF phenotypes were deregressed breeding values (BV), whereas FF phenotypes where NRR DYD in QTL detection study. When the 14 families of the first French QTL programme were analysed with the NRR DYD, no QTL mapping to BTA07 was detected.
The addition of more families to the initial 14 half-sib families allowed us to identify four additional QTL underlying NRR281: two significant QTL (p < 0.01) mapping to BTA02 and BTA03, respectively, and two suggestive QTL (p < 0.05) mapping to BTA20 and BTA23 in the PH breed. To our knowledge, only one study has reported QTL affecting NRR in similar regions. Indeed, Schrooten et al. (2000) reported a female fertility QTL affecting NRR56 mapping to BTA02 (near BM2113) in PH.
Increasing marker information in the most significant QTL regions located on BTA01, BTA02 and BTA03, allowed us to confirm these QTL in the PH breed (at least at the 5% chromosome wise significance threshold).
Determining the NRR phenotype at different stages of gestation made it possible to distinguish between QTL effects during gestation. For example, for the QTL mapping to BTA02, we detected two different QTL affecting respectively both NRR90 and NRR281 (given their similar LA curve profiles and peak location), and NRR56. The QTL reported by Schrooten et al. (2000) and affecting NRR56 is located closer to the one affecting NRR90 and NRR281 in our study. It is more likely that the same QTL is influencing both NRR in our study and the NRR56 used by Schrooten et al. (2000). The addition of new genotyping data might help in the future to distinguish between the effects of the different QTL suggested by our respective studies.
The significant QTL mapping to BTA01 in NO families was not confirmed in our QTL validation analysis, mainly because some families segregating the QTL in the enlarged pedigree were not sampled. In particular, in NO: only one family segregating the BTA01 QTL was retained for the QTL confirmation study. Finally, the results obtained for BTA03 QTL region were in good agreement with previously published data (Guillaume et al. 2007).
The number of heterozygous sires (p < 0.05) was 5 out of 26 (19%) for all the QTL except for NRR90 QTL detected on the BTA02 where four heterozygous sires were identified (15%). Assuming a bi-allelic QTL and HWE in the sire population, the minor allele frequency at the QTL is approximately 10%. Sires 6865 and 3487 were heterozygous for NRR90 (p < 0.05) but not for NRR281, respectively on BTA01 and BTA02. This is probably because that NRR data added between 90 and 281 days after AI are not only due to the identified QTL effects: a second linked QTL responsible for foetal death and carried by these two sires might explain these findings.
The average allele substitution effects of the identified QTL were 4.86, 3.8, 4.84 and 4.72%, respectively for QTL mapping to BTA01, BTA02 (NRR90 and NRR56) and BTA03. Although, these values are approximations (because they suppose a HWE and a biallelic QTL model), they are encouraging with respect to a possible genetic improvement of the NRR traits via MAS.
Since the 12 genomic regions investigated cover only 17% of the whole bovine genome, it is most likely that other QTL affecting NRR and located in regions not covered by the French MAS programme exist. The availability of whole genome scans with dense SNP maps will be useful to identify additional QTL influencing NRR with smaller confidence intervals.
Acknowledgements
We wish to thank the two anonymous referees for their helpful suggestions. The research was partly funded by the UNCEIA and the CASDAR.




